Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Bbx
Z-value: 0.46
Transcription factors associated with Bbx
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Bbx
|
ENSMUSG00000022641.16 | Bbx |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Bbx | mm39_v1_chr16_-_50252703_50252772 | 0.26 | 2.6e-02 | Click! |
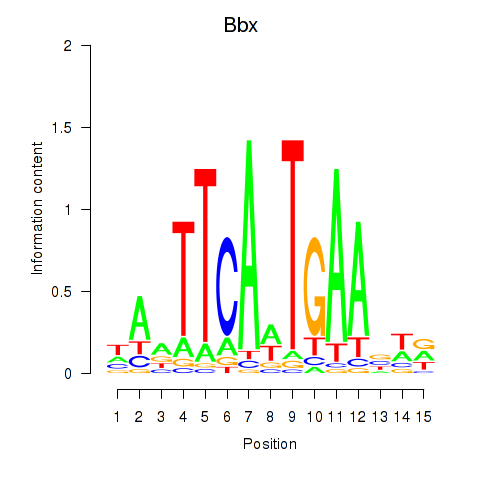
Activity profile of Bbx motif
Sorted Z-values of Bbx motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Bbx
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_46655324 | 3.76 |
ENSMUST00000021802.16
|
Cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr12_+_103354915 | 3.30 |
ENSMUST00000101094.9
ENSMUST00000021620.13 |
Otub2
|
OTU domain, ubiquitin aldehyde binding 2 |
| chr16_+_20511991 | 3.24 |
ENSMUST00000149543.9
ENSMUST00000232207.2 ENSMUST00000118919.9 |
Fam131a
|
family with sequence similarity 131, member A |
| chr13_-_52685305 | 3.01 |
ENSMUST00000057442.8
|
Diras2
|
DIRAS family, GTP-binding RAS-like 2 |
| chr11_-_79418500 | 2.46 |
ENSMUST00000154415.2
|
Evi2a
|
ecotropic viral integration site 2a |
| chr13_+_49761506 | 2.40 |
ENSMUST00000021822.7
|
Ogn
|
osteoglycin |
| chrX_-_149597261 | 2.38 |
ENSMUST00000026302.13
ENSMUST00000129768.8 ENSMUST00000112699.9 |
Maged2
|
MAGE family member D2 |
| chr15_+_91949032 | 2.17 |
ENSMUST00000169825.8
|
Cntn1
|
contactin 1 |
| chr18_+_37554471 | 1.69 |
ENSMUST00000053073.6
|
Pcdhb11
|
protocadherin beta 11 |
| chr1_-_144651157 | 1.62 |
ENSMUST00000027603.4
|
Rgs18
|
regulator of G-protein signaling 18 |
| chr17_+_41121979 | 1.34 |
ENSMUST00000024721.8
ENSMUST00000233740.2 |
Rhag
|
Rhesus blood group-associated A glycoprotein |
| chr10_-_44334683 | 1.32 |
ENSMUST00000105490.3
|
Prdm1
|
PR domain containing 1, with ZNF domain |
| chr8_+_46111703 | 1.26 |
ENSMUST00000134675.8
ENSMUST00000139869.8 ENSMUST00000126067.8 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr11_+_59197746 | 1.24 |
ENSMUST00000000128.10
ENSMUST00000108783.4 |
Wnt9a
|
wingless-type MMTV integration site family, member 9A |
| chr15_+_21111428 | 1.16 |
ENSMUST00000075132.8
|
Cdh12
|
cadherin 12 |
| chr12_-_86125793 | 1.08 |
ENSMUST00000003687.8
|
Tgfb3
|
transforming growth factor, beta 3 |
| chr15_+_30457772 | 0.97 |
ENSMUST00000228282.2
|
Ctnnd2
|
catenin (cadherin associated protein), delta 2 |
| chr15_-_11907267 | 0.96 |
ENSMUST00000228489.2
|
Npr3
|
natriuretic peptide receptor 3 |
| chr10_-_44334711 | 0.92 |
ENSMUST00000039174.11
|
Prdm1
|
PR domain containing 1, with ZNF domain |
| chr2_-_134396268 | 0.91 |
ENSMUST00000028704.3
|
Hao1
|
hydroxyacid oxidase 1, liver |
| chr10_+_58207229 | 0.85 |
ENSMUST00000238939.2
|
Lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr10_+_85857726 | 0.76 |
ENSMUST00000130320.8
|
Fbxo7
|
F-box protein 7 |
| chr13_-_98152768 | 0.69 |
ENSMUST00000238746.2
|
Arhgef28
|
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr8_+_46111778 | 0.67 |
ENSMUST00000143820.8
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr15_-_80929669 | 0.66 |
ENSMUST00000135047.2
|
Mrtfa
|
myocardin related transcription factor A |
| chrX_+_158491589 | 0.63 |
ENSMUST00000080394.13
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr18_-_60724855 | 0.61 |
ENSMUST00000056533.9
|
Myoz3
|
myozenin 3 |
| chr2_-_90301592 | 0.54 |
ENSMUST00000111493.8
|
Ptprj
|
protein tyrosine phosphatase, receptor type, J |
| chr9_-_106448182 | 0.52 |
ENSMUST00000085111.5
|
Iqcf4
|
IQ motif containing F4 |
| chr8_+_71069476 | 0.50 |
ENSMUST00000052437.6
|
Lrrc25
|
leucine rich repeat containing 25 |
| chr8_+_46111310 | 0.50 |
ENSMUST00000153798.8
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chrX_+_77413242 | 0.46 |
ENSMUST00000035626.8
|
4930480E11Rik
|
RIKEN cDNA 4930480E11 gene |
| chr6_+_67890534 | 0.44 |
ENSMUST00000197406.5
ENSMUST00000103311.3 |
Igkv11-125
|
immunoglobulin kappa variable 11-125 |
| chr7_-_97970388 | 0.44 |
ENSMUST00000120520.8
|
Acer3
|
alkaline ceramidase 3 |
| chr2_+_122146153 | 0.40 |
ENSMUST00000099461.4
|
Duox1
|
dual oxidase 1 |
| chr1_-_163552693 | 0.39 |
ENSMUST00000159679.8
|
Mettl11b
|
methyltransferase like 11B |
| chr10_+_85858050 | 0.34 |
ENSMUST00000120344.8
ENSMUST00000117597.2 |
Fbxo7
|
F-box protein 7 |
| chr1_-_144427302 | 0.32 |
ENSMUST00000184189.3
|
Rgs21
|
regulator of G-protein signalling 21 |
| chr10_+_129334361 | 0.29 |
ENSMUST00000203966.3
|
Olfr790
|
olfactory receptor 790 |
| chr2_+_136734088 | 0.27 |
ENSMUST00000099311.9
|
Slx4ip
|
SLX4 interacting protein |
| chrX_+_67805443 | 0.25 |
ENSMUST00000069731.12
ENSMUST00000114647.8 |
Fmr1nb
|
Fmr1 neighbor |
| chr13_-_65200204 | 0.23 |
ENSMUST00000222769.2
|
Prss47
|
protease, serine 47 |
| chr12_-_25147139 | 0.13 |
ENSMUST00000221761.2
|
Id2
|
inhibitor of DNA binding 2 |
| chr2_+_87185159 | 0.12 |
ENSMUST00000215163.3
|
Olfr1120
|
olfactory receptor 1120 |
| chr11_-_87788066 | 0.07 |
ENSMUST00000217095.2
ENSMUST00000215150.2 |
Olfr463
|
olfactory receptor 463 |
| chr10_-_107321938 | 0.05 |
ENSMUST00000000445.2
|
Myf5
|
myogenic factor 5 |
| chr1_-_155908695 | 0.05 |
ENSMUST00000136397.8
|
Tor1aip1
|
torsin A interacting protein 1 |
| chr9_-_38585897 | 0.01 |
ENSMUST00000215461.3
|
Olfr918
|
olfactory receptor 918 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.4 | 1.1 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.3 | 1.3 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.3 | 2.4 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.2 | 3.3 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.2 | 0.9 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.1 | 1.1 | GO:1903207 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.1 | 0.4 | GO:0042335 | cuticle development(GO:0042335) |
| 0.1 | 1.0 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.1 | 0.9 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 1.2 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.1 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 3.8 | GO:0045761 | regulation of adenylate cyclase activity(GO:0045761) |
| 0.0 | 0.5 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.7 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.5 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) |
| 0.0 | 2.4 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.4 | GO:0046512 | sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 2.2 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 1.6 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 3.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.2 | 0.9 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.2 | 1.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.2 | 1.0 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 3.8 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 0.6 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 1.3 | GO:0022842 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.1 | 0.4 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 2.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.5 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 1.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 2.4 | GO:0008083 | growth factor activity(GO:0008083) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.1 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 2.2 | PID NOTCH PATHWAY | Notch signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 3.8 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 2.2 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.3 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 1.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.6 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |