Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Bptf
Z-value: 2.23
Transcription factors associated with Bptf
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Bptf
|
ENSMUSG00000040481.18 | Bptf |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Bptf | mm39_v1_chr11_-_107022748_107022781 | 0.11 | 3.6e-01 | Click! |
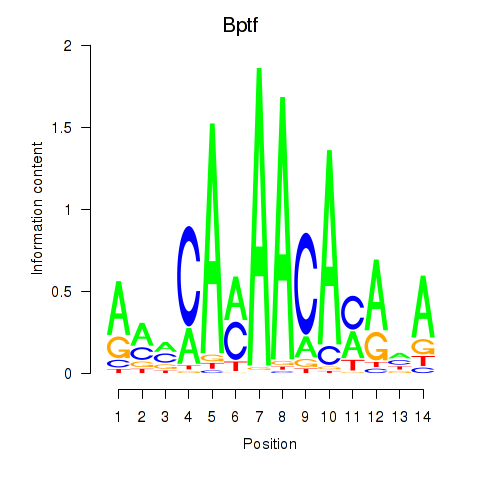
Activity profile of Bptf motif
Sorted Z-values of Bptf motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Bptf
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_128104525 | 18.04 |
ENSMUST00000050901.5
|
Apof
|
apolipoprotein F |
| chr4_-_60618357 | 17.57 |
ENSMUST00000084544.5
ENSMUST00000098046.10 |
Mup11
|
major urinary protein 11 |
| chr12_-_103923145 | 15.68 |
ENSMUST00000085054.5
|
Serpina1e
|
serine (or cysteine) peptidase inhibitor, clade A, member 1E |
| chr12_-_103829810 | 14.71 |
ENSMUST00000085056.8
ENSMUST00000072876.12 ENSMUST00000124717.2 |
Serpina1a
|
serine (or cysteine) peptidase inhibitor, clade A, member 1A |
| chr12_-_103871146 | 14.05 |
ENSMUST00000074051.6
|
Serpina1c
|
serine (or cysteine) peptidase inhibitor, clade A, member 1C |
| chr4_-_61437704 | 13.93 |
ENSMUST00000095051.6
ENSMUST00000107483.8 |
Mup16
|
major urinary protein 16 |
| chr10_-_59277570 | 13.85 |
ENSMUST00000009798.5
|
Oit3
|
oncoprotein induced transcript 3 |
| chr8_-_41668182 | 13.78 |
ENSMUST00000034003.5
|
Fgl1
|
fibrinogen-like protein 1 |
| chr4_-_60377932 | 13.18 |
ENSMUST00000107506.9
ENSMUST00000122381.8 ENSMUST00000118759.8 ENSMUST00000132829.3 |
Mup9
|
major urinary protein 9 |
| chr7_-_12732067 | 13.03 |
ENSMUST00000032539.14
ENSMUST00000120903.8 |
Slc27a5
|
solute carrier family 27 (fatty acid transporter), member 5 |
| chr4_-_60139857 | 12.79 |
ENSMUST00000107490.5
ENSMUST00000074700.9 |
Mup2
|
major urinary protein 2 |
| chr12_-_103704417 | 12.01 |
ENSMUST00000095450.11
ENSMUST00000187220.2 |
Serpina1b
|
serine (or cysteine) preptidase inhibitor, clade A, member 1B |
| chr11_-_110142565 | 11.60 |
ENSMUST00000044003.14
|
Abca6
|
ATP-binding cassette, sub-family A (ABC1), member 6 |
| chr4_-_61592331 | 11.36 |
ENSMUST00000098040.4
|
Mup18
|
major urinary protein 18 |
| chr4_-_61700450 | 10.86 |
ENSMUST00000107477.2
ENSMUST00000080606.9 |
Mup19
|
major urinary protein 19 |
| chr11_-_46581135 | 10.80 |
ENSMUST00000169584.8
|
Timd2
|
T cell immunoglobulin and mucin domain containing 2 |
| chr14_-_66246652 | 10.71 |
ENSMUST00000059970.9
|
Gulo
|
gulonolactone (L-) oxidase |
| chr1_+_160806194 | 10.67 |
ENSMUST00000064725.11
ENSMUST00000191936.2 |
Serpinc1
|
serine (or cysteine) peptidase inhibitor, clade C (antithrombin), member 1 |
| chr4_-_61514108 | 10.63 |
ENSMUST00000107484.2
|
Mup17
|
major urinary protein 17 |
| chr4_-_60777462 | 10.38 |
ENSMUST00000211875.2
|
Mup22
|
major urinary protein 22 |
| chr12_-_81015479 | 10.31 |
ENSMUST00000218162.2
|
Slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 1 |
| chr3_-_86906591 | 10.23 |
ENSMUST00000063869.11
ENSMUST00000029717.4 |
Cd1d1
|
CD1d1 antigen |
| chr1_+_160806241 | 9.98 |
ENSMUST00000195760.2
|
Serpinc1
|
serine (or cysteine) peptidase inhibitor, clade C (antithrombin), member 1 |
| chr4_-_60538151 | 9.63 |
ENSMUST00000098047.3
|
Mup10
|
major urinary protein 10 |
| chr4_-_61357980 | 9.58 |
ENSMUST00000095049.5
|
Mup15
|
major urinary protein 15 |
| chr9_-_103099262 | 9.43 |
ENSMUST00000170904.2
|
Trf
|
transferrin |
| chr1_+_172525613 | 9.12 |
ENSMUST00000038495.5
|
Crp
|
C-reactive protein, pentraxin-related |
| chr4_-_61184270 | 9.11 |
ENSMUST00000072678.6
ENSMUST00000098042.10 |
Mup13
|
major urinary protein 13 |
| chr17_-_56424577 | 8.96 |
ENSMUST00000019808.12
|
Plin5
|
perilipin 5 |
| chr19_-_39801188 | 7.77 |
ENSMUST00000162507.2
ENSMUST00000160476.9 ENSMUST00000239028.2 |
Cyp2c40
|
cytochrome P450, family 2, subfamily c, polypeptide 40 |
| chr11_+_108271990 | 7.72 |
ENSMUST00000146050.2
ENSMUST00000152958.8 |
Apoh
|
apolipoprotein H |
| chr16_-_45975440 | 7.59 |
ENSMUST00000059524.7
|
Gm4737
|
predicted gene 4737 |
| chr19_-_39637489 | 7.51 |
ENSMUST00000067328.7
|
Cyp2c67
|
cytochrome P450, family 2, subfamily c, polypeptide 67 |
| chr3_-_116762617 | 7.27 |
ENSMUST00000143611.2
ENSMUST00000040097.14 |
Palmd
|
palmdelphin |
| chr1_-_73055043 | 7.14 |
ENSMUST00000027374.7
|
Tnp1
|
transition protein 1 |
| chr2_-_34990689 | 7.08 |
ENSMUST00000226631.2
ENSMUST00000045776.5 ENSMUST00000226972.2 |
AI182371
|
expressed sequence AI182371 |
| chr15_+_6474808 | 6.99 |
ENSMUST00000022749.17
ENSMUST00000239466.2 |
C9
|
complement component 9 |
| chr4_-_60457902 | 6.89 |
ENSMUST00000084548.11
ENSMUST00000103012.10 ENSMUST00000107499.4 |
Mup1
|
major urinary protein 1 |
| chr4_-_61972348 | 6.72 |
ENSMUST00000074018.4
|
Mup20
|
major urinary protein 20 |
| chr7_+_26819334 | 6.28 |
ENSMUST00000003100.10
|
Cyp2f2
|
cytochrome P450, family 2, subfamily f, polypeptide 2 |
| chr6_-_128503666 | 6.02 |
ENSMUST00000143664.2
ENSMUST00000112132.8 |
Pzp
|
PZP, alpha-2-macroglobulin like |
| chr4_-_49408040 | 5.87 |
ENSMUST00000081541.9
|
Acnat2
|
acyl-coenzyme A amino acid N-acyltransferase 2 |
| chr3_+_86893869 | 5.78 |
ENSMUST00000041920.5
|
Cd1d2
|
CD1d2 antigen |
| chr4_-_96552349 | 5.76 |
ENSMUST00000030299.8
|
Cyp2j5
|
cytochrome P450, family 2, subfamily j, polypeptide 5 |
| chr6_-_3988835 | 5.67 |
ENSMUST00000203257.2
|
Tfpi2
|
tissue factor pathway inhibitor 2 |
| chr9_-_71070506 | 5.58 |
ENSMUST00000074465.9
|
Aqp9
|
aquaporin 9 |
| chr9_-_103165423 | 5.58 |
ENSMUST00000123530.8
|
1300017J02Rik
|
RIKEN cDNA 1300017J02 gene |
| chr8_-_3770642 | 5.58 |
ENSMUST00000062037.7
|
Clec4g
|
C-type lectin domain family 4, member g |
| chr12_-_103739847 | 5.58 |
ENSMUST00000078869.6
|
Serpina1d
|
serine (or cysteine) peptidase inhibitor, clade A, member 1D |
| chr11_+_3981769 | 5.58 |
ENSMUST00000019512.8
|
Sec14l4
|
SEC14-like lipid binding 4 |
| chr11_-_60702081 | 5.53 |
ENSMUST00000018744.15
|
Shmt1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr6_-_142418801 | 5.51 |
ENSMUST00000032371.8
|
Gys2
|
glycogen synthase 2 |
| chr2_-_25517945 | 5.46 |
ENSMUST00000028307.9
|
Fcna
|
ficolin A |
| chr8_-_25314032 | 5.44 |
ENSMUST00000050300.15
ENSMUST00000209935.2 |
Adam5
|
a disintegrin and metallopeptidase domain 5 |
| chr8_+_105775224 | 5.40 |
ENSMUST00000093222.13
ENSMUST00000093223.5 |
Ces3a
|
carboxylesterase 3A |
| chr6_-_3988900 | 5.40 |
ENSMUST00000183682.3
|
Tfpi2
|
tissue factor pathway inhibitor 2 |
| chr1_-_139487951 | 5.39 |
ENSMUST00000023965.8
|
Cfhr1
|
complement factor H-related 1 |
| chr11_+_7147779 | 5.39 |
ENSMUST00000020704.8
|
Igfbp1
|
insulin-like growth factor binding protein 1 |
| chr1_-_139786421 | 5.38 |
ENSMUST00000194186.6
ENSMUST00000094489.5 ENSMUST00000239380.2 |
Cfhr2
|
complement factor H-related 2 |
| chr4_-_60697274 | 5.38 |
ENSMUST00000117932.2
|
Mup12
|
major urinary protein 12 |
| chr15_-_31453708 | 5.31 |
ENSMUST00000110408.3
|
Ropn1l
|
ropporin 1-like |
| chr18_+_64473091 | 5.24 |
ENSMUST00000175965.10
|
Onecut2
|
one cut domain, family member 2 |
| chr7_+_46401214 | 5.21 |
ENSMUST00000210769.2
ENSMUST00000210272.2 ENSMUST00000075982.4 |
Saa2
|
serum amyloid A 2 |
| chr19_-_40175709 | 5.18 |
ENSMUST00000051846.13
|
Cyp2c70
|
cytochrome P450, family 2, subfamily c, polypeptide 70 |
| chr8_-_45747883 | 4.91 |
ENSMUST00000026907.6
|
Klkb1
|
kallikrein B, plasma 1 |
| chr9_-_71075939 | 4.87 |
ENSMUST00000113570.8
|
Aqp9
|
aquaporin 9 |
| chr15_+_9335636 | 4.84 |
ENSMUST00000072403.7
|
Ugt3a2
|
UDP glycosyltransferases 3 family, polypeptide A2 |
| chr12_-_72675624 | 4.83 |
ENSMUST00000208039.2
ENSMUST00000207585.2 |
Gm4756
|
predicted gene 4756 |
| chr10_+_87694117 | 4.82 |
ENSMUST00000122386.8
|
Igf1
|
insulin-like growth factor 1 |
| chr3_+_59989282 | 4.77 |
ENSMUST00000029326.6
|
Sucnr1
|
succinate receptor 1 |
| chr1_+_163889551 | 4.70 |
ENSMUST00000192047.6
ENSMUST00000027871.13 |
Sell
|
selectin, lymphocyte |
| chr10_-_93375832 | 4.62 |
ENSMUST00000016034.3
|
Amdhd1
|
amidohydrolase domain containing 1 |
| chr4_-_6275629 | 4.61 |
ENSMUST00000029905.2
|
Cyp7a1
|
cytochrome P450, family 7, subfamily a, polypeptide 1 |
| chr18_+_14839202 | 4.60 |
ENSMUST00000040860.3
ENSMUST00000234680.2 |
Psma8
|
proteasome subunit alpha 8 |
| chr14_-_59380335 | 4.53 |
ENSMUST00000022548.10
ENSMUST00000162674.8 ENSMUST00000159858.2 ENSMUST00000162271.2 |
1700129C05Rik
|
RIKEN cDNA 1700129C05 gene |
| chr10_-_127587576 | 4.51 |
ENSMUST00000079692.6
|
Gpr182
|
G protein-coupled receptor 182 |
| chr4_+_115375461 | 4.44 |
ENSMUST00000058785.10
ENSMUST00000094886.4 |
Cyp4a10
|
cytochrome P450, family 4, subfamily a, polypeptide 10 |
| chr2_-_164670452 | 4.17 |
ENSMUST00000017911.4
|
Spata25
|
spermatogenesis associated 25 |
| chr10_-_88520877 | 4.15 |
ENSMUST00000138734.2
|
Spic
|
Spi-C transcription factor (Spi-1/PU.1 related) |
| chr1_-_192923816 | 4.13 |
ENSMUST00000160929.8
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr10_+_87694924 | 4.12 |
ENSMUST00000095360.11
|
Igf1
|
insulin-like growth factor 1 |
| chr1_+_175459559 | 3.97 |
ENSMUST00000040250.15
|
Kmo
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr1_-_162812087 | 3.89 |
ENSMUST00000028010.9
|
Fmo3
|
flavin containing monooxygenase 3 |
| chr11_+_78389913 | 3.79 |
ENSMUST00000017488.5
|
Vtn
|
vitronectin |
| chr1_+_88062508 | 3.78 |
ENSMUST00000113134.8
ENSMUST00000140092.8 |
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A |
| chr9_+_77824646 | 3.77 |
ENSMUST00000034904.14
|
Elovl5
|
ELOVL family member 5, elongation of long chain fatty acids (yeast) |
| chr8_+_46080746 | 3.74 |
ENSMUST00000145458.9
ENSMUST00000134321.8 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr19_-_39875192 | 3.70 |
ENSMUST00000168838.3
|
Cyp2c69
|
cytochrome P450, family 2, subfamily c, polypeptide 69 |
| chr3_-_121056944 | 3.69 |
ENSMUST00000128909.8
ENSMUST00000029777.14 |
Tlcd4
|
TLC domain containing 4 |
| chr13_-_64514830 | 3.68 |
ENSMUST00000222971.2
|
Ctsl
|
cathepsin L |
| chr2_+_172821620 | 3.66 |
ENSMUST00000109125.8
ENSMUST00000109126.5 ENSMUST00000050442.15 |
Spo11
|
SPO11 initiator of meiotic double stranded breaks |
| chr16_-_16647139 | 3.62 |
ENSMUST00000023468.6
|
Spag6l
|
sperm associated antigen 6-like |
| chr9_+_92157655 | 3.59 |
ENSMUST00000034932.14
ENSMUST00000180154.8 |
Plscr2
|
phospholipid scramblase 2 |
| chr16_+_22769844 | 3.57 |
ENSMUST00000232422.2
|
Hrg
|
histidine-rich glycoprotein |
| chr5_+_130058119 | 3.54 |
ENSMUST00000026608.11
ENSMUST00000202163.4 ENSMUST00000202756.2 |
Crcp
|
calcitonin gene-related peptide-receptor component protein |
| chr8_+_106154266 | 3.52 |
ENSMUST00000067305.8
|
Lrrc36
|
leucine rich repeat containing 36 |
| chr15_+_76178542 | 3.49 |
ENSMUST00000163991.4
|
Smpd5
|
sphingomyelin phosphodiesterase 5 |
| chr17_-_47748289 | 3.46 |
ENSMUST00000061885.9
|
1700001C19Rik
|
RIKEN cDNA 1700001C19 gene |
| chr3_+_20011201 | 3.40 |
ENSMUST00000091309.12
ENSMUST00000108329.8 ENSMUST00000003714.13 |
Cp
|
ceruloplasmin |
| chr3_-_146200870 | 3.39 |
ENSMUST00000093951.3
|
Spata1
|
spermatogenesis associated 1 |
| chr4_-_49506538 | 3.39 |
ENSMUST00000043056.9
|
Baat
|
bile acid-Coenzyme A: amino acid N-acyltransferase |
| chr1_-_44140820 | 3.38 |
ENSMUST00000152239.8
|
Tex30
|
testis expressed 30 |
| chr13_+_55300453 | 3.33 |
ENSMUST00000005452.6
|
Fgfr4
|
fibroblast growth factor receptor 4 |
| chr9_+_107457316 | 3.32 |
ENSMUST00000093785.6
|
Naa80
|
N(alpha)-acetyltransferase 80, NatH catalytic subunit |
| chr3_+_90011435 | 3.31 |
ENSMUST00000029548.9
ENSMUST00000200410.2 |
Nup210l
|
nucleoporin 210-like |
| chr2_+_122065230 | 3.24 |
ENSMUST00000110551.4
|
Sord
|
sorbitol dehydrogenase |
| chr18_+_74416161 | 3.24 |
ENSMUST00000114895.4
|
Cfap53
|
cilia and flagella associated protein 53 |
| chr10_+_33733706 | 3.23 |
ENSMUST00000218204.2
|
Sult3a1
|
sulfotransferase family 3A, member 1 |
| chr4_+_144619397 | 3.22 |
ENSMUST00000105744.8
ENSMUST00000171001.8 |
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr16_-_23339329 | 3.16 |
ENSMUST00000230040.2
ENSMUST00000229619.2 |
Masp1
|
mannan-binding lectin serine peptidase 1 |
| chr19_-_58442866 | 3.15 |
ENSMUST00000169850.8
|
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr5_-_123185073 | 3.14 |
ENSMUST00000031437.14
|
Morn3
|
MORN repeat containing 3 |
| chr14_+_25942963 | 3.12 |
ENSMUST00000184016.3
|
Cphx1
|
cytoplasmic polyadenylated homeobox 1 |
| chr17_-_56424265 | 3.09 |
ENSMUST00000113072.3
|
Plin5
|
perilipin 5 |
| chr7_-_89552785 | 3.07 |
ENSMUST00000041195.6
|
Ccdc81
|
coiled-coil domain containing 81 |
| chr3_-_98767090 | 3.06 |
ENSMUST00000107016.10
ENSMUST00000029465.10 |
Hsd3b1
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1 |
| chr3_+_20011405 | 3.03 |
ENSMUST00000108325.9
|
Cp
|
ceruloplasmin |
| chr4_-_132459762 | 3.01 |
ENSMUST00000045550.5
|
Xkr8
|
X-linked Kx blood group related 8 |
| chr7_+_67925718 | 2.98 |
ENSMUST00000210558.2
|
Fam169b
|
family with sequence similarity 169, member B |
| chr7_+_131012330 | 2.98 |
ENSMUST00000015829.15
ENSMUST00000117518.2 ENSMUST00000133277.4 |
Acadsb
|
acyl-Coenzyme A dehydrogenase, short/branched chain |
| chr19_-_44017637 | 2.97 |
ENSMUST00000026211.10
ENSMUST00000211830.2 |
Cyp2c23
|
cytochrome P450, family 2, subfamily c, polypeptide 23 |
| chr13_+_81034214 | 2.95 |
ENSMUST00000161441.2
|
Arrdc3
|
arrestin domain containing 3 |
| chr11_-_105346120 | 2.94 |
ENSMUST00000138977.8
|
Marchf10
|
membrane associated ring-CH-type finger 10 |
| chr5_-_130053120 | 2.93 |
ENSMUST00000161640.8
ENSMUST00000161884.2 ENSMUST00000161094.8 |
Asl
|
argininosuccinate lyase |
| chr3_+_20011251 | 2.93 |
ENSMUST00000108328.8
|
Cp
|
ceruloplasmin |
| chr6_+_124489364 | 2.90 |
ENSMUST00000068593.9
|
C1ra
|
complement component 1, r subcomponent A |
| chr4_+_141473983 | 2.90 |
ENSMUST00000038161.5
|
Agmat
|
agmatine ureohydrolase (agmatinase) |
| chr12_+_84098888 | 2.89 |
ENSMUST00000120927.8
ENSMUST00000021653.8 |
Acot3
|
acyl-CoA thioesterase 3 |
| chr16_+_23043474 | 2.80 |
ENSMUST00000023601.14
|
St6gal1
|
beta galactoside alpha 2,6 sialyltransferase 1 |
| chr11_+_46701619 | 2.78 |
ENSMUST00000068877.7
|
Timd4
|
T cell immunoglobulin and mucin domain containing 4 |
| chr12_-_72675605 | 2.78 |
ENSMUST00000222413.2
|
Gm4756
|
predicted gene 4756 |
| chr4_+_133280680 | 2.77 |
ENSMUST00000042706.3
|
Nr0b2
|
nuclear receptor subfamily 0, group B, member 2 |
| chr8_+_46945826 | 2.77 |
ENSMUST00000110371.8
|
Acsl1
|
acyl-CoA synthetase long-chain family member 1 |
| chr3_+_66892979 | 2.72 |
ENSMUST00000162362.8
ENSMUST00000065074.14 ENSMUST00000065047.13 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr6_+_120440159 | 2.72 |
ENSMUST00000002976.5
|
Il17ra
|
interleukin 17 receptor A |
| chr7_-_46365108 | 2.72 |
ENSMUST00000006956.9
ENSMUST00000210913.2 |
Saa3
|
serum amyloid A 3 |
| chr5_-_82271183 | 2.71 |
ENSMUST00000186079.2
ENSMUST00000185607.2 |
1700031L13Rik
|
RIKEN cDNA 1700031L13 gene |
| chr5_-_123185029 | 2.68 |
ENSMUST00000045843.15
|
Morn3
|
MORN repeat containing 3 |
| chr8_-_118398264 | 2.68 |
ENSMUST00000037955.14
|
Sdr42e1
|
short chain dehydrogenase/reductase family 42E, member 1 |
| chr7_-_101494472 | 2.65 |
ENSMUST00000211566.2
ENSMUST00000094141.7 ENSMUST00000209329.2 |
Folr2
|
folate receptor 2 (fetal) |
| chr18_+_35963353 | 2.65 |
ENSMUST00000235169.2
|
Cxxc5
|
CXXC finger 5 |
| chr18_+_52779281 | 2.62 |
ENSMUST00000118724.8
ENSMUST00000091904.6 |
1700034E13Rik
|
RIKEN cDNA 1700034E13 gene |
| chr7_+_46510627 | 2.62 |
ENSMUST00000014545.11
|
Ldhc
|
lactate dehydrogenase C |
| chr8_-_96302083 | 2.61 |
ENSMUST00000213096.2
ENSMUST00000052690.13 |
Prss54
|
protease, serine 54 |
| chr7_-_24134919 | 2.60 |
ENSMUST00000080594.8
|
Irgc1
|
immunity-related GTPase family, cinema 1 |
| chr7_+_46510677 | 2.60 |
ENSMUST00000211784.2
ENSMUST00000210585.2 ENSMUST00000148565.8 |
Ldhc
|
lactate dehydrogenase C |
| chr11_-_69652789 | 2.59 |
ENSMUST00000102586.5
|
Slc35g3
|
solute carrier family 35, member G3 |
| chr3_-_145813802 | 2.59 |
ENSMUST00000160285.2
|
Dnai3
|
dynein axonemal intermediate chain 3 |
| chr19_+_12775938 | 2.57 |
ENSMUST00000025601.8
|
Lpxn
|
leupaxin |
| chr6_+_82029288 | 2.56 |
ENSMUST00000149023.2
|
Eva1a
|
eva-1 homolog A (C. elegans) |
| chr11_+_44508137 | 2.56 |
ENSMUST00000109268.2
ENSMUST00000101326.10 ENSMUST00000081265.12 |
Ebf1
|
early B cell factor 1 |
| chr1_+_9868332 | 2.54 |
ENSMUST00000166384.8
ENSMUST00000168907.8 |
Sgk3
|
serum/glucocorticoid regulated kinase 3 |
| chr7_+_46633328 | 2.54 |
ENSMUST00000070660.11
ENSMUST00000188312.2 |
Gm9999
|
predicted gene 9999 |
| chr11_-_69652670 | 2.53 |
ENSMUST00000231829.2
|
Slc35g3
|
solute carrier family 35, member G3 |
| chr10_+_51367052 | 2.51 |
ENSMUST00000217705.2
ENSMUST00000078778.5 ENSMUST00000220182.2 ENSMUST00000220226.2 |
Lilrb4a
|
leukocyte immunoglobulin-like receptor, subfamily B, member 4A |
| chr2_-_103858632 | 2.50 |
ENSMUST00000056170.4
|
4931422A03Rik
|
RIKEN cDNA 4931422A03 gene |
| chrX_-_74918122 | 2.47 |
ENSMUST00000033547.14
|
Pls3
|
plastin 3 (T-isoform) |
| chr12_-_72675670 | 2.44 |
ENSMUST00000209038.2
|
Gm4756
|
predicted gene 4756 |
| chr15_+_41652777 | 2.42 |
ENSMUST00000230778.2
ENSMUST00000022918.15 ENSMUST00000090095.13 |
Oxr1
|
oxidation resistance 1 |
| chr17_-_32643067 | 2.42 |
ENSMUST00000237130.2
|
Pglyrp2
|
peptidoglycan recognition protein 2 |
| chr2_+_153621851 | 2.42 |
ENSMUST00000126656.4
|
Efcab8
|
EF-hand calcium binding domain 8 |
| chr3_+_134986982 | 2.40 |
ENSMUST00000120397.8
ENSMUST00000029817.11 |
Bdh2
|
3-hydroxybutyrate dehydrogenase, type 2 |
| chr12_-_104010690 | 2.40 |
ENSMUST00000043915.4
|
Serpina12
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 12 |
| chr17_-_40519480 | 2.38 |
ENSMUST00000033585.7
|
Pgk2
|
phosphoglycerate kinase 2 |
| chr14_-_59416198 | 2.38 |
ENSMUST00000066558.4
|
Gm5142
|
predicted gene 5142 |
| chr12_+_71184614 | 2.37 |
ENSMUST00000045907.16
|
2700049A03Rik
|
RIKEN cDNA 2700049A03 gene |
| chr15_-_36165017 | 2.36 |
ENSMUST00000058643.4
|
Fbxo43
|
F-box protein 43 |
| chr1_+_21419819 | 2.35 |
ENSMUST00000088407.4
|
Khdc1a
|
KH domain containing 1A |
| chr11_+_80749184 | 2.35 |
ENSMUST00000103223.8
ENSMUST00000103222.4 |
Spaca3
|
sperm acrosome associated 3 |
| chr4_-_108075119 | 2.35 |
ENSMUST00000223127.2
ENSMUST00000043793.7 ENSMUST00000106690.9 |
Zyg11a
|
zyg-11 family member A, cell cycle regulator |
| chr4_+_144619696 | 2.33 |
ENSMUST00000142808.8
|
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr7_-_80020830 | 2.32 |
ENSMUST00000205436.2
ENSMUST00000098346.5 |
Man2a2
|
mannosidase 2, alpha 2 |
| chr17_-_32643131 | 2.32 |
ENSMUST00000236386.2
|
Pglyrp2
|
peptidoglycan recognition protein 2 |
| chr14_-_24537067 | 2.31 |
ENSMUST00000026322.9
|
Polr3a
|
polymerase (RNA) III (DNA directed) polypeptide A |
| chr17_-_31852128 | 2.31 |
ENSMUST00000236909.2
|
Cbs
|
cystathionine beta-synthase |
| chr19_-_7780025 | 2.30 |
ENSMUST00000065634.8
|
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr18_+_84106188 | 2.30 |
ENSMUST00000060223.4
|
Zadh2
|
zinc binding alcohol dehydrogenase, domain containing 2 |
| chr3_-_95264467 | 2.30 |
ENSMUST00000107171.10
ENSMUST00000015841.12 ENSMUST00000107170.3 |
Setdb1
|
SET domain, bifurcated 1 |
| chr18_-_3281752 | 2.30 |
ENSMUST00000140332.8
ENSMUST00000147138.8 |
Crem
|
cAMP responsive element modulator |
| chr4_+_108336164 | 2.30 |
ENSMUST00000155068.2
|
Tut4
|
terminal uridylyl transferase 4 |
| chr4_+_144619647 | 2.28 |
ENSMUST00000154208.8
|
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr9_+_68561042 | 2.28 |
ENSMUST00000034766.14
|
Rora
|
RAR-related orphan receptor alpha |
| chr18_+_35962832 | 2.27 |
ENSMUST00000060722.8
|
Cxxc5
|
CXXC finger 5 |
| chr7_-_100164007 | 2.27 |
ENSMUST00000207405.2
|
Dnajb13
|
DnaJ heat shock protein family (Hsp40) member B13 |
| chr8_+_95161006 | 2.26 |
ENSMUST00000211816.2
|
Nlrc5
|
NLR family, CARD domain containing 5 |
| chr4_-_53159885 | 2.26 |
ENSMUST00000030010.4
|
Abca1
|
ATP-binding cassette, sub-family A (ABC1), member 1 |
| chr9_+_108437485 | 2.26 |
ENSMUST00000081111.14
ENSMUST00000193421.2 |
Impdh2
|
inosine monophosphate dehydrogenase 2 |
| chr6_-_125262974 | 2.26 |
ENSMUST00000088246.6
|
Tuba3a
|
tubulin, alpha 3A |
| chr5_-_105130522 | 2.25 |
ENSMUST00000031239.13
|
Abcg3
|
ATP binding cassette subfamily G member 3 |
| chr2_+_67935015 | 2.25 |
ENSMUST00000042456.4
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr9_+_119186447 | 2.25 |
ENSMUST00000039610.10
|
Xylb
|
xylulokinase homolog (H. influenzae) |
| chr11_-_109986804 | 2.25 |
ENSMUST00000100287.9
|
Abca8a
|
ATP-binding cassette, sub-family A (ABC1), member 8a |
| chr2_-_151318073 | 2.22 |
ENSMUST00000080132.3
|
4921509C19Rik
|
RIKEN cDNA 4921509C19 gene |
| chr11_-_100713348 | 2.21 |
ENSMUST00000107358.9
|
Stat5b
|
signal transducer and activator of transcription 5B |
| chr13_+_24599086 | 2.20 |
ENSMUST00000224657.2
|
Cmah
|
cytidine monophospho-N-acetylneuraminic acid hydroxylase |
| chr3_+_5283606 | 2.20 |
ENSMUST00000026284.13
|
Zfhx4
|
zinc finger homeodomain 4 |
| chr11_+_87685032 | 2.17 |
ENSMUST00000121303.8
|
Mpo
|
myeloperoxidase |
| chr7_-_44465998 | 2.16 |
ENSMUST00000209072.2
ENSMUST00000047356.11 |
Atf5
|
activating transcription factor 5 |
| chr2_-_23939401 | 2.16 |
ENSMUST00000051416.12
|
Hnmt
|
histamine N-methyltransferase |
| chr3_+_66893031 | 2.16 |
ENSMUST00000046542.13
ENSMUST00000162693.8 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 33.3 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 3.4 | 10.2 | GO:0048007 | positive regulation of interleukin-4 biosynthetic process(GO:0045404) antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 3.3 | 13.0 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 2.9 | 5.8 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) |
| 2.4 | 12.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 2.1 | 10.7 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 2.1 | 6.3 | GO:0090420 | naphthalene metabolic process(GO:0018931) naphthalene-containing compound metabolic process(GO:0090420) |
| 2.1 | 24.7 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 1.8 | 5.5 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 1.8 | 7.1 | GO:0015855 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 1.6 | 3.2 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 1.4 | 7.1 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 1.4 | 4.1 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 1.3 | 10.3 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 1.3 | 8.9 | GO:1904075 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 1.3 | 8.8 | GO:0052805 | imidazole-containing compound catabolic process(GO:0052805) |
| 1.2 | 3.7 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 1.2 | 3.6 | GO:0097037 | heme export(GO:0097037) |
| 1.2 | 9.4 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 1.1 | 3.4 | GO:0051439 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051439) |
| 1.1 | 6.7 | GO:0008355 | olfactory learning(GO:0008355) |
| 1.1 | 3.3 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 1.0 | 5.2 | GO:0019516 | lactate oxidation(GO:0019516) |
| 1.0 | 2.9 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 1.0 | 2.9 | GO:0032788 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.9 | 4.6 | GO:1904253 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.9 | 5.4 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.9 | 2.7 | GO:1904736 | negative regulation of electron carrier activity(GO:1904733) regulation of fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:1904735) negative regulation of fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:1904736) |
| 0.9 | 23.6 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.9 | 28.4 | GO:0080184 | response to phenylpropanoid(GO:0080184) |
| 0.8 | 3.4 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.8 | 2.4 | GO:1904092 | regulation of autophagic cell death(GO:1904092) negative regulation of autophagic cell death(GO:1904093) |
| 0.8 | 1.6 | GO:0046226 | coumarin catabolic process(GO:0046226) |
| 0.8 | 2.3 | GO:0006535 | cysteine biosynthetic process from serine(GO:0006535) |
| 0.8 | 2.3 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.7 | 3.0 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.7 | 3.0 | GO:0090327 | negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.7 | 4.4 | GO:0032827 | negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.7 | 2.1 | GO:0001807 | regulation of type IV hypersensitivity(GO:0001807) |
| 0.7 | 2.1 | GO:0070104 | negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 0.7 | 2.7 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.7 | 0.7 | GO:0019343 | cysteine biosynthetic process via cystathionine(GO:0019343) |
| 0.7 | 2.0 | GO:1903920 | positive regulation of actin filament severing(GO:1903920) |
| 0.7 | 2.0 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.7 | 8.6 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.7 | 4.0 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.7 | 2.6 | GO:0060849 | regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.6 | 0.6 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 0.6 | 4.1 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.6 | 1.8 | GO:0032685 | negative regulation of granulocyte macrophage colony-stimulating factor production(GO:0032685) |
| 0.6 | 2.3 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.6 | 2.9 | GO:0050787 | detoxification of mercury ion(GO:0050787) |
| 0.6 | 8.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.6 | 2.3 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.6 | 2.8 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.6 | 2.2 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.6 | 1.7 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 0.5 | 1.6 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) |
| 0.5 | 2.2 | GO:2000473 | regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 0.5 | 3.7 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.5 | 1.6 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.5 | 2.6 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.5 | 3.2 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.5 | 1.5 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.5 | 1.5 | GO:0045660 | positive regulation of neutrophil differentiation(GO:0045660) |
| 0.5 | 11.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.5 | 1.5 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.5 | 2.0 | GO:0001905 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 0.5 | 1.5 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.5 | 2.9 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.5 | 1.4 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.5 | 1.9 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.5 | 1.4 | GO:0002148 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 0.5 | 1.4 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.4 | 4.3 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.4 | 1.7 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.4 | 2.5 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.4 | 2.1 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.4 | 1.2 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.4 | 1.2 | GO:0021664 | rhombomere formation(GO:0021594) rhombomere 3 formation(GO:0021660) rhombomere 5 morphogenesis(GO:0021664) rhombomere 5 formation(GO:0021666) |
| 0.4 | 1.2 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.4 | 1.6 | GO:2000373 | regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.4 | 0.8 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.4 | 3.5 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.4 | 1.2 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.4 | 3.8 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.4 | 2.3 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.4 | 1.9 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.4 | 8.5 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.4 | 1.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.4 | 7.3 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.4 | 1.5 | GO:0090346 | nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.4 | 2.2 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.4 | 5.0 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.4 | 3.2 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.4 | 2.8 | GO:0042167 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.4 | 0.7 | GO:0010705 | meiotic DNA double-strand break processing involved in reciprocal meiotic recombination(GO:0010705) |
| 0.3 | 1.4 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.3 | 10.3 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.3 | 1.7 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.3 | 1.0 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.3 | 1.3 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.3 | 3.6 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.3 | 1.3 | GO:0030091 | protein repair(GO:0030091) |
| 0.3 | 2.3 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.3 | 2.6 | GO:0070669 | response to interleukin-2(GO:0070669) |
| 0.3 | 1.0 | GO:0031554 | regulation of DNA-templated transcription, termination(GO:0031554) |
| 0.3 | 17.9 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.3 | 1.3 | GO:0032382 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) |
| 0.3 | 1.6 | GO:0003342 | proepicardium development(GO:0003342) septum transversum development(GO:0003343) |
| 0.3 | 3.8 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.3 | 0.6 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.3 | 1.2 | GO:0000239 | pachytene(GO:0000239) |
| 0.3 | 3.3 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.3 | 3.0 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.3 | 9.3 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.3 | 2.1 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.3 | 1.2 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.3 | 0.9 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.3 | 1.8 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.3 | 1.2 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.3 | 0.6 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.3 | 2.0 | GO:0046218 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) |
| 0.3 | 3.7 | GO:0042637 | catagen(GO:0042637) |
| 0.3 | 1.7 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.3 | 1.7 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.3 | 5.6 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.3 | 1.7 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.3 | 1.4 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.3 | 0.5 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.3 | 0.8 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.3 | 1.6 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.3 | 1.0 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 0.3 | 5.2 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.3 | 1.3 | GO:0071455 | cellular response to hyperoxia(GO:0071455) |
| 0.3 | 0.8 | GO:1901536 | negative regulation of DNA demethylation(GO:1901536) |
| 0.3 | 0.8 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.3 | 2.3 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.3 | 0.8 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.3 | 0.8 | GO:0034240 | negative regulation of macrophage fusion(GO:0034240) |
| 0.2 | 3.7 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.2 | 1.0 | GO:0002461 | tolerance induction dependent upon immune response(GO:0002461) |
| 0.2 | 8.2 | GO:0097286 | iron ion import(GO:0097286) |
| 0.2 | 1.5 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.2 | 3.4 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.2 | 1.2 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.2 | 2.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.2 | 11.2 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.2 | 2.3 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.2 | 0.2 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.2 | 0.9 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.2 | 22.2 | GO:0030317 | sperm motility(GO:0030317) |
| 0.2 | 2.9 | GO:0015747 | urate transport(GO:0015747) |
| 0.2 | 0.7 | GO:0021558 | trochlear nerve development(GO:0021558) |
| 0.2 | 2.0 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.2 | 0.7 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.2 | 0.7 | GO:0060197 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) cloacal septation(GO:0060197) |
| 0.2 | 0.7 | GO:0045829 | negative regulation of isotype switching(GO:0045829) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.2 | 1.3 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.2 | 1.5 | GO:0032466 | negative regulation of cytokinesis(GO:0032466) |
| 0.2 | 2.4 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.2 | 2.6 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.2 | 1.9 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.2 | 1.3 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.2 | 4.2 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.2 | 1.5 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.2 | 1.7 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.2 | 2.3 | GO:2000188 | regulation of cholesterol homeostasis(GO:2000188) |
| 0.2 | 0.8 | GO:0097298 | regulation of nucleus size(GO:0097298) |
| 0.2 | 1.7 | GO:0060022 | hard palate development(GO:0060022) |
| 0.2 | 0.6 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.2 | 0.6 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.2 | 0.6 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.2 | 1.2 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.2 | 1.6 | GO:0035655 | interleukin-18-mediated signaling pathway(GO:0035655) cellular response to interleukin-18(GO:0071351) |
| 0.2 | 3.6 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.2 | 1.0 | GO:0009173 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.2 | 0.8 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.2 | 0.6 | GO:1904826 | regulation of hydrogen sulfide biosynthetic process(GO:1904826) positive regulation of hydrogen sulfide biosynthetic process(GO:1904828) |
| 0.2 | 2.9 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.2 | 0.6 | GO:0033306 | sesquiterpenoid metabolic process(GO:0006714) phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 0.2 | 0.6 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.2 | 2.1 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.2 | 2.5 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.2 | 0.8 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.2 | 1.7 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.2 | 4.1 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.2 | 0.6 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.2 | 2.4 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.2 | 2.0 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.2 | 0.5 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 0.2 | 1.3 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.2 | 0.9 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.2 | 0.9 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.2 | 3.7 | GO:0034204 | lipid translocation(GO:0034204) |
| 0.2 | 0.9 | GO:0042758 | long-chain fatty acid catabolic process(GO:0042758) |
| 0.2 | 1.8 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.2 | 7.5 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.2 | 1.6 | GO:0072642 | type I interferon secretion(GO:0072641) interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.2 | 1.6 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.2 | 0.7 | GO:0080163 | regulation of protein serine/threonine phosphatase activity(GO:0080163) |
| 0.2 | 2.4 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.2 | 1.0 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.2 | 0.2 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.2 | 2.6 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.2 | 2.0 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.2 | 0.5 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.2 | 1.7 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.2 | 2.2 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.2 | 1.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.2 | 0.5 | GO:0030221 | basophil differentiation(GO:0030221) |
| 0.2 | 0.5 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.2 | 0.6 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.2 | 1.1 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.2 | 0.2 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.2 | 1.3 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.2 | 4.4 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.2 | 0.3 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.2 | 0.3 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.2 | 1.4 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.2 | 0.8 | GO:0044330 | canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) |
| 0.2 | 0.5 | GO:0060023 | soft palate development(GO:0060023) |
| 0.1 | 0.9 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.1 | 0.4 | GO:0031999 | negative regulation of fatty acid beta-oxidation(GO:0031999) |
| 0.1 | 3.7 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 1.0 | GO:0046013 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 0.4 | GO:0033577 | protein glycosylation in endoplasmic reticulum(GO:0033577) |
| 0.1 | 1.4 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 0.7 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.1 | 1.5 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 1.1 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.1 | 2.2 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.1 | 1.8 | GO:0046606 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.1 | 2.0 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.1 | 1.1 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.1 | 1.0 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.1 | 0.9 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.1 | 0.4 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.1 | 0.9 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.1 | 2.5 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 0.5 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.1 | 1.0 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.1 | 1.0 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.8 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.3 | GO:0002859 | negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) negative regulation of natural killer cell mediated immune response to tumor cell(GO:0002856) negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002859) |
| 0.1 | 0.6 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) |
| 0.1 | 0.2 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.1 | 0.6 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 | 0.6 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 0.4 | GO:1990428 | miRNA transport(GO:1990428) |
| 0.1 | 0.4 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.1 | 0.9 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.1 | 0.6 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.1 | 2.3 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.1 | 1.3 | GO:1900107 | regulation of nodal signaling pathway(GO:1900107) |
| 0.1 | 0.7 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.1 | 0.8 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.2 | GO:0019230 | proprioception(GO:0019230) |
| 0.1 | 2.0 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.1 | 2.2 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 2.7 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 4.4 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 1.8 | GO:0032740 | positive regulation of interleukin-17 production(GO:0032740) |
| 0.1 | 0.7 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 0.6 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 | 1.1 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 1.8 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 0.3 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 | 0.2 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.1 | 2.4 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 1.4 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 1.9 | GO:0043320 | natural killer cell degranulation(GO:0043320) |
| 0.1 | 0.2 | GO:2000911 | regulation of cholesterol import(GO:0060620) positive regulation of cholesterol import(GO:1904109) regulation of sterol import(GO:2000909) positive regulation of sterol import(GO:2000911) |
| 0.1 | 1.5 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.1 | 0.7 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.1 | 0.6 | GO:0051105 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) |
| 0.1 | 0.4 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 2.1 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.4 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 0.3 | GO:0016132 | brassinosteroid metabolic process(GO:0016131) brassinosteroid biosynthetic process(GO:0016132) |
| 0.1 | 1.0 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 1.0 | GO:2000143 | negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.1 | 0.7 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 0.1 | 2.4 | GO:0019184 | nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.1 | 0.5 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.1 | 2.2 | GO:0051923 | sulfation(GO:0051923) |
| 0.1 | 1.0 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.1 | 1.1 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.1 | 1.4 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 0.8 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.1 | 1.1 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 0.8 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 0.1 | 3.5 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.1 | 2.0 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 0.6 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.1 | 0.6 | GO:0044838 | cell quiescence(GO:0044838) |
| 0.1 | 0.4 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.1 | 1.5 | GO:0003283 | atrial septum development(GO:0003283) |
| 0.1 | 0.3 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.1 | 1.0 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.1 | 0.6 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.1 | 0.7 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 1.1 | GO:0032825 | positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.1 | 5.1 | GO:0003341 | cilium movement(GO:0003341) |
| 0.1 | 0.4 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.1 | 0.8 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.1 | 4.4 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 4.8 | GO:0035036 | sperm-egg recognition(GO:0035036) |
| 0.1 | 2.5 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 0.3 | GO:2000813 | actin filament uncapping(GO:0051695) negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.1 | 33.8 | GO:0043434 | response to peptide hormone(GO:0043434) |
| 0.1 | 2.1 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 0.5 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.1 | 0.3 | GO:0071033 | nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.1 | 0.3 | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.1 | 2.1 | GO:0050927 | positive regulation of positive chemotaxis(GO:0050927) |
| 0.1 | 0.6 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.1 | 0.2 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.1 | 1.6 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.1 | 3.0 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.1 | 1.4 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.4 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.1 | 0.7 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.1 | 0.3 | GO:0090155 | negative regulation of sphingolipid biosynthetic process(GO:0090155) cellular sphingolipid homeostasis(GO:0090156) negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.1 | 1.0 | GO:0043144 | snoRNA processing(GO:0043144) |
| 0.1 | 0.6 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.1 | 0.4 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.9 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 0.6 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.1 | 2.8 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 | 1.5 | GO:0051307 | meiotic chromosome separation(GO:0051307) |
| 0.1 | 2.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.5 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 | 0.9 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.1 | 1.5 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.1 | 1.0 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.1 | 1.1 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.1 | 0.6 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 0.8 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 0.3 | GO:0002155 | thyroid hormone mediated signaling pathway(GO:0002154) regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.1 | 5.5 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.1 | 0.1 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.1 | 0.3 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 1.0 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.1 | 1.3 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.1 | 2.1 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.1 | 1.2 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.1 | 0.4 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.1 | 0.3 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.1 | 1.2 | GO:0032042 | mitochondrial DNA metabolic process(GO:0032042) |
| 0.1 | 0.2 | GO:0015734 | beta-alanine transport(GO:0001762) taurine transport(GO:0015734) |
| 0.1 | 0.5 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.1 | 0.8 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 0.3 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.1 | 3.2 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.3 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.1 | 0.8 | GO:0072431 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) |
| 0.1 | 0.3 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.1 | 0.7 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 0.2 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 0.5 | GO:0042640 | anagen(GO:0042640) hair cycle phase(GO:0044851) |
| 0.1 | 0.4 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 2.7 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.1 | 4.1 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.1 | 0.7 | GO:0015838 | amino-acid betaine transport(GO:0015838) |
| 0.1 | 0.6 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 1.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 0.2 | GO:0050755 | chemokine metabolic process(GO:0050755) |
| 0.1 | 0.5 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 1.2 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.1 | 0.6 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 | 2.7 | GO:0051438 | regulation of ubiquitin-protein transferase activity(GO:0051438) |
| 0.1 | 0.2 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.1 | 0.9 | GO:0033687 | osteoblast proliferation(GO:0033687) |
| 0.1 | 1.0 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.1 | 0.4 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.1 | 1.4 | GO:0070229 | negative regulation of lymphocyte apoptotic process(GO:0070229) |
| 0.1 | 0.9 | GO:0071451 | removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) |
| 0.1 | 0.5 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.2 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.1 | 0.6 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 0.3 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.1 | 0.3 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.1 | 2.0 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.1 | 0.3 | GO:0014717 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.1 | 0.9 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.1 | 0.9 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.1 | 0.5 | GO:0048820 | hair follicle maturation(GO:0048820) |
| 0.0 | 0.5 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.0 | 1.0 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.5 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.0 | 1.4 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.7 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.6 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.3 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.4 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 1.0 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.0 | 0.7 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.5 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.6 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.3 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.3 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 4.8 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 1.4 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 3.9 | GO:1903955 | positive regulation of establishment of protein localization to mitochondrion(GO:1903749) positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 1.2 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.3 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 1.6 | GO:1901099 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.4 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 1.2 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.2 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.0 | 0.2 | GO:0090205 | positive regulation of cholesterol biosynthetic process(GO:0045542) positive regulation of cholesterol metabolic process(GO:0090205) regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.2 | GO:1904306 | positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 0.0 | 0.4 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.5 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 1.0 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 1.3 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.9 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 1.0 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 2.7 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 5.1 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.2 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.0 | 1.1 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.3 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.7 | GO:1903427 | negative regulation of reactive oxygen species biosynthetic process(GO:1903427) |
| 0.0 | 0.6 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 1.2 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.3 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.7 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.0 | 0.2 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 1.3 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.5 | GO:0044065 | regulation of respiratory system process(GO:0044065) |
| 0.0 | 0.3 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.0 | 1.1 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.4 | GO:0044247 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.7 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.3 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.1 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 0.2 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.0 | 0.5 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.0 | 1.0 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.2 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.5 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.0 | 0.2 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.3 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.3 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.4 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 1.2 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.3 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.6 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.1 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.3 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 1.0 | GO:0007569 | cell aging(GO:0007569) |
| 0.0 | 0.3 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.1 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.2 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.2 | GO:0071549 | cellular response to dexamethasone stimulus(GO:0071549) |
| 0.0 | 0.2 | GO:0032264 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.0 | 2.5 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.0 | 0.4 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) |
| 0.0 | 0.2 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.4 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.1 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.0 | 0.8 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.1 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 | 1.0 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 0.2 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.6 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.1 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.0 | 0.3 | GO:0035584 | calcium-mediated signaling using intracellular calcium source(GO:0035584) |
| 0.0 | 1.2 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.5 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 0.9 | GO:0030218 | erythrocyte differentiation(GO:0030218) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.2 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 1.3 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.1 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.0 | 0.0 | GO:0042262 | DNA protection(GO:0042262) |
| 0.0 | 0.8 | GO:0003279 | cardiac septum development(GO:0003279) |
| 0.0 | 0.1 | GO:0015961 | diadenosine polyphosphate catabolic process(GO:0015961) |
| 0.0 | 0.2 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 0.1 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.0 | 0.4 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.0 | 0.3 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.0 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 1.2 | 7.0 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 1.0 | 10.7 | GO:0097433 | dense body(GO:0097433) |
| 0.9 | 3.6 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.9 | 36.0 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.8 | 4.6 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.8 | 7.7 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.7 | 5.2 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.7 | 2.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.7 | 2.0 | GO:0000811 | GINS complex(GO:0000811) |
| 0.5 | 1.6 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.5 | 5.5 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.5 | 1.4 | GO:0060187 | cell pole(GO:0060187) |
| 0.4 | 1.7 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.4 | 10.4 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.4 | 2.6 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.4 | 1.4 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.3 | 3.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.3 | 2.0 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.3 | 1.8 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.3 | 2.0 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.3 | 1.1 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.3 | 0.8 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.3 | 1.6 | GO:0071547 | piP-body(GO:0071547) |
| 0.3 | 4.7 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.3 | 1.0 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.3 | 2.3 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.2 | 1.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.2 | 8.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.2 | 3.7 | GO:0032797 | SMN complex(GO:0032797) |
| 0.2 | 1.4 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.2 | 0.7 | GO:0034066 | RIC1-RGP1 guanyl-nucleotide exchange factor complex(GO:0034066) |
| 0.2 | 14.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.2 | 1.7 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.2 | 2.8 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.2 | 0.6 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.2 | 4.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 1.3 | GO:0072487 | MSL complex(GO:0072487) |
| 0.2 | 0.6 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.2 | 0.9 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.2 | 1.8 | GO:0002177 | manchette(GO:0002177) |
| 0.2 | 3.0 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.2 | 1.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.2 | 0.8 | GO:0031251 | PAN complex(GO:0031251) |
| 0.2 | 1.9 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.2 | 4.3 | GO:0097346 | INO80-type complex(GO:0097346) |
| 0.2 | 2.6 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.2 | 4.8 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 3.7 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.9 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.1 | 1.5 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.6 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 2.0 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 1.4 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 15.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 1.1 | GO:0031313 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.5 | GO:1990429 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.1 | 2.1 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 1.1 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 1.4 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 1.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 1.4 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 1.5 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 4.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 1.0 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 0.4 | GO:0036501 | UFD1-NPL4 complex(GO:0036501) |
| 0.1 | 0.5 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.1 | 0.8 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.9 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 1.8 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 3.1 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 1.0 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.7 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 1.6 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.8 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 0.5 | GO:1990923 | PET complex(GO:1990923) |
| 0.1 | 20.2 | GO:0031514 | motile cilium(GO:0031514) |
| 0.1 | 1.8 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 1.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.5 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.1 | 1.5 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 3.7 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 1.4 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 15.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 2.8 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 0.4 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 0.3 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.1 | 1.8 | GO:0030430 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.1 | 0.4 | GO:0032444 | activin responsive factor complex(GO:0032444) SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 4.1 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 0.2 | GO:0002095 | caveolar macromolecular signaling complex(GO:0002095) |
| 0.1 | 16.4 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 1.8 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.4 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 1.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.6 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 1.6 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 0.7 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.1 | 0.3 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.1 | 0.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 0.4 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 1.6 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.1 | 0.9 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 1.3 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 1.0 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 0.9 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.1 | 1.6 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.4 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 0.5 | GO:0043073 | female germ cell nucleus(GO:0001674) germ cell nucleus(GO:0043073) |
| 0.1 | 0.5 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 0.7 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 0.5 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 0.3 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 1.9 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 0.8 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 0.8 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 0.4 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.1 | 0.7 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 0.2 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.1 | 0.4 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 2.2 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 0.2 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 0.2 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 0.6 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.1 | 0.8 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 0.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 3.5 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 0.8 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.3 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 0.4 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 0.3 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.1 | 2.0 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.9 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 0.6 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 97.0 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 1.0 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.5 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.5 | GO:0030663 | COPI vesicle coat(GO:0030126) COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 3.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.3 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.9 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 2.0 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.5 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.7 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 1.0 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.7 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.3 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.9 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.2 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 1.2 | GO:0005684 | U2-type spliceosomal complex(GO:0005684) |
| 0.0 | 8.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 2.7 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 3.5 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.2 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.1 | GO:0070992 | translation initiation complex(GO:0070992) |
| 0.0 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 1.6 | GO:1903293 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 0.3 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 15.2 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.0 | 0.1 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.2 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 5.6 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 2.9 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.5 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 8.0 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.1 | GO:0034448 | EGO complex(GO:0034448) |
| 0.0 | 19.4 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.4 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 1.2 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.3 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.7 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 1.9 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 2.3 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 2.8 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 11.3 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 2.3 | 6.9 | GO:0005009 | insulin-activated receptor activity(GO:0005009) |
| 2.2 | 10.8 | GO:0070287 | ferritin receptor activity(GO:0070287) |
| 1.9 | 9.5 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 1.8 | 5.5 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) |
| 1.8 | 5.5 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 1.4 | 10.0 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 1.4 | 4.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 1.3 | 7.7 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 1.3 | 8.8 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 1.2 | 9.9 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 1.1 | 9.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 1.1 | 8.0 | GO:0050051 | alkane 1-monooxygenase activity(GO:0018685) leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 1.1 | 7.8 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 1.1 | 3.3 | GO:0032190 | acrosin binding(GO:0032190) |
| 1.1 | 12.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.9 | 29.9 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.9 | 3.7 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.9 | 3.7 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.9 | 4.5 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.9 | 8.0 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.9 | 2.6 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.9 | 13.0 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.8 | 6.7 | GO:0005186 | pheromone activity(GO:0005186) |
| 0.8 | 3.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.8 | 2.4 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.8 | 3.2 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.8 | 2.3 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.8 | 2.3 | GO:0070026 | cystathionine beta-synthase activity(GO:0004122) oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) nitrite reductase (NO-forming) activity(GO:0050421) carbon monoxide binding(GO:0070025) nitric oxide binding(GO:0070026) |
| 0.8 | 2.3 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.7 | 3.0 | GO:0031699 | beta-3 adrenergic receptor binding(GO:0031699) |
| 0.7 | 2.2 | GO:0030338 | CMP-N-acetylneuraminate monooxygenase activity(GO:0030338) |
| 0.7 | 4.4 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.7 | 2.1 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.7 | 8.4 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.7 | 2.1 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.7 | 2.0 | GO:0042602 | riboflavin reductase (NADPH) activity(GO:0042602) |
| 0.7 | 2.0 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.6 | 98.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.6 | 1.7 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.6 | 2.3 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.6 | 2.3 | GO:0018455 | alcohol dehydrogenase [NAD(P)+] activity(GO:0018455) |
| 0.6 | 2.8 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.6 | 1.7 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.5 | 1.6 | GO:0000401 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.5 | 2.6 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.5 | 4.6 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.5 | 2.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.5 | 1.9 | GO:0036470 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 0.5 | 2.8 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.5 | 2.8 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.5 | 1.9 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.5 | 4.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.4 | 4.9 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.4 | 0.4 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.4 | 3.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.4 | 2.1 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.4 | 1.2 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.4 | 1.2 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.4 | 1.5 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.4 | 2.3 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.4 | 8.2 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.4 | 1.9 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.4 | 1.5 | GO:0042008 | interleukin-18 receptor activity(GO:0042008) |
| 0.4 | 1.5 | GO:0008941 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) iron-cytochrome-c reductase activity(GO:0047726) |
| 0.3 | 1.4 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.3 | 0.3 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.3 | 2.1 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.3 | 2.7 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.3 | 1.0 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.3 | 3.9 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.3 | 1.0 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.3 | 1.3 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.3 | 1.3 | GO:0033814 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.3 | 3.8 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.3 | 6.3 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.3 | 9.1 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.3 | 5.8 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.3 | 2.7 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.3 | 3.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.3 | 2.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.3 | 0.9 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.3 | 1.8 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.3 | 1.7 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.3 | 0.8 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.3 | 1.9 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.3 | 2.7 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.3 | 1.9 | GO:0070404 | NADH binding(GO:0070404) |
| 0.3 | 0.8 | GO:0015152 | glucose-6-phosphate transmembrane transporter activity(GO:0015152) |
| 0.3 | 2.9 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.3 | 1.6 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.3 | 1.0 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.3 | 1.0 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.3 | 1.8 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.3 | 0.8 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.2 | 6.1 | GO:0051861 | glycolipid binding(GO:0051861) |
| 0.2 | 1.2 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.2 | 4.9 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.2 | 9.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 4.6 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.2 | 2.4 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.2 | 9.5 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.2 | 1.3 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.2 | 2.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.2 | 2.7 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.2 | 6.1 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.2 | 0.6 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.2 | 1.0 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.2 | 1.2 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.2 | 0.6 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 0.2 | 3.8 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.2 | 0.8 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.2 | 3.6 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.2 | 0.7 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.2 | 2.0 | GO:0043426 | MRF binding(GO:0043426) |
| 0.2 | 0.7 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.2 | 2.9 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.2 | 0.5 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.2 | 1.0 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.2 | 1.0 | GO:0052724 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.2 | 0.5 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.2 | 0.5 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.2 | 6.4 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.2 | 1.7 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.2 | 0.5 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.2 | 2.4 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 1.9 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.1 | 0.6 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.1 | 4.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 3.8 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 2.2 | GO:0015093 | iron ion transmembrane transporter activity(GO:0005381) ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 3.0 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 2.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 1.0 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 1.3 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.1 | 2.4 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.1 | 2.0 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 0.6 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.1 | 1.0 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.1 | 2.6 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 1.1 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 1.6 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 2.3 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 1.8 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 3.0 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 1.3 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.6 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 3.2 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.1 | 5.4 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 1.0 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 0.7 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.1 | 1.6 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 2.6 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 0.4 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.1 | 2.7 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.9 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 0.3 | GO:0009918 | sterol delta7 reductase activity(GO:0009918) 7-dehydrocholesterol reductase activity(GO:0047598) |
| 0.1 | 2.3 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.1 | 0.8 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 2.1 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.1 | 2.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 2.6 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 1.0 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 1.8 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 6.7 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 4.8 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.1 | 1.9 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 1.4 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 0.5 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.1 | 0.7 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.1 | 3.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 1.8 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.1 | 0.9 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.1 | 1.4 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 4.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 1.2 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 0.6 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 0.6 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 1.3 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.1 | 0.4 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.1 | 7.1 | GO:0004386 | helicase activity(GO:0004386) |
| 0.1 | 3.7 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 0.4 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.1 | 1.3 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.1 | 0.2 | GO:0070905 | serine binding(GO:0070905) |
| 0.1 | 0.4 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) |
| 0.1 | 1.7 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 0.1 | 1.9 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.1 | 2.3 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 1.9 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 1.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 0.8 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.1 | 1.9 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 0.9 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 1.1 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.1 | 7.7 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.1 | 1.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 0.2 | GO:0005368 | beta-alanine transmembrane transporter activity(GO:0001761) taurine transmembrane transporter activity(GO:0005368) taurine:sodium symporter activity(GO:0005369) |
| 0.1 | 0.2 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.1 | 0.6 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 6.7 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.1 | 0.2 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.1 | 0.9 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 0.2 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 2.9 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 0.7 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.4 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 0.2 | GO:0031370 | eukaryotic initiation factor 4G binding(GO:0031370) |
| 0.1 | 0.9 | GO:0015923 | alpha-mannosidase activity(GO:0004559) mannosidase activity(GO:0015923) |
| 0.1 | 0.6 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 3.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.2 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 0.9 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 2.5 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 0.8 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.3 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.1 | 1.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.5 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 0.4 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.1 | 0.3 | GO:0055056 | dehydroascorbic acid transporter activity(GO:0033300) D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.3 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 2.2 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.1 | 1.2 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.1 | 2.1 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 2.8 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 1.2 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 1.0 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 6.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.3 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 1.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 1.8 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 1.3 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.5 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.6 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 1.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 3.1 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.4 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 1.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 2.2 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.4 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.8 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 2.2 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 0.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 1.1 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.5 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 1.4 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.6 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.4 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 9.5 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 0.5 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 2.1 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 1.0 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 1.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.3 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 1.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 3.0 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.7 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.3 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.0 | 0.1 | GO:0008124 | 4-alpha-hydroxytetrahydrobiopterin dehydratase activity(GO:0008124) |
| 0.0 | 1.7 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.6 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 1.1 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 0.5 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.0 | 1.8 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.5 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 2.2 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.5 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 1.1 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.7 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.1 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.0 | 0.6 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 5.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.7 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.9 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.5 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.8 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.1 | GO:0016453 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.1 | GO:0004420 | hydroxymethylglutaryl-CoA reductase (NADPH) activity(GO:0004420) hydroxymethylglutaryl-CoA reductase activity(GO:0042282) |
| 0.0 | 2.3 | GO:0005506 | iron ion binding(GO:0005506) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.4 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0035325 | Toll-like receptor binding(GO:0035325) |
| 0.0 | 0.6 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.3 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.2 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.4 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.5 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.4 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.1 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.0 | GO:0035539 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.0 | 0.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.0 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.4 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 0.9 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.4 | 6.8 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.4 | 20.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.2 | 7.7 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.2 | 13.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 2.8 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 7.2 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.1 | 11.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 31.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 0.7 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 2.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 3.9 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 2.9 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 2.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 8.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 2.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 7.1 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 6.3 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 4.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 2.1 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 2.9 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 1.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 19.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.3 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 3.5 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.1 | 6.5 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.0 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 1.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 3.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.6 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 3.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.5 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 1.5 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.4 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.8 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.3 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.1 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.6 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.1 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.1 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.4 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.3 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 26.7 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 1.6 | 12.9 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 1.2 | 20.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.7 | 9.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.5 | 2.2 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.4 | 8.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.4 | 6.0 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.4 | 10.0 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.4 | 6.5 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.3 | 6.6 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.2 | 5.9 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.2 | 4.9 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.2 | 3.8 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 3.3 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.2 | 9.0 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.2 | 3.6 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.2 | 5.5 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.2 | 2.8 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.2 | 2.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 1.9 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 4.9 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 4.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 2.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.2 | 3.7 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.2 | 2.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 2.9 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.2 | 2.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.2 | 4.1 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 1.0 | REACTOME AQUAPORIN MEDIATED TRANSPORT | Genes involved in Aquaporin-mediated transport |
| 0.1 | 2.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 2.1 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 7.5 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 6.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 2.8 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 2.5 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 1.9 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 2.8 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 0.9 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 1.9 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 0.5 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.1 | 1.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 0.4 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.1 | 2.6 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 1.0 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.1 | 3.2 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 1.8 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 2.1 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.1 | 3.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 1.5 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 6.2 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 2.3 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 4.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 1.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 0.7 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 1.3 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 3.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 0.9 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 3.8 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 2.4 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 1.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 1.2 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF MITOTIC PROTEINS | Genes involved in APC/C:Cdc20 mediated degradation of mitotic proteins |
| 0.1 | 0.1 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.1 | 3.1 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 3.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 2.1 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 5.0 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.1 | 6.1 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 0.5 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 0.9 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 1.0 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 3.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 2.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.6 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.5 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 1.3 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.4 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.7 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.5 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.3 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 1.5 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.3 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.0 | 0.5 | REACTOME CDT1 ASSOCIATION WITH THE CDC6 ORC ORIGIN COMPLEX | Genes involved in CDT1 association with the CDC6:ORC:origin complex |
| 0.0 | 0.4 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.2 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.5 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 3.0 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 1.0 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 1.0 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.3 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 4.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.2 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.6 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |