Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
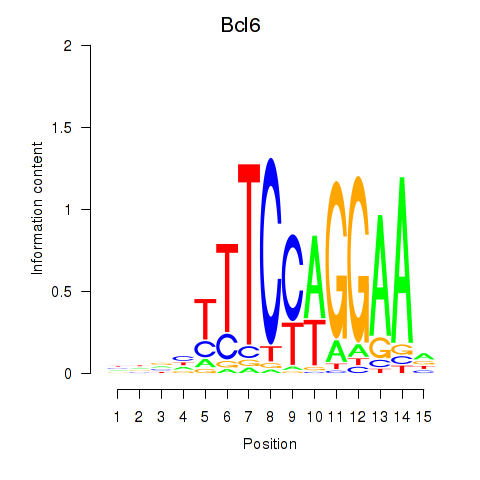
Results for Bcl6
Z-value: 0.95
Transcription factors associated with Bcl6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Bcl6
|
ENSMUSG00000022508.6 | Bcl6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Bcl6 | mm39_v1_chr16_-_23807602_23807602 | 0.09 | 4.5e-01 | Click! |
Activity profile of Bcl6 motif
Sorted Z-values of Bcl6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Bcl6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_15881256 | 7.74 |
ENSMUST00000029876.2
|
Calb1
|
calbindin 1 |
| chrX_-_60229164 | 6.52 |
ENSMUST00000166381.3
|
Cdr1
|
cerebellar degeneration related antigen 1 |
| chr8_+_23525101 | 5.81 |
ENSMUST00000117662.8
ENSMUST00000117296.8 ENSMUST00000141784.9 |
Ank1
|
ankyrin 1, erythroid |
| chr6_-_5496261 | 5.43 |
ENSMUST00000203347.3
ENSMUST00000019721.7 |
Pdk4
|
pyruvate dehydrogenase kinase, isoenzyme 4 |
| chr9_+_107173907 | 5.15 |
ENSMUST00000168260.2
|
Cish
|
cytokine inducible SH2-containing protein |
| chrX_+_163219983 | 4.79 |
ENSMUST00000036858.11
|
Asb11
|
ankyrin repeat and SOCS box-containing 11 |
| chr9_+_50664288 | 4.07 |
ENSMUST00000214962.2
ENSMUST00000216755.2 |
Cryab
|
crystallin, alpha B |
| chr9_+_50664207 | 3.78 |
ENSMUST00000034562.9
|
Cryab
|
crystallin, alpha B |
| chr1_-_66984178 | 3.69 |
ENSMUST00000027151.12
|
Myl1
|
myosin, light polypeptide 1 |
| chr1_-_66984521 | 3.69 |
ENSMUST00000160100.2
|
Myl1
|
myosin, light polypeptide 1 |
| chr11_+_101136821 | 3.52 |
ENSMUST00000129680.8
|
Ramp2
|
receptor (calcitonin) activity modifying protein 2 |
| chr1_-_169575203 | 3.52 |
ENSMUST00000027991.12
ENSMUST00000111357.2 |
Rgs4
|
regulator of G-protein signaling 4 |
| chr4_+_120523758 | 3.40 |
ENSMUST00000094814.6
|
Cited4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 |
| chr9_+_107174081 | 3.33 |
ENSMUST00000167072.2
|
Cish
|
cytokine inducible SH2-containing protein |
| chr9_-_121621544 | 3.32 |
ENSMUST00000035110.11
|
Hhatl
|
hedgehog acyltransferase-like |
| chr14_+_70694887 | 3.21 |
ENSMUST00000003561.10
|
Phyhip
|
phytanoyl-CoA hydroxylase interacting protein |
| chr4_+_152358648 | 3.17 |
ENSMUST00000105650.8
ENSMUST00000105651.8 |
Gpr153
|
G protein-coupled receptor 153 |
| chr8_+_69333143 | 3.04 |
ENSMUST00000015712.15
|
Lpl
|
lipoprotein lipase |
| chr11_-_3864664 | 2.89 |
ENSMUST00000109995.2
ENSMUST00000051207.2 |
Slc35e4
|
solute carrier family 35, member E4 |
| chr3_+_19698631 | 2.74 |
ENSMUST00000029139.9
|
Trim55
|
tripartite motif-containing 55 |
| chr10_+_86136236 | 2.73 |
ENSMUST00000020234.14
|
Timp3
|
tissue inhibitor of metalloproteinase 3 |
| chr14_-_70864448 | 2.69 |
ENSMUST00000110984.4
|
Dmtn
|
dematin actin binding protein |
| chr14_-_70864666 | 2.69 |
ENSMUST00000022694.17
|
Dmtn
|
dematin actin binding protein |
| chr18_+_61178211 | 2.68 |
ENSMUST00000025522.11
ENSMUST00000115274.2 |
Pdgfrb
|
platelet derived growth factor receptor, beta polypeptide |
| chr3_-_152687877 | 2.55 |
ENSMUST00000044278.6
|
St6galnac5
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| chr17_+_71326510 | 2.50 |
ENSMUST00000073211.13
ENSMUST00000024847.14 |
Myom1
|
myomesin 1 |
| chr3_-_126918491 | 2.43 |
ENSMUST00000238781.2
|
Ank2
|
ankyrin 2, brain |
| chr1_-_165955445 | 2.41 |
ENSMUST00000085992.4
ENSMUST00000192369.6 |
Dusp27
|
dual specificity phosphatase 27 (putative) |
| chr9_-_50663571 | 2.40 |
ENSMUST00000042790.5
|
Hspb2
|
heat shock protein 2 |
| chr3_+_14011445 | 2.36 |
ENSMUST00000192209.6
ENSMUST00000171075.8 ENSMUST00000108372.4 |
Ralyl
|
RALY RNA binding protein-like |
| chr14_+_66581745 | 2.26 |
ENSMUST00000152093.8
ENSMUST00000074523.13 |
Stmn4
|
stathmin-like 4 |
| chr9_-_58066484 | 2.25 |
ENSMUST00000041477.15
|
Islr
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr3_-_87170903 | 2.19 |
ENSMUST00000090986.11
|
Fcrls
|
Fc receptor-like S, scavenger receptor |
| chr8_+_12965876 | 2.18 |
ENSMUST00000110876.9
ENSMUST00000110879.9 |
Mcf2l
|
mcf.2 transforming sequence-like |
| chr7_-_81143412 | 2.15 |
ENSMUST00000238438.2
ENSMUST00000238711.2 |
Ap3b2
|
adaptor-related protein complex 3, beta 2 subunit |
| chr2_-_18042548 | 2.14 |
ENSMUST00000066163.3
|
A930004D18Rik
|
RIKEN cDNA A930004D18 gene |
| chr4_+_114916703 | 2.10 |
ENSMUST00000162489.2
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr2_+_32518402 | 2.10 |
ENSMUST00000156578.8
|
Ak1
|
adenylate kinase 1 |
| chr9_+_77543776 | 2.08 |
ENSMUST00000057781.8
|
Klhl31
|
kelch-like 31 |
| chr7_+_45055077 | 2.07 |
ENSMUST00000107774.3
|
Kcna7
|
potassium voltage-gated channel, shaker-related subfamily, member 7 |
| chr19_+_56450085 | 2.04 |
ENSMUST00000225909.2
|
Plekhs1
|
pleckstrin homology domain containing, family S member 1 |
| chr18_+_32087883 | 1.99 |
ENSMUST00000223753.2
|
Lims2
|
LIM and senescent cell antigen like domains 2 |
| chr3_+_37402795 | 1.96 |
ENSMUST00000200585.5
ENSMUST00000038885.10 |
Fgf2
|
fibroblast growth factor 2 |
| chr17_+_71326542 | 1.95 |
ENSMUST00000179759.3
|
Myom1
|
myomesin 1 |
| chr3_+_95406567 | 1.94 |
ENSMUST00000015664.5
|
Ctsk
|
cathepsin K |
| chr12_-_100486950 | 1.90 |
ENSMUST00000223020.2
ENSMUST00000062957.8 |
Ttc7b
|
tetratricopeptide repeat domain 7B |
| chr19_+_56450062 | 1.83 |
ENSMUST00000178590.9
ENSMUST00000039666.8 |
Plekhs1
|
pleckstrin homology domain containing, family S member 1 |
| chr7_-_142215027 | 1.83 |
ENSMUST00000105936.8
|
Igf2
|
insulin-like growth factor 2 |
| chr19_-_10847121 | 1.81 |
ENSMUST00000120524.2
ENSMUST00000025645.14 |
Tmem132a
|
transmembrane protein 132A |
| chr13_+_21679387 | 1.81 |
ENSMUST00000104942.2
|
AK157302
|
cDNA sequence AK157302 |
| chr7_+_110367375 | 1.80 |
ENSMUST00000170374.8
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr16_-_34083549 | 1.79 |
ENSMUST00000114949.8
ENSMUST00000114954.8 |
Kalrn
|
kalirin, RhoGEF kinase |
| chr3_-_57202301 | 1.79 |
ENSMUST00000171384.8
|
Tm4sf1
|
transmembrane 4 superfamily member 1 |
| chr16_-_36605286 | 1.77 |
ENSMUST00000168279.2
ENSMUST00000164579.9 ENSMUST00000023616.11 |
Slc15a2
|
solute carrier family 15 (H+/peptide transporter), member 2 |
| chr9_-_73876182 | 1.76 |
ENSMUST00000184666.8
|
Unc13c
|
unc-13 homolog C |
| chr3_-_57202546 | 1.75 |
ENSMUST00000196506.2
|
Tm4sf1
|
transmembrane 4 superfamily member 1 |
| chr9_+_123822000 | 1.74 |
ENSMUST00000039171.9
|
Ccr3
|
chemokine (C-C motif) receptor 3 |
| chr8_-_11329656 | 1.74 |
ENSMUST00000208095.2
|
Col4a1
|
collagen, type IV, alpha 1 |
| chr4_-_133981387 | 1.70 |
ENSMUST00000060050.6
|
Grrp1
|
glycine/arginine rich protein 1 |
| chr13_+_83720457 | 1.69 |
ENSMUST00000196730.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr7_+_110372860 | 1.68 |
ENSMUST00000143786.2
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr14_+_84680993 | 1.65 |
ENSMUST00000071370.7
|
Pcdh17
|
protocadherin 17 |
| chr8_-_105350898 | 1.64 |
ENSMUST00000212882.2
ENSMUST00000163783.4 |
Cdh16
|
cadherin 16 |
| chr9_-_50663648 | 1.63 |
ENSMUST00000217159.2
|
Hspb2
|
heat shock protein 2 |
| chr11_-_55310724 | 1.61 |
ENSMUST00000108858.8
ENSMUST00000141530.2 |
Sparc
|
secreted acidic cysteine rich glycoprotein |
| chr14_-_56181993 | 1.60 |
ENSMUST00000022834.7
ENSMUST00000226280.2 |
Cma1
|
chymase 1, mast cell |
| chr6_+_42015319 | 1.55 |
ENSMUST00000117406.8
ENSMUST00000024059.5 |
Sva
|
seminal vesicle antigen |
| chr1_-_133681419 | 1.55 |
ENSMUST00000125659.8
ENSMUST00000048953.14 ENSMUST00000165602.9 |
Atp2b4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr9_-_79884920 | 1.55 |
ENSMUST00000239133.2
|
Filip1
|
filamin A interacting protein 1 |
| chr11_-_90528888 | 1.55 |
ENSMUST00000020858.14
ENSMUST00000107875.8 ENSMUST00000107872.8 ENSMUST00000143203.8 |
Stxbp4
|
syntaxin binding protein 4 |
| chr6_+_83162928 | 1.54 |
ENSMUST00000113907.2
|
Dctn1
|
dynactin 1 |
| chr13_+_83720484 | 1.53 |
ENSMUST00000196207.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr5_+_34153328 | 1.53 |
ENSMUST00000056355.9
|
Nat8l
|
N-acetyltransferase 8-like |
| chr16_-_34083315 | 1.52 |
ENSMUST00000114953.8
|
Kalrn
|
kalirin, RhoGEF kinase |
| chr4_+_33062999 | 1.51 |
ENSMUST00000108162.8
ENSMUST00000024035.9 |
Gabrr2
|
gamma-aminobutyric acid (GABA) C receptor, subunit rho 2 |
| chr3_-_88417251 | 1.45 |
ENSMUST00000149068.2
|
Lmna
|
lamin A |
| chr9_-_79885063 | 1.44 |
ENSMUST00000093811.11
|
Filip1
|
filamin A interacting protein 1 |
| chr1_-_134883577 | 1.44 |
ENSMUST00000168381.8
|
Ppp1r12b
|
protein phosphatase 1, regulatory subunit 12B |
| chr14_-_22039543 | 1.43 |
ENSMUST00000043409.9
|
Zfp503
|
zinc finger protein 503 |
| chr9_+_122980006 | 1.42 |
ENSMUST00000026890.6
|
Clec3b
|
C-type lectin domain family 3, member b |
| chr10_+_69761597 | 1.39 |
ENSMUST00000182269.8
ENSMUST00000183261.8 ENSMUST00000183074.8 |
Ank3
|
ankyrin 3, epithelial |
| chr6_-_124746510 | 1.37 |
ENSMUST00000149652.2
ENSMUST00000112476.8 ENSMUST00000004378.15 |
Eno2
|
enolase 2, gamma neuronal |
| chr11_-_118246566 | 1.36 |
ENSMUST00000155707.3
|
Timp2
|
tissue inhibitor of metalloproteinase 2 |
| chr9_-_49479791 | 1.36 |
ENSMUST00000194252.6
|
Ncam1
|
neural cell adhesion molecule 1 |
| chr16_-_34083200 | 1.36 |
ENSMUST00000114947.2
|
Kalrn
|
kalirin, RhoGEF kinase |
| chr15_-_27788693 | 1.35 |
ENSMUST00000226287.2
|
Trio
|
triple functional domain (PTPRF interacting) |
| chr8_-_105350881 | 1.34 |
ENSMUST00000211903.2
|
Cdh16
|
cadherin 16 |
| chr10_+_69234810 | 1.32 |
ENSMUST00000218680.2
|
Ank3
|
ankyrin 3, epithelial |
| chrX_+_72719098 | 1.31 |
ENSMUST00000171398.2
|
Slc6a8
|
solute carrier family 6 (neurotransmitter transporter, creatine), member 8 |
| chr9_-_39515420 | 1.30 |
ENSMUST00000042485.11
ENSMUST00000141370.8 |
AW551984
|
expressed sequence AW551984 |
| chr1_-_134883645 | 1.30 |
ENSMUST00000045665.13
ENSMUST00000086444.6 ENSMUST00000112163.2 |
Ppp1r12b
|
protein phosphatase 1, regulatory subunit 12B |
| chr18_-_36859732 | 1.29 |
ENSMUST00000061829.8
|
Cd14
|
CD14 antigen |
| chr2_+_70393782 | 1.29 |
ENSMUST00000123330.3
|
Gad1
|
glutamate decarboxylase 1 |
| chr15_-_8739893 | 1.29 |
ENSMUST00000157065.2
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr1_+_21288799 | 1.27 |
ENSMUST00000027065.12
|
Tmem14a
|
transmembrane protein 14A |
| chr4_+_42922253 | 1.26 |
ENSMUST00000139100.2
|
Phf24
|
PHD finger protein 24 |
| chr18_+_37453427 | 1.26 |
ENSMUST00000078271.4
|
Pcdhb5
|
protocadherin beta 5 |
| chr16_-_36605147 | 1.25 |
ENSMUST00000165531.9
|
Slc15a2
|
solute carrier family 15 (H+/peptide transporter), member 2 |
| chr11_+_81926394 | 1.24 |
ENSMUST00000000193.6
|
Ccl2
|
chemokine (C-C motif) ligand 2 |
| chr11_+_68582923 | 1.23 |
ENSMUST00000018887.15
|
Myh10
|
myosin, heavy polypeptide 10, non-muscle |
| chr2_-_64806106 | 1.22 |
ENSMUST00000156765.2
|
Grb14
|
growth factor receptor bound protein 14 |
| chr5_+_37332834 | 1.20 |
ENSMUST00000208827.2
ENSMUST00000207619.2 |
Gm1043
|
predicted gene 1043 |
| chrX_+_72108393 | 1.18 |
ENSMUST00000060418.8
|
Pnma3
|
paraneoplastic antigen MA3 |
| chr15_-_54783357 | 1.18 |
ENSMUST00000167541.3
ENSMUST00000171545.9 ENSMUST00000041591.16 ENSMUST00000173516.8 |
Enpp2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr8_-_105350842 | 1.17 |
ENSMUST00000212324.2
|
Cdh16
|
cadherin 16 |
| chr1_+_173986288 | 1.17 |
ENSMUST00000068403.4
|
Olfr420
|
olfactory receptor 420 |
| chr9_+_102597486 | 1.17 |
ENSMUST00000130602.2
|
Amotl2
|
angiomotin-like 2 |
| chr14_+_52122439 | 1.17 |
ENSMUST00000167984.2
|
Mettl17
|
methyltransferase like 17 |
| chr8_-_105350816 | 1.16 |
ENSMUST00000212447.2
|
Cdh16
|
cadherin 16 |
| chr3_-_14843512 | 1.16 |
ENSMUST00000094365.11
|
Car1
|
carbonic anhydrase 1 |
| chr10_+_99851679 | 1.16 |
ENSMUST00000130190.8
ENSMUST00000218200.2 ENSMUST00000020129.8 |
Kitl
|
kit ligand |
| chr10_+_69761784 | 1.16 |
ENSMUST00000181974.8
ENSMUST00000182795.8 ENSMUST00000182437.8 |
Ank3
|
ankyrin 3, epithelial |
| chr15_-_8740218 | 1.15 |
ENSMUST00000005493.14
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr7_+_65511482 | 1.14 |
ENSMUST00000176199.8
|
Pcsk6
|
proprotein convertase subtilisin/kexin type 6 |
| chr3_-_89000591 | 1.12 |
ENSMUST00000090929.12
ENSMUST00000052539.13 |
Rusc1
|
RUN and SH3 domain containing 1 |
| chr2_+_70393195 | 1.11 |
ENSMUST00000130998.8
|
Gad1
|
glutamate decarboxylase 1 |
| chr10_+_29019645 | 1.10 |
ENSMUST00000092629.4
|
Soga3
|
SOGA family member 3 |
| chr3_+_38941089 | 1.10 |
ENSMUST00000061260.8
|
Fat4
|
FAT atypical cadherin 4 |
| chr17_+_28910302 | 1.08 |
ENSMUST00000004990.14
ENSMUST00000114754.8 ENSMUST00000062694.16 |
Mapk14
|
mitogen-activated protein kinase 14 |
| chr9_+_105520154 | 1.07 |
ENSMUST00000190358.2
ENSMUST00000191268.7 ENSMUST00000065778.13 ENSMUST00000188784.2 |
Pik3r4
|
phosphoinositide-3-kinase regulatory subunit 4 |
| chr9_-_109025043 | 1.06 |
ENSMUST00000061456.8
|
Fbxw13
|
F-box and WD-40 domain protein 13 |
| chr12_+_88920169 | 1.06 |
ENSMUST00000057634.14
|
Nrxn3
|
neurexin III |
| chr2_+_123931879 | 1.06 |
ENSMUST00000103241.8
|
Sema6d
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| chr12_-_112637998 | 1.05 |
ENSMUST00000128300.9
|
Akt1
|
thymoma viral proto-oncogene 1 |
| chr15_-_93417380 | 1.04 |
ENSMUST00000109255.3
|
Prickle1
|
prickle planar cell polarity protein 1 |
| chr12_-_111933328 | 1.03 |
ENSMUST00000222874.2
ENSMUST00000223191.2 ENSMUST00000021719.7 |
Atp5mpl
|
ATP synthase membrane subunit 6.8PL |
| chr11_+_97732108 | 1.02 |
ENSMUST00000155954.3
ENSMUST00000164364.2 ENSMUST00000170806.2 |
B230217C12Rik
|
RIKEN cDNA B230217C12 gene |
| chr18_+_44403169 | 1.02 |
ENSMUST00000042747.4
|
Npy6r
|
neuropeptide Y receptor Y6 |
| chr6_-_131655849 | 1.01 |
ENSMUST00000076756.3
|
Tas2r106
|
taste receptor, type 2, member 106 |
| chrX_-_50031587 | 1.00 |
ENSMUST00000060650.7
|
Frmd7
|
FERM domain containing 7 |
| chr5_+_102629240 | 1.00 |
ENSMUST00000073302.12
ENSMUST00000094559.9 |
Arhgap24
|
Rho GTPase activating protein 24 |
| chr8_-_29709455 | 0.99 |
ENSMUST00000211448.2
ENSMUST00000210298.2 ENSMUST00000209401.2 ENSMUST00000210785.2 |
Unc5d
|
unc-5 netrin receptor D |
| chr2_-_7086018 | 0.99 |
ENSMUST00000114923.3
ENSMUST00000182706.8 |
Celf2
|
CUGBP, Elav-like family member 2 |
| chr7_-_30614249 | 0.98 |
ENSMUST00000190950.7
ENSMUST00000187137.7 ENSMUST00000190638.7 |
Mag
|
myelin-associated glycoprotein |
| chr7_-_102408573 | 0.97 |
ENSMUST00000210453.3
ENSMUST00000232246.3 ENSMUST00000239110.2 ENSMUST00000060187.15 ENSMUST00000168007.3 |
Olfr560
Olfr78
|
olfactory receptor 560 olfactory receptor 78 |
| chr15_-_37459570 | 0.97 |
ENSMUST00000119730.8
ENSMUST00000120746.8 |
Ncald
|
neurocalcin delta |
| chr2_-_13798843 | 0.96 |
ENSMUST00000003509.10
|
St8sia6
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 6 |
| chrX_+_100342749 | 0.95 |
ENSMUST00000118111.8
ENSMUST00000130555.8 ENSMUST00000151528.8 |
Nlgn3
|
neuroligin 3 |
| chr14_+_63284438 | 0.95 |
ENSMUST00000067990.8
ENSMUST00000111203.2 |
Defb42
|
defensin beta 42 |
| chr9_-_109168710 | 0.94 |
ENSMUST00000196351.2
ENSMUST00000200156.5 ENSMUST00000112040.9 ENSMUST00000112039.7 |
Fbxw28
|
F-box and WD-40 domain protein 28 |
| chr6_+_41498716 | 0.93 |
ENSMUST00000070380.5
|
Prss2
|
protease, serine 2 |
| chr17_+_28910393 | 0.93 |
ENSMUST00000124886.9
ENSMUST00000114758.9 |
Mapk14
|
mitogen-activated protein kinase 14 |
| chr8_-_96580129 | 0.91 |
ENSMUST00000212628.2
ENSMUST00000040481.4 ENSMUST00000212270.2 |
Slc38a7
|
solute carrier family 38, member 7 |
| chr16_-_4698148 | 0.90 |
ENSMUST00000037843.7
|
Ubald1
|
UBA-like domain containing 1 |
| chr8_-_18791557 | 0.90 |
ENSMUST00000033846.7
|
Angpt2
|
angiopoietin 2 |
| chrX_-_72974357 | 0.90 |
ENSMUST00000155597.2
ENSMUST00000114379.8 |
Renbp
|
renin binding protein |
| chr7_-_12096691 | 0.88 |
ENSMUST00000086228.3
|
Vmn1r84
|
vomeronasal 1 receptor 84 |
| chrX_-_72974440 | 0.88 |
ENSMUST00000116578.8
|
Renbp
|
renin binding protein |
| chr2_-_76671491 | 0.87 |
ENSMUST00000137854.8
|
Ttn
|
titin |
| chr9_-_109116739 | 0.86 |
ENSMUST00000112041.6
ENSMUST00000198844.5 |
Fbxw14
|
F-box and WD-40 domain protein 14 |
| chr17_-_56428968 | 0.86 |
ENSMUST00000041357.9
|
Lrg1
|
leucine-rich alpha-2-glycoprotein 1 |
| chr7_-_81356557 | 0.86 |
ENSMUST00000207983.2
|
Homer2
|
homer scaffolding protein 2 |
| chr10_+_69761630 | 0.85 |
ENSMUST00000182029.8
|
Ank3
|
ankyrin 3, epithelial |
| chr5_-_114127800 | 0.85 |
ENSMUST00000077689.14
|
Ssh1
|
slingshot protein phosphatase 1 |
| chr6_-_13838423 | 0.84 |
ENSMUST00000115492.2
|
Gpr85
|
G protein-coupled receptor 85 |
| chr14_-_51384236 | 0.84 |
ENSMUST00000080126.4
|
Rnase1
|
ribonuclease, RNase A family, 1 (pancreatic) |
| chr3_+_37402495 | 0.84 |
ENSMUST00000138563.9
|
Fgf2
|
fibroblast growth factor 2 |
| chrX_+_100342813 | 0.83 |
ENSMUST00000065858.3
|
Nlgn3
|
neuroligin 3 |
| chr9_+_90045219 | 0.83 |
ENSMUST00000147250.8
ENSMUST00000113060.3 |
Adamts7
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 7 |
| chr15_-_37458768 | 0.82 |
ENSMUST00000116445.9
|
Ncald
|
neurocalcin delta |
| chr11_-_78948432 | 0.82 |
ENSMUST00000129463.2
|
Ksr1
|
kinase suppressor of ras 1 |
| chr6_-_57938488 | 0.82 |
ENSMUST00000228097.2
|
Vmn1r24
|
vomeronasal 1 receptor 24 |
| chr17_+_25339734 | 0.81 |
ENSMUST00000054930.7
ENSMUST00000232902.2 ENSMUST00000233502.2 |
Ptx4
|
pentraxin 4 |
| chr7_+_6463510 | 0.81 |
ENSMUST00000056120.5
|
Olfr1336
|
olfactory receptor 1336 |
| chr1_+_21288886 | 0.80 |
ENSMUST00000027064.8
|
Tmem14a
|
transmembrane protein 14A |
| chr14_+_118374511 | 0.80 |
ENSMUST00000022728.4
|
Gpr180
|
G protein-coupled receptor 180 |
| chr3_+_79791798 | 0.80 |
ENSMUST00000118853.8
ENSMUST00000145992.2 |
Gask1b
|
golgi associated kinase 1B |
| chr4_-_135000109 | 0.80 |
ENSMUST00000037099.9
|
Clic4
|
chloride intracellular channel 4 (mitochondrial) |
| chr2_+_177834868 | 0.77 |
ENSMUST00000103065.2
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr2_+_119863909 | 0.76 |
ENSMUST00000126150.2
|
Pla2g4b
|
phospholipase A2, group IVB (cytosolic) |
| chr6_-_41354538 | 0.75 |
ENSMUST00000096003.7
|
Prss3
|
protease, serine 3 |
| chr5_+_134212836 | 0.75 |
ENSMUST00000016086.10
|
Gtf2ird2
|
GTF2I repeat domain containing 2 |
| chr2_-_25498459 | 0.75 |
ENSMUST00000058137.9
|
Rabl6
|
RAB, member RAS oncogene family-like 6 |
| chr5_-_6926523 | 0.74 |
ENSMUST00000164784.2
|
Zfp804b
|
zinc finger protein 804B |
| chr7_+_81508741 | 0.74 |
ENSMUST00000041890.8
|
Tm6sf1
|
transmembrane 6 superfamily member 1 |
| chr12_-_114160354 | 0.73 |
ENSMUST00000103476.2
ENSMUST00000191693.2 |
Ighv3-3
|
immunoglobulin heavy variable V3-3 |
| chr8_-_29709652 | 0.73 |
ENSMUST00000168630.4
|
Unc5d
|
unc-5 netrin receptor D |
| chr6_+_66706723 | 0.73 |
ENSMUST00000227923.2
ENSMUST00000226886.2 |
Vmn1r37
|
vomeronasal 1 receptor 37 |
| chr2_+_101716577 | 0.73 |
ENSMUST00000028584.8
|
Commd9
|
COMM domain containing 9 |
| chr8_-_88531018 | 0.73 |
ENSMUST00000165770.9
|
Zfp423
|
zinc finger protein 423 |
| chr2_+_164174660 | 0.72 |
ENSMUST00000017148.8
|
Svs5
|
seminal vesicle secretory protein 5 |
| chr12_-_114073050 | 0.72 |
ENSMUST00000103472.4
|
Ighv9-2
|
immunoglobulin heavy variable V9-2 |
| chr4_-_130068484 | 0.72 |
ENSMUST00000132545.3
ENSMUST00000175992.8 ENSMUST00000105999.9 |
Tinagl1
|
tubulointerstitial nephritis antigen-like 1 |
| chr8_-_41469332 | 0.71 |
ENSMUST00000131965.2
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chrX_-_135641869 | 0.71 |
ENSMUST00000166930.8
ENSMUST00000113095.8 |
Morf4l2
|
mortality factor 4 like 2 |
| chr8_-_37420293 | 0.71 |
ENSMUST00000179501.2
|
Dlc1
|
deleted in liver cancer 1 |
| chr10_+_97442727 | 0.70 |
ENSMUST00000105286.4
|
Kera
|
keratocan |
| chr2_+_111329683 | 0.69 |
ENSMUST00000219064.3
|
Olfr1291-ps1
|
olfactory receptor 1291, pseudogene 1 |
| chrX_+_92718695 | 0.69 |
ENSMUST00000045898.4
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr19_-_12206908 | 0.69 |
ENSMUST00000180978.3
|
Olfr1432
|
olfactory receptor 1432 |
| chr16_-_16345197 | 0.69 |
ENSMUST00000069284.14
|
Fgd4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr9_+_44398524 | 0.69 |
ENSMUST00000218183.2
|
Bcl9l
|
B cell CLL/lymphoma 9-like |
| chr15_-_101262452 | 0.69 |
ENSMUST00000230909.2
|
Krt80
|
keratin 80 |
| chr11_-_58346806 | 0.69 |
ENSMUST00000055204.6
|
Olfr30
|
olfactory receptor 30 |
| chr1_+_173868569 | 0.68 |
ENSMUST00000052975.6
|
Olfr433
|
olfactory receptor 433 |
| chr8_+_85628557 | 0.68 |
ENSMUST00000067060.10
ENSMUST00000239392.2 |
Klf1
|
Kruppel-like factor 1 (erythroid) |
| chr13_-_23417289 | 0.67 |
ENSMUST00000077116.5
|
Vmn1r222
|
vomeronasal 1 receptor 222 |
| chr1_-_169938298 | 0.67 |
ENSMUST00000192312.6
|
Ddr2
|
discoidin domain receptor family, member 2 |
| chrX_-_135642025 | 0.67 |
ENSMUST00000155207.8
ENSMUST00000080411.13 ENSMUST00000169418.8 |
Morf4l2
|
mortality factor 4 like 2 |
| chr19_+_12647803 | 0.66 |
ENSMUST00000207341.3
ENSMUST00000208494.3 ENSMUST00000208657.3 |
Olfr1442
|
olfactory receptor 1442 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.7 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 1.4 | 5.4 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 1.1 | 3.3 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.9 | 2.8 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.9 | 5.4 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.9 | 2.7 | GO:0072223 | cell migration involved in vasculogenesis(GO:0035441) metanephric glomerular mesangium development(GO:0072223) metanephric glomerular mesangial cell proliferation involved in metanephros development(GO:0072262) |
| 0.7 | 7.8 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.7 | 2.1 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.7 | 4.8 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.7 | 2.7 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.7 | 5.9 | GO:0098885 | modification of postsynaptic actin cytoskeleton(GO:0098885) |
| 0.6 | 1.8 | GO:2000331 | regulation of terminal button organization(GO:2000331) |
| 0.5 | 1.6 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) |
| 0.5 | 1.5 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.5 | 1.4 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.4 | 3.5 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.4 | 1.3 | GO:0015881 | creatine transport(GO:0015881) |
| 0.4 | 3.0 | GO:0055095 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.4 | 1.3 | GO:0071724 | response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) |
| 0.4 | 3.0 | GO:0042938 | antibiotic transport(GO:0042891) dipeptide transport(GO:0042938) |
| 0.4 | 1.2 | GO:0090264 | immune complex clearance by monocytes and macrophages(GO:0002436) monocyte homeostasis(GO:0035702) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) |
| 0.4 | 1.2 | GO:1903537 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.4 | 1.5 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.4 | 1.5 | GO:0051586 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.4 | 4.0 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.4 | 4.7 | GO:1900827 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.4 | 2.1 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.3 | 3.5 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.3 | 1.0 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 0.3 | 1.4 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.3 | 1.0 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.3 | 5.3 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.3 | 0.6 | GO:1900133 | regulation of renin secretion into blood stream(GO:1900133) |
| 0.3 | 1.4 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.3 | 0.9 | GO:0098976 | excitatory chemical synaptic transmission(GO:0098976) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.3 | 1.1 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.3 | 2.4 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.3 | 3.2 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.3 | 1.0 | GO:0060709 | glycogen cell differentiation involved in embryonic placenta development(GO:0060709) |
| 0.3 | 1.8 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.3 | 2.0 | GO:2001184 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.2 | 1.8 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.2 | 8.5 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.2 | 1.7 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.2 | 0.8 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.2 | 1.1 | GO:0032902 | nerve growth factor production(GO:0032902) |
| 0.2 | 2.0 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.2 | 0.9 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.2 | 0.9 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.2 | 2.3 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.2 | 1.2 | GO:1902035 | mast cell proliferation(GO:0070662) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.2 | 1.8 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.2 | 1.4 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.2 | 0.5 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.2 | 1.8 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.2 | 5.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 1.2 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 1.5 | GO:1904177 | regulation of adipose tissue development(GO:1904177) |
| 0.1 | 0.6 | GO:0051673 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) membrane disruption in other organism(GO:0051673) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.1 | 0.8 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 5.8 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.1 | 1.8 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 3.5 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.1 | 0.4 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.1 | 0.6 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.1 | 0.4 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.1 | 0.5 | GO:0035483 | gastric motility(GO:0035482) gastric emptying(GO:0035483) |
| 0.1 | 1.0 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 | 1.3 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.1 | 0.5 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.1 | 1.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.3 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 0.4 | GO:0072233 | ascending thin limb development(GO:0072021) thick ascending limb development(GO:0072023) metanephric ascending thin limb development(GO:0072218) metanephric thick ascending limb development(GO:0072233) |
| 0.1 | 2.1 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 0.4 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.1 | 2.3 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 0.7 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 0.4 | GO:0075044 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.1 | 0.8 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 0.4 | GO:1902966 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.1 | 9.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 0.3 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.1 | 0.6 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.1 | 2.8 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 1.3 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.1 | 1.7 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) |
| 0.1 | 0.2 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.1 | 0.7 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 | 1.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.7 | GO:2000980 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 | 1.9 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.3 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 1.0 | GO:0048711 | positive regulation of astrocyte differentiation(GO:0048711) |
| 0.0 | 0.7 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 0.4 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.0 | 0.1 | GO:1900248 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.0 | 1.1 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.7 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.4 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 1.7 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 2.6 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 0.2 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.8 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 1.9 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.5 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.0 | 0.3 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.7 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.0 | 0.3 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 1.1 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.7 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.2 | GO:1990839 | response to endothelin(GO:1990839) |
| 0.0 | 0.8 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 1.2 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.6 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 1.0 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.2 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.8 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.0 | 0.9 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 1.2 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 6.0 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.0 | 2.4 | GO:0043627 | response to estrogen(GO:0043627) |
| 0.0 | 0.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.4 | GO:0061615 | glycolytic process through fructose-6-phosphate(GO:0061615) |
| 0.0 | 0.2 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.6 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.0 | 0.7 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.7 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 | 1.4 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 2.9 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.0 | 0.3 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.2 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.7 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.0 | 0.8 | GO:0090501 | RNA phosphodiester bond hydrolysis(GO:0090501) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 5.4 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.7 | 2.1 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 0.6 | 3.5 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.5 | 10.5 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.4 | 7.8 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.4 | 1.6 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.3 | 1.8 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.2 | 1.7 | GO:0098651 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.2 | 5.7 | GO:0032982 | myosin filament(GO:0032982) |
| 0.2 | 1.1 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.2 | 1.5 | GO:0005638 | lamin filament(GO:0005638) |
| 0.2 | 1.4 | GO:0001652 | granular component(GO:0001652) |
| 0.2 | 1.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.2 | 1.2 | GO:0044299 | C-fiber(GO:0044299) |
| 0.2 | 2.1 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 2.1 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.1 | 0.7 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 3.0 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 1.0 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.1 | 3.3 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 1.5 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 1.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.6 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.1 | 6.2 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 2.7 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 4.0 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 1.5 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.2 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.0 | 7.7 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.6 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 1.4 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.6 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.5 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.4 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 1.5 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.4 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.6 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.2 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.4 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.2 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.5 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.6 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 1.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.7 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 1.1 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.2 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 6.6 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.1 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 2.5 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 4.2 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.6 | GO:0014704 | intercalated disc(GO:0014704) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.4 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 1.0 | 3.0 | GO:0042936 | dipeptide transporter activity(GO:0042936) |
| 1.0 | 7.7 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.9 | 2.7 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.6 | 3.5 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.6 | 1.8 | GO:0001566 | phorbol ester receptor activity(GO:0001565) non-kinase phorbol ester receptor activity(GO:0001566) |
| 0.6 | 5.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.4 | 1.3 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.4 | 2.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.4 | 11.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.3 | 3.5 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.3 | 2.4 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.3 | 1.2 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.3 | 2.5 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.3 | 5.3 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.2 | 1.2 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.2 | 1.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.2 | 0.9 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.2 | 0.6 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.2 | 0.8 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.2 | 4.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 1.3 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.2 | 1.3 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.2 | 0.5 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.2 | 0.7 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.2 | 0.6 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.2 | 0.5 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.2 | 12.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 2.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 1.0 | GO:0001602 | peptide YY receptor activity(GO:0001601) pancreatic polypeptide receptor activity(GO:0001602) |
| 0.1 | 3.0 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 2.6 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.1 | 0.6 | GO:0086077 | gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.1 | 4.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 1.7 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 1.7 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 3.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 1.2 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 0.3 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.1 | 2.0 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.1 | 0.8 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.1 | 1.8 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 0.6 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.1 | 1.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.3 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 0.1 | 0.3 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 0.8 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.1 | 1.0 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 1.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.8 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 2.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 1.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 0.9 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 8.3 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.1 | 2.8 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 1.8 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.1 | 0.9 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.5 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.1 | 0.2 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.1 | 0.3 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.1 | 0.3 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.1 | 0.8 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 0.7 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.3 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.1 | 1.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 2.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 1.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 3.3 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 3.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 1.8 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 1.9 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.1 | 1.0 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 0.6 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 1.8 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 1.8 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 1.5 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.5 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 2.0 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 3.4 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.4 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.2 | GO:0005223 | intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.6 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.0 | 0.2 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 2.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.2 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.0 | 0.6 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 2.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.2 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 1.0 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.7 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.4 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 1.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.6 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.5 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 1.0 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 2.1 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.3 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.3 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.0 | 0.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.4 | GO:0001614 | purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) |
| 0.0 | 0.1 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 12.3 | GO:0004984 | olfactory receptor activity(GO:0004984) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 8.5 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 2.0 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 4.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 3.7 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.1 | 2.8 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 2.3 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.4 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 2.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 2.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 2.0 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.6 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.2 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.8 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 4.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.4 | ST GA13 PATHWAY | G alpha 13 Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.4 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.2 | 11.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 2.1 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 3.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 2.8 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 3.1 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.1 | 3.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 7.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.3 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 5.5 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 1.2 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.1 | 1.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 3.2 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 2.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 2.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 6.4 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 6.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 3.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 1.7 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 2.1 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 0.7 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.2 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 3.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.1 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.5 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 1.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.0 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.5 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 1.0 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 2.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 2.7 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.6 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.4 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 1.7 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 3.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.4 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 4.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |