Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Batf3
Z-value: 0.72
Transcription factors associated with Batf3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Batf3
|
ENSMUSG00000026630.10 | Batf3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Batf3 | mm39_v1_chr1_+_190830522_190830633 | 0.44 | 1.3e-04 | Click! |
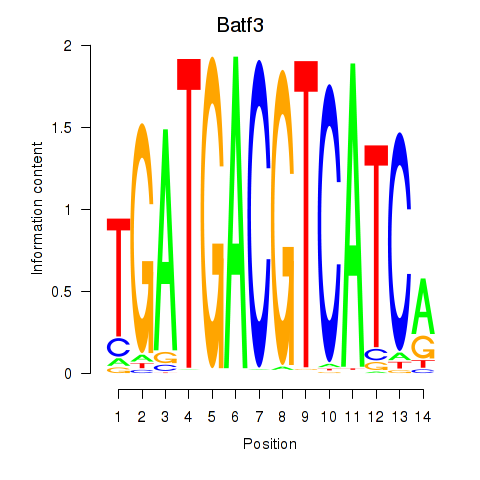
Activity profile of Batf3 motif
Sorted Z-values of Batf3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Batf3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_17823736 | 4.54 |
ENSMUST00000037879.8
|
Heca
|
hdc homolog, cell cycle regulator |
| chr3_-_88455556 | 3.96 |
ENSMUST00000131775.2
ENSMUST00000008745.13 |
Rab25
|
RAB25, member RAS oncogene family |
| chr19_-_61215743 | 3.86 |
ENSMUST00000237386.2
|
Csf2ra
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
| chr7_+_3706992 | 3.75 |
ENSMUST00000006496.15
ENSMUST00000108623.8 ENSMUST00000139818.2 ENSMUST00000108625.8 |
Rps9
|
ribosomal protein S9 |
| chrX_+_73352694 | 3.66 |
ENSMUST00000130581.2
|
Gdi1
|
guanosine diphosphate (GDP) dissociation inhibitor 1 |
| chr2_+_120439858 | 3.39 |
ENSMUST00000124187.8
|
Haus2
|
HAUS augmin-like complex, subunit 2 |
| chr17_+_56199093 | 3.38 |
ENSMUST00000224379.2
ENSMUST00000054780.10 |
Zfp959
|
zinc finger protein 959 |
| chr11_+_83553400 | 3.33 |
ENSMUST00000019074.4
|
Ccl4
|
chemokine (C-C motif) ligand 4 |
| chr15_+_79231720 | 3.30 |
ENSMUST00000096350.11
|
Maff
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian) |
| chr14_-_51045182 | 3.28 |
ENSMUST00000227614.2
|
Ccnb1ip1
|
cyclin B1 interacting protein 1 |
| chr3_+_134918298 | 3.23 |
ENSMUST00000062893.12
|
Cenpe
|
centromere protein E |
| chr12_-_69205882 | 3.13 |
ENSMUST00000037023.9
|
Rps29
|
ribosomal protein S29 |
| chr4_+_141028539 | 3.01 |
ENSMUST00000006614.3
|
Epha2
|
Eph receptor A2 |
| chr17_-_26727437 | 2.95 |
ENSMUST00000236661.2
ENSMUST00000025025.7 |
Dusp1
|
dual specificity phosphatase 1 |
| chr12_+_73333641 | 2.82 |
ENSMUST00000153941.8
ENSMUST00000122920.8 ENSMUST00000101313.4 |
Slc38a6
|
solute carrier family 38, member 6 |
| chr7_+_90091937 | 2.80 |
ENSMUST00000061767.5
|
Crebzf
|
CREB/ATF bZIP transcription factor |
| chr4_+_32238712 | 2.80 |
ENSMUST00000108180.9
|
Bach2
|
BTB and CNC homology, basic leucine zipper transcription factor 2 |
| chr4_-_89229256 | 2.77 |
ENSMUST00000097981.6
|
Cdkn2b
|
cyclin dependent kinase inhibitor 2B |
| chr14_+_51045298 | 2.61 |
ENSMUST00000036126.7
|
Parp2
|
poly (ADP-ribose) polymerase family, member 2 |
| chr15_+_79232137 | 2.59 |
ENSMUST00000163691.3
|
Maff
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian) |
| chr7_+_144854542 | 2.58 |
ENSMUST00000033386.12
|
Mrgprf
|
MAS-related GPR, member F |
| chr1_+_75145275 | 2.57 |
ENSMUST00000162768.8
ENSMUST00000160439.8 ENSMUST00000027394.12 |
Zfand2b
|
zinc finger, AN1 type domain 2B |
| chr5_-_124170305 | 2.53 |
ENSMUST00000040967.9
|
Vps37b
|
vacuolar protein sorting 37B |
| chr3_-_85648696 | 2.39 |
ENSMUST00000094148.6
|
Fam160a1
|
family with sequence similarity 160, member A1 |
| chr16_+_38383182 | 2.38 |
ENSMUST00000163948.8
|
Tmem39a
|
transmembrane protein 39a |
| chr16_+_38383154 | 2.36 |
ENSMUST00000171687.8
ENSMUST00000002924.15 |
Tmem39a
|
transmembrane protein 39a |
| chr17_-_56185930 | 2.33 |
ENSMUST00000079642.8
|
Zfp119a
|
zinc finger protein 119a |
| chr7_+_144854627 | 2.33 |
ENSMUST00000117718.2
|
Mrgprf
|
MAS-related GPR, member F |
| chr8_-_70169087 | 2.24 |
ENSMUST00000238761.2
ENSMUST00000238460.2 |
Zfp869
|
zinc finger protein 869 |
| chr8_-_70169500 | 2.11 |
ENSMUST00000238408.2
ENSMUST00000238418.2 ENSMUST00000238753.2 |
Zfp869
|
zinc finger protein 869 |
| chr17_-_56252259 | 2.09 |
ENSMUST00000056147.14
|
Zfp119b
|
zinc finger protein 119b |
| chrX_-_153999440 | 2.03 |
ENSMUST00000026318.15
|
Sat1
|
spermidine/spermine N1-acetyl transferase 1 |
| chrX_-_153999333 | 2.03 |
ENSMUST00000112551.4
|
Sat1
|
spermidine/spermine N1-acetyl transferase 1 |
| chr8_-_70169474 | 2.02 |
ENSMUST00000238657.2
ENSMUST00000238292.2 |
Zfp869
|
zinc finger protein 869 |
| chr7_+_112278534 | 1.96 |
ENSMUST00000106638.10
|
Tead1
|
TEA domain family member 1 |
| chr9_+_102595628 | 1.89 |
ENSMUST00000156485.2
ENSMUST00000145937.2 ENSMUST00000134483.2 ENSMUST00000190047.7 |
Amotl2
|
angiomotin-like 2 |
| chr6_-_86646118 | 1.86 |
ENSMUST00000001184.10
|
Mxd1
|
MAX dimerization protein 1 |
| chr2_+_162859274 | 1.84 |
ENSMUST00000018002.13
ENSMUST00000150396.3 |
Ift52
|
intraflagellar transport 52 |
| chr12_+_73333553 | 1.82 |
ENSMUST00000140523.8
ENSMUST00000126488.8 |
Slc38a6
|
solute carrier family 38, member 6 |
| chr7_+_112278520 | 1.80 |
ENSMUST00000084705.13
ENSMUST00000239442.2 ENSMUST00000239404.2 ENSMUST00000059768.18 |
Tead1
|
TEA domain family member 1 |
| chr7_-_138447933 | 1.75 |
ENSMUST00000118810.2
ENSMUST00000075667.5 ENSMUST00000119664.2 |
Mapk1ip1
|
mitogen-activated protein kinase 1 interacting protein 1 |
| chr13_-_67508655 | 1.75 |
ENSMUST00000081582.8
|
Zfp953
|
zinc finger protein 953 |
| chr4_+_130253925 | 1.63 |
ENSMUST00000105994.4
|
Snrnp40
|
small nuclear ribonucleoprotein 40 (U5) |
| chr4_+_32238950 | 1.60 |
ENSMUST00000037416.13
|
Bach2
|
BTB and CNC homology, basic leucine zipper transcription factor 2 |
| chr17_-_71833752 | 1.54 |
ENSMUST00000232863.2
ENSMUST00000024851.10 |
Ndc80
|
NDC80 kinetochore complex component |
| chrX_-_8042129 | 1.46 |
ENSMUST00000143984.2
|
Tbc1d25
|
TBC1 domain family, member 25 |
| chr12_+_24701273 | 1.37 |
ENSMUST00000020982.7
|
Klf11
|
Kruppel-like factor 11 |
| chr9_-_119151428 | 1.35 |
ENSMUST00000040853.11
|
Oxsr1
|
oxidative-stress responsive 1 |
| chr12_-_86931529 | 1.27 |
ENSMUST00000038422.8
|
Irf2bpl
|
interferon regulatory factor 2 binding protein-like |
| chrX_-_55643429 | 1.24 |
ENSMUST00000059899.3
|
Mmgt1
|
membrane magnesium transporter 1 |
| chr11_+_20151406 | 1.21 |
ENSMUST00000020358.12
ENSMUST00000109602.8 ENSMUST00000109601.8 ENSMUST00000163483.2 |
Rab1a
|
RAB1A, member RAS oncogene family |
| chr13_-_67872383 | 1.20 |
ENSMUST00000127979.9
ENSMUST00000223829.2 ENSMUST00000130891.3 |
Zfp65
|
zinc finger protein 65 |
| chr11_+_93935156 | 1.14 |
ENSMUST00000024979.15
|
Spag9
|
sperm associated antigen 9 |
| chr6_-_83030759 | 1.13 |
ENSMUST00000134606.8
|
Htra2
|
HtrA serine peptidase 2 |
| chr11_+_93935066 | 1.07 |
ENSMUST00000103168.10
|
Spag9
|
sperm associated antigen 9 |
| chr10_+_44144346 | 1.06 |
ENSMUST00000039286.5
|
Atg5
|
autophagy related 5 |
| chr9_+_106048116 | 1.06 |
ENSMUST00000020490.13
|
Wdr82
|
WD repeat domain containing 82 |
| chr14_-_87378641 | 1.03 |
ENSMUST00000168889.3
ENSMUST00000022599.14 |
Diaph3
|
diaphanous related formin 3 |
| chr15_-_76906150 | 0.99 |
ENSMUST00000230031.2
|
Mb
|
myoglobin |
| chr11_+_19874354 | 0.95 |
ENSMUST00000093299.13
|
Spred2
|
sprouty-related EVH1 domain containing 2 |
| chr17_-_36290571 | 0.93 |
ENSMUST00000173724.2
ENSMUST00000172900.8 ENSMUST00000174849.8 |
Prr3
|
proline-rich polypeptide 3 |
| chr5_+_25427860 | 0.92 |
ENSMUST00000045737.14
|
Galnt11
|
polypeptide N-acetylgalactosaminyltransferase 11 |
| chr3_+_40754489 | 0.92 |
ENSMUST00000203295.3
|
Plk4
|
polo like kinase 4 |
| chr4_-_130253703 | 0.90 |
ENSMUST00000134159.3
|
Zcchc17
|
zinc finger, CCHC domain containing 17 |
| chr11_+_93934940 | 0.84 |
ENSMUST00000132079.8
|
Spag9
|
sperm associated antigen 9 |
| chr7_+_97437709 | 0.83 |
ENSMUST00000206984.2
|
Pak1
|
p21 (RAC1) activated kinase 1 |
| chr11_+_93935021 | 0.83 |
ENSMUST00000075695.13
ENSMUST00000092777.11 |
Spag9
|
sperm associated antigen 9 |
| chr8_+_106738105 | 0.82 |
ENSMUST00000034375.11
|
Dus2
|
dihydrouridine synthase 2 |
| chr13_-_34261975 | 0.82 |
ENSMUST00000056427.10
|
Tubb2a
|
tubulin, beta 2A class IIA |
| chr12_-_73333472 | 0.73 |
ENSMUST00000116420.4
ENSMUST00000221189.2 |
Trmt5
|
TRM5 tRNA methyltransferase 5 |
| chr1_-_126758369 | 0.65 |
ENSMUST00000112583.8
ENSMUST00000094609.10 |
Nckap5
|
NCK-associated protein 5 |
| chr17_-_33979442 | 0.63 |
ENSMUST00000057373.14
|
Rab11b
|
RAB11B, member RAS oncogene family |
| chr11_+_19874403 | 0.62 |
ENSMUST00000093298.12
|
Spred2
|
sprouty-related EVH1 domain containing 2 |
| chr1_-_126758520 | 0.62 |
ENSMUST00000162646.8
|
Nckap5
|
NCK-associated protein 5 |
| chr11_+_69214971 | 0.62 |
ENSMUST00000108662.2
|
Trappc1
|
trafficking protein particle complex 1 |
| chr7_-_79974166 | 0.59 |
ENSMUST00000047362.11
ENSMUST00000121882.8 |
Rccd1
|
RCC1 domain containing 1 |
| chr17_-_24926589 | 0.59 |
ENSMUST00000126319.8
|
Tbl3
|
transducin (beta)-like 3 |
| chr10_-_82077105 | 0.59 |
ENSMUST00000041264.15
|
Zfp938
|
zinc finger protein 938 |
| chr14_-_55950939 | 0.58 |
ENSMUST00000168729.8
ENSMUST00000228123.2 ENSMUST00000178034.9 |
Tgm1
|
transglutaminase 1, K polypeptide |
| chr11_+_69214895 | 0.56 |
ENSMUST00000060956.13
|
Trappc1
|
trafficking protein particle complex 1 |
| chr7_-_79973863 | 0.54 |
ENSMUST00000123189.2
|
Rccd1
|
RCC1 domain containing 1 |
| chr5_+_130284431 | 0.53 |
ENSMUST00000044204.12
|
Tyw1
|
tRNA-yW synthesizing protein 1 homolog (S. cerevisiae) |
| chr17_-_56783376 | 0.51 |
ENSMUST00000223859.2
|
Ptprs
|
protein tyrosine phosphatase, receptor type, S |
| chr11_-_70128587 | 0.51 |
ENSMUST00000108576.10
|
0610010K14Rik
|
RIKEN cDNA 0610010K14 gene |
| chr5_-_130284366 | 0.49 |
ENSMUST00000026387.11
|
Sbds
|
SBDS ribosome maturation factor |
| chr2_-_120439826 | 0.47 |
ENSMUST00000102497.10
|
Lrrc57
|
leucine rich repeat containing 57 |
| chr7_+_130294262 | 0.47 |
ENSMUST00000033141.7
|
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr7_+_15853792 | 0.45 |
ENSMUST00000006178.5
|
Kptn
|
kaptin |
| chr11_+_69214883 | 0.41 |
ENSMUST00000102601.10
|
Trappc1
|
trafficking protein particle complex 1 |
| chr7_+_130294403 | 0.41 |
ENSMUST00000207282.2
|
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr2_-_124965537 | 0.35 |
ENSMUST00000142718.8
ENSMUST00000152367.8 ENSMUST00000067780.10 ENSMUST00000147105.8 |
Myef2
|
myelin basic protein expression factor 2, repressor |
| chr11_+_69214789 | 0.34 |
ENSMUST00000102602.8
|
Trappc1
|
trafficking protein particle complex 1 |
| chr10_+_81019076 | 0.34 |
ENSMUST00000219133.2
ENSMUST00000047665.7 |
Dapk3
|
death-associated protein kinase 3 |
| chr10_+_81019117 | 0.32 |
ENSMUST00000218157.2
|
Dapk3
|
death-associated protein kinase 3 |
| chr5_-_25428151 | 0.31 |
ENSMUST00000066954.2
|
E130116L18Rik
|
RIKEN cDNA E130116L18 gene |
| chr5_+_142946098 | 0.29 |
ENSMUST00000031565.15
ENSMUST00000198017.5 |
Fscn1
|
fascin actin-bundling protein 1 |
| chr11_-_70128462 | 0.28 |
ENSMUST00000100950.10
|
0610010K14Rik
|
RIKEN cDNA 0610010K14 gene |
| chr8_-_61407760 | 0.27 |
ENSMUST00000110302.8
|
Clcn3
|
chloride channel, voltage-sensitive 3 |
| chr2_-_120439764 | 0.14 |
ENSMUST00000102496.8
|
Lrrc57
|
leucine rich repeat containing 57 |
| chr10_+_81018834 | 0.12 |
ENSMUST00000178422.9
|
Dapk3
|
death-associated protein kinase 3 |
| chr6_-_83031358 | 0.09 |
ENSMUST00000113962.8
ENSMUST00000089645.13 ENSMUST00000113963.8 |
Htra2
|
HtrA serine peptidase 2 |
| chr6_+_83031502 | 0.05 |
ENSMUST00000092618.9
|
Aup1
|
ancient ubiquitous protein 1 |
| chr19_+_8828132 | 0.03 |
ENSMUST00000235683.2
ENSMUST00000096257.3 |
Lrrn4cl
|
LRRN4 C-terminal like |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 6.4 | GO:0070893 | transposon integration(GO:0070893) regulation of transposon integration(GO:0070894) negative regulation of transposon integration(GO:0070895) |
| 1.0 | 3.0 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.9 | 2.6 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.8 | 3.3 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.8 | 4.1 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.6 | 1.8 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.5 | 2.5 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.5 | 3.9 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.5 | 3.8 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.5 | 1.4 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.4 | 1.2 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.4 | 2.6 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.4 | 3.2 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.3 | 1.1 | GO:0035973 | aggrephagy(GO:0035973) protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.2 | 0.7 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.2 | 3.7 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.2 | 3.3 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.2 | 4.0 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.2 | 3.8 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.2 | 3.9 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) |
| 0.2 | 1.5 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.1 | 0.8 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.1 | 2.8 | GO:0060253 | negative regulation of glial cell proliferation(GO:0060253) |
| 0.1 | 0.9 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.1 | 0.9 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.1 | 1.0 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 1.3 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.1 | 1.5 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.1 | 0.5 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.1 | 0.8 | GO:0031038 | myosin II filament organization(GO:0031038) regulation of myosin II filament organization(GO:0043519) |
| 0.1 | 1.4 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.3 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.1 | 1.2 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.1 | 5.9 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 4.6 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 1.6 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.1 | 0.5 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.1 | 1.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.6 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.1 | 1.1 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.1 | 3.0 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 1.9 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.9 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.0 | 0.2 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.3 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 3.4 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.5 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 4.5 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 | 1.9 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.6 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 1.3 | GO:0008033 | tRNA processing(GO:0008033) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.8 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.4 | 1.5 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.4 | 3.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.4 | 1.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.3 | 3.2 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.2 | 4.0 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.2 | 2.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.2 | 1.8 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 0.9 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 1.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 6.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.6 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 1.9 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 1.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 0.3 | GO:0044393 | microspike(GO:0044393) |
| 0.1 | 1.6 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 0.8 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 4.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.8 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 1.2 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 3.7 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 3.3 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.5 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 1.5 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 4.9 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.3 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 3.0 | GO:0031256 | leading edge membrane(GO:0031256) |
| 0.0 | 0.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 1.1 | 3.3 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 1.0 | 4.1 | GO:0019809 | spermidine binding(GO:0019809) |
| 1.0 | 3.0 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.9 | 3.8 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.9 | 3.7 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.5 | 3.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.4 | 1.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.3 | 1.6 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.3 | 2.6 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.3 | 4.4 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.2 | 3.9 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.2 | 0.8 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.2 | 3.8 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.1 | 4.2 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 2.8 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 1.2 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.1 | 3.0 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.1 | 1.0 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 0.7 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 0.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.6 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.5 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.8 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 1.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 1.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 4.6 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.5 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 2.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 3.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.6 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 2.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.4 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 1.9 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.5 | GO:0019843 | rRNA binding(GO:0019843) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.9 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 3.8 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 6.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 4.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 3.0 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 3.9 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 4.5 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 3.3 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 2.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.0 | PID CDC42 PATHWAY | CDC42 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.1 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.2 | 2.5 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 6.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 3.8 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 2.8 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 3.9 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 0.8 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 3.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.9 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 0.8 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 0.8 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 4.3 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 5.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.6 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 1.1 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 3.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.9 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |