Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Brca1
Z-value: 1.09
Transcription factors associated with Brca1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Brca1
|
ENSMUSG00000017146.13 | Brca1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Brca1 | mm39_v1_chr11_-_101442663_101442719 | -0.23 | 5.1e-02 | Click! |
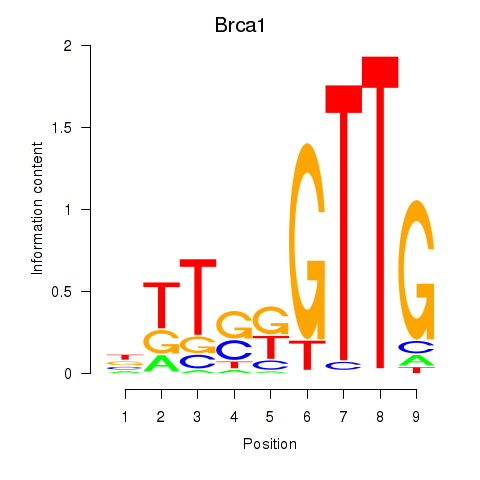
Activity profile of Brca1 motif
Sorted Z-values of Brca1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Brca1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_107107558 | 11.44 |
ENSMUST00000105280.5
|
Lin7a
|
lin-7 homolog A (C. elegans) |
| chr16_+_20514925 | 7.75 |
ENSMUST00000128273.2
|
Fam131a
|
family with sequence similarity 131, member A |
| chr2_-_73605387 | 7.15 |
ENSMUST00000166199.9
|
Chn1
|
chimerin 1 |
| chr13_+_42834989 | 5.83 |
ENSMUST00000149235.8
|
Phactr1
|
phosphatase and actin regulator 1 |
| chrX_+_165127688 | 5.72 |
ENSMUST00000112223.8
ENSMUST00000112224.8 ENSMUST00000112229.9 ENSMUST00000112228.8 ENSMUST00000112227.9 ENSMUST00000112226.3 |
Gpm6b
|
glycoprotein m6b |
| chr3_-_121076745 | 5.07 |
ENSMUST00000135818.8
ENSMUST00000137234.2 |
Tlcd4
|
TLC domain containing 4 |
| chr13_+_68745542 | 5.02 |
ENSMUST00000222604.2
|
1700001L19Rik
|
RIKEN cDNA 1700001L19 gene |
| chr10_+_107107477 | 4.83 |
ENSMUST00000020057.16
|
Lin7a
|
lin-7 homolog A (C. elegans) |
| chr13_+_68745528 | 4.73 |
ENSMUST00000022007.8
|
1700001L19Rik
|
RIKEN cDNA 1700001L19 gene |
| chr2_-_57942844 | 4.69 |
ENSMUST00000090940.6
|
Ermn
|
ermin, ERM-like protein |
| chr15_-_96917804 | 4.35 |
ENSMUST00000231039.2
|
Slc38a4
|
solute carrier family 38, member 4 |
| chrX_+_134894573 | 4.27 |
ENSMUST00000058119.9
|
Arxes2
|
adipocyte-related X-chromosome expressed sequence 2 |
| chr15_-_96918203 | 4.14 |
ENSMUST00000166223.2
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr5_+_16139683 | 4.12 |
ENSMUST00000167946.9
|
Cacna2d1
|
calcium channel, voltage-dependent, alpha2/delta subunit 1 |
| chr9_+_89791943 | 4.00 |
ENSMUST00000189545.2
ENSMUST00000034909.11 ENSMUST00000034912.6 |
Rasgrf1
|
RAS protein-specific guanine nucleotide-releasing factor 1 |
| chr4_+_98919183 | 4.00 |
ENSMUST00000030280.7
|
Angptl3
|
angiopoietin-like 3 |
| chr6_-_122778598 | 3.91 |
ENSMUST00000165884.8
|
Slc2a3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chrX_+_149377416 | 3.76 |
ENSMUST00000112713.3
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr7_-_101749140 | 3.57 |
ENSMUST00000106934.2
|
Art5
|
ADP-ribosyltransferase 5 |
| chr11_+_104441489 | 3.51 |
ENSMUST00000018800.9
|
Myl4
|
myosin, light polypeptide 4 |
| chr4_-_138123700 | 3.43 |
ENSMUST00000105032.4
|
Fam43b
|
family with sequence similarity 43, member B |
| chr4_-_11966367 | 3.38 |
ENSMUST00000056050.5
ENSMUST00000108299.2 ENSMUST00000108297.3 |
Pdp1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr12_-_100486950 | 3.37 |
ENSMUST00000223020.2
ENSMUST00000062957.8 |
Ttc7b
|
tetratricopeptide repeat domain 7B |
| chr13_+_93441447 | 3.28 |
ENSMUST00000109497.8
ENSMUST00000109498.8 ENSMUST00000060490.11 ENSMUST00000109492.9 ENSMUST00000109496.8 ENSMUST00000109495.8 |
Homer1
|
homer scaffolding protein 1 |
| chr16_+_41353212 | 3.24 |
ENSMUST00000078873.11
|
Lsamp
|
limbic system-associated membrane protein |
| chr5_+_35214110 | 3.21 |
ENSMUST00000101298.9
ENSMUST00000114270.8 ENSMUST00000133381.2 |
Dok7
|
docking protein 7 |
| chr5_+_20112500 | 3.17 |
ENSMUST00000101558.10
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr11_-_41891111 | 3.13 |
ENSMUST00000109290.2
|
Gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 2 |
| chr7_-_101749433 | 3.12 |
ENSMUST00000106937.8
|
Art5
|
ADP-ribosyltransferase 5 |
| chr2_-_45000389 | 3.07 |
ENSMUST00000201804.4
ENSMUST00000028229.13 ENSMUST00000202187.4 ENSMUST00000153561.6 ENSMUST00000201490.2 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr13_+_4486105 | 3.06 |
ENSMUST00000156277.2
|
Akr1c6
|
aldo-keto reductase family 1, member C6 |
| chr18_+_37085673 | 3.05 |
ENSMUST00000192512.6
ENSMUST00000192295.2 ENSMUST00000115661.5 |
Pcdha4
Gm42416
|
protocadherin alpha 4 predicted gene, 42416 |
| chr3_-_107838895 | 3.04 |
ENSMUST00000133947.9
ENSMUST00000124215.2 ENSMUST00000106688.8 ENSMUST00000106687.9 |
Gstm7
|
glutathione S-transferase, mu 7 |
| chr11_-_41891359 | 2.98 |
ENSMUST00000070735.10
|
Gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 2 |
| chr17_-_90395568 | 2.94 |
ENSMUST00000173222.2
|
Nrxn1
|
neurexin I |
| chr1_-_46893206 | 2.92 |
ENSMUST00000027131.6
|
Slc39a10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr6_-_122317457 | 2.92 |
ENSMUST00000160843.8
|
Phc1
|
polyhomeotic 1 |
| chr14_-_78970160 | 2.84 |
ENSMUST00000226342.3
|
Dgkh
|
diacylglycerol kinase, eta |
| chr19_+_4771089 | 2.81 |
ENSMUST00000238976.3
|
Sptbn2
|
spectrin beta, non-erythrocytic 2 |
| chr13_-_95386776 | 2.80 |
ENSMUST00000162153.9
ENSMUST00000160957.9 ENSMUST00000159598.2 ENSMUST00000162412.8 |
Pde8b
|
phosphodiesterase 8B |
| chr9_+_43996236 | 2.77 |
ENSMUST00000065461.9
ENSMUST00000176416.8 |
Usp2
|
ubiquitin specific peptidase 2 |
| chr17_+_71511642 | 2.70 |
ENSMUST00000126681.8
|
Lpin2
|
lipin 2 |
| chr1_-_132635078 | 2.66 |
ENSMUST00000187861.7
|
Nfasc
|
neurofascin |
| chr1_-_132635042 | 2.66 |
ENSMUST00000043189.14
|
Nfasc
|
neurofascin |
| chr5_-_87240405 | 2.66 |
ENSMUST00000132667.2
ENSMUST00000145617.8 ENSMUST00000094649.11 |
Ugt2b36
|
UDP glucuronosyltransferase 2 family, polypeptide B36 |
| chr1_+_37068387 | 2.65 |
ENSMUST00000067178.14
ENSMUST00000238500.2 |
Vwa3b
|
von Willebrand factor A domain containing 3B |
| chr10_-_33500583 | 2.63 |
ENSMUST00000161692.2
ENSMUST00000160299.2 ENSMUST00000019920.13 |
Clvs2
|
clavesin 2 |
| chr6_-_142418801 | 2.62 |
ENSMUST00000032371.8
|
Gys2
|
glycogen synthase 2 |
| chr7_-_112968533 | 2.61 |
ENSMUST00000047091.14
ENSMUST00000119278.8 |
Btbd10
|
BTB (POZ) domain containing 10 |
| chr7_+_51528715 | 2.59 |
ENSMUST00000051912.13
|
Gas2
|
growth arrest specific 2 |
| chr9_+_110948492 | 2.54 |
ENSMUST00000217341.3
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr2_-_113659360 | 2.54 |
ENSMUST00000024005.8
|
Scg5
|
secretogranin V |
| chr2_+_25178147 | 2.53 |
ENSMUST00000028340.4
|
Tmem210
|
transmembrane protein 210 |
| chr10_+_86599836 | 2.48 |
ENSMUST00000218802.2
|
Gm49358
|
predicted gene, 49358 |
| chr6_-_122317484 | 2.48 |
ENSMUST00000112600.9
|
Phc1
|
polyhomeotic 1 |
| chr2_+_67935015 | 2.46 |
ENSMUST00000042456.4
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr6_-_133830613 | 2.42 |
ENSMUST00000203168.2
ENSMUST00000048032.5 |
Kap
|
kidney androgen regulated protein |
| chr8_+_12965876 | 2.38 |
ENSMUST00000110876.9
ENSMUST00000110879.9 |
Mcf2l
|
mcf.2 transforming sequence-like |
| chr7_-_27252543 | 2.37 |
ENSMUST00000127240.8
ENSMUST00000117095.8 ENSMUST00000117611.8 |
Pld3
|
phospholipase D family, member 3 |
| chr5_+_67126304 | 2.34 |
ENSMUST00000132991.5
|
Limch1
|
LIM and calponin homology domains 1 |
| chr8_+_55024446 | 2.33 |
ENSMUST00000239166.2
ENSMUST00000239106.2 ENSMUST00000239152.2 |
Asb5
|
ankyrin repeat and SOCs box-containing 5 |
| chr14_-_20706556 | 2.32 |
ENSMUST00000090469.8
|
Myoz1
|
myozenin 1 |
| chr10_+_40446326 | 2.26 |
ENSMUST00000078314.14
|
Slc22a16
|
solute carrier family 22 (organic cation transporter), member 16 |
| chr2_-_104324035 | 2.23 |
ENSMUST00000111124.8
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chr16_+_34471558 | 2.21 |
ENSMUST00000023530.5
|
Ropn1
|
ropporin, rhophilin associated protein 1 |
| chr7_+_27252658 | 2.21 |
ENSMUST00000067386.14
ENSMUST00000191126.7 ENSMUST00000187960.7 |
2310022A10Rik
|
RIKEN cDNA 2310022A10 gene |
| chr1_+_93271340 | 2.20 |
ENSMUST00000185498.7
|
Ppp1r7
|
protein phosphatase 1, regulatory subunit 7 |
| chr4_+_148085179 | 2.18 |
ENSMUST00000103230.5
|
Nppa
|
natriuretic peptide type A |
| chr2_+_109523908 | 2.17 |
ENSMUST00000111045.9
|
Bdnf
|
brain derived neurotrophic factor |
| chr10_+_40446605 | 2.15 |
ENSMUST00000019978.9
|
Slc22a16
|
solute carrier family 22 (organic cation transporter), member 16 |
| chr2_-_120681078 | 2.10 |
ENSMUST00000028740.11
|
Ttbk2
|
tau tubulin kinase 2 |
| chr3_+_20011405 | 2.04 |
ENSMUST00000108325.9
|
Cp
|
ceruloplasmin |
| chrX_+_111150171 | 1.99 |
ENSMUST00000164272.3
ENSMUST00000132037.2 |
4933403O08Rik
|
RIKEN cDNA 4933403O08 gene |
| chr12_+_113507528 | 1.96 |
ENSMUST00000053086.3
|
Adam6a
|
a disintegrin and metallopeptidase domain 6A |
| chr9_-_108911438 | 1.93 |
ENSMUST00000192801.2
|
Tma7
|
translational machinery associated 7 |
| chr6_+_17693941 | 1.91 |
ENSMUST00000115420.8
ENSMUST00000115419.8 |
St7
|
suppression of tumorigenicity 7 |
| chr17_+_43671314 | 1.85 |
ENSMUST00000226087.2
|
Adgrf5
|
adhesion G protein-coupled receptor F5 |
| chr14_+_32555912 | 1.81 |
ENSMUST00000104926.3
|
Fam170b
|
family with sequence similarity 170, member B |
| chr5_+_77151902 | 1.78 |
ENSMUST00000071199.9
|
Arl9
|
ADP-ribosylation factor-like 9 |
| chr6_-_40917431 | 1.75 |
ENSMUST00000122181.8
ENSMUST00000031935.10 |
1700074P13Rik
|
RIKEN cDNA 1700074P13 gene |
| chrM_+_9459 | 1.73 |
ENSMUST00000082411.1
|
mt-Nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr4_+_10874498 | 1.71 |
ENSMUST00000080517.14
|
2610301B20Rik
|
RIKEN cDNA 2610301B20 gene |
| chr4_+_10874537 | 1.69 |
ENSMUST00000101504.3
|
2610301B20Rik
|
RIKEN cDNA 2610301B20 gene |
| chr6_-_141892686 | 1.68 |
ENSMUST00000042119.6
|
Slco1a1
|
solute carrier organic anion transporter family, member 1a1 |
| chr13_+_100788087 | 1.67 |
ENSMUST00000190165.7
ENSMUST00000185767.7 |
Taf9
|
TATA-box binding protein associated factor 9 |
| chr9_+_64718596 | 1.67 |
ENSMUST00000038890.6
|
Dennd4a
|
DENN/MADD domain containing 4A |
| chr2_-_134486039 | 1.66 |
ENSMUST00000038228.11
|
Tmx4
|
thioredoxin-related transmembrane protein 4 |
| chr7_-_55669702 | 1.66 |
ENSMUST00000052204.6
|
Nipa1
|
non imprinted in Prader-Willi/Angelman syndrome 1 homolog (human) |
| chrX_+_168468186 | 1.65 |
ENSMUST00000112107.8
ENSMUST00000112104.8 |
Mid1
|
midline 1 |
| chr18_+_37138256 | 1.64 |
ENSMUST00000115658.6
|
Pcdha11
|
protocadherin alpha 11 |
| chr19_-_6191440 | 1.64 |
ENSMUST00000025893.7
|
Arl2
|
ADP-ribosylation factor-like 2 |
| chr11_-_70537878 | 1.63 |
ENSMUST00000014750.15
|
Slc25a11
|
solute carrier family 25 (mitochondrial carrier oxoglutarate carrier), member 11 |
| chr12_+_108145802 | 1.63 |
ENSMUST00000221167.2
|
Ccnk
|
cyclin K |
| chr2_-_134485936 | 1.62 |
ENSMUST00000110120.2
|
Tmx4
|
thioredoxin-related transmembrane protein 4 |
| chr2_-_45000250 | 1.61 |
ENSMUST00000201211.4
ENSMUST00000177302.8 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr8_-_68514879 | 1.57 |
ENSMUST00000212505.2
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr9_+_108270020 | 1.57 |
ENSMUST00000035234.6
|
1700102P08Rik
|
RIKEN cDNA 1700102P08 gene |
| chr10_-_79422946 | 1.55 |
ENSMUST00000077433.13
|
Theg
|
testicular haploid expressed gene |
| chr2_+_120459593 | 1.55 |
ENSMUST00000180041.9
|
Stard9
|
START domain containing 9 |
| chr13_-_95359955 | 1.55 |
ENSMUST00000159608.8
|
Pde8b
|
phosphodiesterase 8B |
| chr10_-_30531768 | 1.54 |
ENSMUST00000092610.12
|
Ncoa7
|
nuclear receptor coactivator 7 |
| chr5_+_20112704 | 1.51 |
ENSMUST00000115267.7
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr2_-_76045696 | 1.51 |
ENSMUST00000144892.2
|
Pde11a
|
phosphodiesterase 11A |
| chrX_-_74886623 | 1.47 |
ENSMUST00000114057.8
|
Pls3
|
plastin 3 (T-isoform) |
| chr18_-_43610829 | 1.47 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr6_-_141892517 | 1.45 |
ENSMUST00000168119.8
|
Slco1a1
|
solute carrier organic anion transporter family, member 1a1 |
| chr4_+_115641996 | 1.44 |
ENSMUST00000177280.8
ENSMUST00000176047.8 |
Atpaf1
|
ATP synthase mitochondrial F1 complex assembly factor 1 |
| chr7_+_50248918 | 1.44 |
ENSMUST00000119710.3
|
4933405O20Rik
|
RIKEN cDNA 4933405O20 gene |
| chr9_+_108269992 | 1.43 |
ENSMUST00000192995.6
|
1700102P08Rik
|
RIKEN cDNA 1700102P08 gene |
| chr1_+_179788675 | 1.43 |
ENSMUST00000076687.12
ENSMUST00000097450.10 ENSMUST00000212756.2 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr6_-_55109826 | 1.41 |
ENSMUST00000164012.3
ENSMUST00000212633.2 |
Crhr2
|
corticotropin releasing hormone receptor 2 |
| chr7_+_129193581 | 1.36 |
ENSMUST00000084519.7
|
Wdr11
|
WD repeat domain 11 |
| chr3_+_20011251 | 1.36 |
ENSMUST00000108328.8
|
Cp
|
ceruloplasmin |
| chr2_-_134485999 | 1.35 |
ENSMUST00000110119.2
|
Tmx4
|
thioredoxin-related transmembrane protein 4 |
| chr10_+_69761630 | 1.33 |
ENSMUST00000182029.8
|
Ank3
|
ankyrin 3, epithelial |
| chr7_-_100581314 | 1.32 |
ENSMUST00000107032.3
|
Arhgef17
|
Rho guanine nucleotide exchange factor (GEF) 17 |
| chr2_-_77000936 | 1.31 |
ENSMUST00000164114.9
ENSMUST00000049544.14 |
Ccdc141
|
coiled-coil domain containing 141 |
| chr17_-_47922374 | 1.30 |
ENSMUST00000024783.9
|
Bysl
|
bystin-like |
| chr4_+_108336164 | 1.28 |
ENSMUST00000155068.2
|
Tut4
|
terminal uridylyl transferase 4 |
| chr7_-_45343785 | 1.26 |
ENSMUST00000040636.9
|
Sec1
|
secretory blood group 1 |
| chr6_-_55109960 | 1.24 |
ENSMUST00000003568.15
|
Crhr2
|
corticotropin releasing hormone receptor 2 |
| chr3_-_51184895 | 1.24 |
ENSMUST00000108051.8
ENSMUST00000108053.9 |
Elf2
|
E74-like factor 2 |
| chr13_+_100787841 | 1.21 |
ENSMUST00000084721.8
ENSMUST00000190729.2 |
Ak6
|
adenylate kinase 6 |
| chr17_+_14087827 | 1.20 |
ENSMUST00000239324.2
|
Afdn
|
afadin, adherens junction formation factor |
| chr7_-_81216687 | 1.19 |
ENSMUST00000042318.6
|
Fsd2
|
fibronectin type III and SPRY domain containing 2 |
| chr10_+_69761597 | 1.19 |
ENSMUST00000182269.8
ENSMUST00000183261.8 ENSMUST00000183074.8 |
Ank3
|
ankyrin 3, epithelial |
| chr13_+_55517545 | 1.19 |
ENSMUST00000063771.14
|
Rgs14
|
regulator of G-protein signaling 14 |
| chr7_-_127048280 | 1.19 |
ENSMUST00000053392.11
|
Zfp689
|
zinc finger protein 689 |
| chr4_-_116565509 | 1.18 |
ENSMUST00000030453.5
|
Mmachc
|
methylmalonic aciduria cblC type, with homocystinuria |
| chr7_+_25016492 | 1.18 |
ENSMUST00000128119.2
|
Megf8
|
multiple EGF-like-domains 8 |
| chr16_-_58930996 | 1.17 |
ENSMUST00000076262.4
|
Olfr193
|
olfactory receptor 193 |
| chr17_-_34962823 | 1.17 |
ENSMUST00000069507.9
|
C4b
|
complement component 4B (Chido blood group) |
| chr7_-_127048165 | 1.17 |
ENSMUST00000106299.2
|
Zfp689
|
zinc finger protein 689 |
| chr7_+_5083212 | 1.13 |
ENSMUST00000098845.10
ENSMUST00000146317.8 ENSMUST00000153169.2 ENSMUST00000045277.7 |
Epn1
|
epsin 1 |
| chr6_-_122317156 | 1.11 |
ENSMUST00000159384.8
|
Phc1
|
polyhomeotic 1 |
| chr16_-_59459745 | 1.11 |
ENSMUST00000099646.10
|
Arl6
|
ADP-ribosylation factor-like 6 |
| chr1_-_170695328 | 1.09 |
ENSMUST00000027974.7
|
Atf6
|
activating transcription factor 6 |
| chr17_+_36152559 | 1.07 |
ENSMUST00000174124.2
|
Mdc1
|
mediator of DNA damage checkpoint 1 |
| chr5_+_67125902 | 1.05 |
ENSMUST00000127184.8
|
Limch1
|
LIM and calponin homology domains 1 |
| chr9_+_64718708 | 1.04 |
ENSMUST00000213926.2
|
Dennd4a
|
DENN/MADD domain containing 4A |
| chr6_-_68857658 | 1.00 |
ENSMUST00000198756.2
|
Gm42543
|
predicted gene 42543 |
| chr17_+_36134398 | 1.00 |
ENSMUST00000173493.8
ENSMUST00000173147.8 |
Flot1
|
flotillin 1 |
| chr10_-_30531832 | 0.99 |
ENSMUST00000217138.2
ENSMUST00000217644.2 ENSMUST00000216172.2 |
Ncoa7
|
nuclear receptor coactivator 7 |
| chr1_+_171975625 | 0.98 |
ENSMUST00000191689.6
ENSMUST00000192704.6 ENSMUST00000074144.11 |
Dcaf8
|
DDB1 and CUL4 associated factor 8 |
| chr12_-_108145498 | 0.97 |
ENSMUST00000071095.14
|
Setd3
|
SET domain containing 3 |
| chr3_-_51184730 | 0.93 |
ENSMUST00000195432.2
ENSMUST00000091144.11 ENSMUST00000156983.3 |
Elf2
|
E74-like factor 2 |
| chrM_+_7758 | 0.91 |
ENSMUST00000082407.1
|
mt-Atp8
|
mitochondrially encoded ATP synthase 8 |
| chr11_-_69712970 | 0.89 |
ENSMUST00000045771.7
|
Spem1
|
sperm maturation 1 |
| chr7_+_101545547 | 0.88 |
ENSMUST00000035395.14
ENSMUST00000106973.8 ENSMUST00000144207.9 |
Anapc15
|
anaphase promoting complex C subunit 15 |
| chr10_-_13264574 | 0.87 |
ENSMUST00000079698.7
|
Phactr2
|
phosphatase and actin regulator 2 |
| chrM_+_7779 | 0.87 |
ENSMUST00000082408.1
|
mt-Atp6
|
mitochondrially encoded ATP synthase 6 |
| chr8_-_43760017 | 0.85 |
ENSMUST00000082120.5
|
Zfp42
|
zinc finger protein 42 |
| chr16_+_44215136 | 0.85 |
ENSMUST00000099742.9
|
Cfap44
|
cilia and flagella associated protein 44 |
| chr12_-_86773160 | 0.85 |
ENSMUST00000021682.9
|
Angel1
|
angel homolog 1 |
| chr17_+_36152383 | 0.84 |
ENSMUST00000082337.13
|
Mdc1
|
mediator of DNA damage checkpoint 1 |
| chr11_+_78717398 | 0.83 |
ENSMUST00000147875.9
ENSMUST00000141321.2 |
Lyrm9
|
LYR motif containing 9 |
| chr15_+_76264695 | 0.82 |
ENSMUST00000096385.11
ENSMUST00000160728.8 |
Mroh1
|
maestro heat-like repeat family member 1 |
| chr5_-_5564730 | 0.82 |
ENSMUST00000115445.8
ENSMUST00000179804.8 ENSMUST00000125110.2 ENSMUST00000115446.8 |
Cldn12
|
claudin 12 |
| chr2_-_29677634 | 0.79 |
ENSMUST00000177467.8
ENSMUST00000113807.10 |
Trub2
|
TruB pseudouridine (psi) synthase family member 2 |
| chr5_+_92535705 | 0.78 |
ENSMUST00000138687.2
ENSMUST00000124509.2 |
Art3
|
ADP-ribosyltransferase 3 |
| chr4_+_109092459 | 0.78 |
ENSMUST00000106631.9
|
Calr4
|
calreticulin 4 |
| chr11_+_70861007 | 0.78 |
ENSMUST00000018593.10
|
Rpain
|
RPA interacting protein |
| chr4_-_25242858 | 0.76 |
ENSMUST00000029922.14
ENSMUST00000108204.2 |
Fhl5
|
four and a half LIM domains 5 |
| chr13_+_24054267 | 0.75 |
ENSMUST00000006785.8
|
Slc17a1
|
solute carrier family 17 (sodium phosphate), member 1 |
| chr12_-_44256843 | 0.74 |
ENSMUST00000220421.2
|
Dnajb9
|
DnaJ heat shock protein family (Hsp40) member B9 |
| chr2_-_31700592 | 0.74 |
ENSMUST00000057407.3
|
Qrfp
|
pyroglutamylated RFamide peptide |
| chr7_-_64522741 | 0.73 |
ENSMUST00000094331.5
|
Nsmce3
|
NSE3 homolog, SMC5-SMC6 complex component |
| chr8_-_85696369 | 0.73 |
ENSMUST00000109736.9
ENSMUST00000140561.8 |
Rnaseh2a
|
ribonuclease H2, large subunit |
| chr10_+_77697785 | 0.72 |
ENSMUST00000218443.2
|
Gm3285
|
predicted gene 3285 |
| chr7_-_105282751 | 0.70 |
ENSMUST00000057525.14
|
Trim3
|
tripartite motif-containing 3 |
| chr2_-_77000878 | 0.70 |
ENSMUST00000111833.3
|
Ccdc141
|
coiled-coil domain containing 141 |
| chr18_+_65183987 | 0.69 |
ENSMUST00000236103.2
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like |
| chr4_-_155445818 | 0.68 |
ENSMUST00000030922.15
|
Prkcz
|
protein kinase C, zeta |
| chr10_+_69761314 | 0.68 |
ENSMUST00000182692.8
ENSMUST00000092433.12 |
Ank3
|
ankyrin 3, epithelial |
| chr7_-_29605949 | 0.68 |
ENSMUST00000159920.2
ENSMUST00000162592.8 |
Zfp27
|
zinc finger protein 27 |
| chr16_-_22676264 | 0.68 |
ENSMUST00000232075.2
ENSMUST00000004576.8 |
Tbccd1
|
TBCC domain containing 1 |
| chr7_-_12829100 | 0.67 |
ENSMUST00000209822.3
ENSMUST00000235753.2 |
Vmn1r85
|
vomeronasal 1 receptor 85 |
| chr12_+_83567240 | 0.66 |
ENSMUST00000021645.9
|
Dcaf4
|
DDB1 and CUL4 associated factor 4 |
| chrX_+_149372903 | 0.66 |
ENSMUST00000080884.11
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr5_-_123804745 | 0.65 |
ENSMUST00000149410.2
|
Clip1
|
CAP-GLY domain containing linker protein 1 |
| chr12_-_12990584 | 0.65 |
ENSMUST00000130990.2
|
Mycn
|
v-myc avian myelocytomatosis viral related oncogene, neuroblastoma derived |
| chr8_+_25507227 | 0.63 |
ENSMUST00000033961.7
ENSMUST00000210536.2 |
Tm2d2
|
TM2 domain containing 2 |
| chr4_-_98705774 | 0.62 |
ENSMUST00000102790.4
|
Kank4
|
KN motif and ankyrin repeat domains 4 |
| chr1_+_37068429 | 0.61 |
ENSMUST00000162449.7
|
Vwa3b
|
von Willebrand factor A domain containing 3B |
| chr6_+_115578863 | 0.59 |
ENSMUST00000000449.9
|
Mkrn2
|
makorin, ring finger protein, 2 |
| chrX_+_73352694 | 0.59 |
ENSMUST00000130581.2
|
Gdi1
|
guanosine diphosphate (GDP) dissociation inhibitor 1 |
| chr19_+_36061096 | 0.59 |
ENSMUST00000025714.9
|
Rpp30
|
ribonuclease P/MRP 30 subunit |
| chr8_+_41128099 | 0.59 |
ENSMUST00000051614.5
|
Adam24
|
a disintegrin and metallopeptidase domain 24 (testase 1) |
| chr2_+_85606930 | 0.57 |
ENSMUST00000076250.2
|
Olfr1014
|
olfactory receptor 1014 |
| chr3_+_5283606 | 0.57 |
ENSMUST00000026284.13
|
Zfhx4
|
zinc finger homeodomain 4 |
| chr11_-_104441218 | 0.56 |
ENSMUST00000106962.9
ENSMUST00000106961.2 ENSMUST00000093923.9 |
Cdc27
|
cell division cycle 27 |
| chr11_+_70350436 | 0.55 |
ENSMUST00000039093.10
|
Zmynd15
|
zinc finger, MYND-type containing 15 |
| chr19_+_46140942 | 0.55 |
ENSMUST00000026254.14
|
Gbf1
|
golgi-specific brefeldin A-resistance factor 1 |
| chr4_-_123507494 | 0.53 |
ENSMUST00000238866.2
|
Macf1
|
microtubule-actin crosslinking factor 1 |
| chr2_+_29533799 | 0.52 |
ENSMUST00000238899.2
|
Rapgef1
|
Rap guanine nucleotide exchange factor (GEF) 1 |
| chr11_-_93776580 | 0.51 |
ENSMUST00000066888.10
|
Utp18
|
UTP18 small subunit processome component |
| chr12_+_83567303 | 0.51 |
ENSMUST00000222502.2
|
Dcaf4
|
DDB1 and CUL4 associated factor 4 |
| chr11_+_87684548 | 0.49 |
ENSMUST00000143021.9
|
Mpo
|
myeloperoxidase |
| chr7_+_27878894 | 0.48 |
ENSMUST00000085901.13
ENSMUST00000172761.8 |
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr6_+_57086439 | 0.46 |
ENSMUST00000228270.2
|
Vmn1r10
|
vomeronasal 1 receptor 10 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 16.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 1.0 | 3.1 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.8 | 4.1 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.8 | 3.9 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.7 | 2.2 | GO:0061193 | taste bud development(GO:0061193) |
| 0.7 | 5.7 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.7 | 4.7 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.7 | 4.0 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.7 | 2.7 | GO:2000293 | regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.6 | 7.2 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.6 | 4.6 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.5 | 2.2 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 0.5 | 5.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.5 | 2.5 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.5 | 3.0 | GO:0070458 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.4 | 4.4 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.4 | 4.4 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.4 | 1.8 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.4 | 1.4 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.3 | 1.2 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.3 | 2.5 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.3 | 6.1 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.3 | 2.9 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.3 | 3.5 | GO:1900827 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 | 1.1 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.2 | 5.1 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.2 | 3.4 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 | 1.6 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.2 | 1.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.2 | 1.2 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.2 | 0.6 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.2 | 0.3 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.2 | 4.0 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.2 | 0.5 | GO:0002148 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 0.2 | 7.5 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 7.2 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 1.0 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.1 | 1.7 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 1.6 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 1.6 | GO:1901374 | acetylcholine transport(GO:0015870) acetate ester transport(GO:1901374) |
| 0.1 | 0.7 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 2.5 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 0.7 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.1 | 2.8 | GO:0046473 | phosphatidic acid metabolic process(GO:0046473) |
| 0.1 | 3.7 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.1 | 2.2 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 2.8 | GO:0021692 | cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.1 | 0.3 | GO:0000451 | rRNA 2'-O-methylation(GO:0000451) |
| 0.1 | 2.1 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.1 | 6.2 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 0.9 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.1 | 2.8 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.1 | 3.2 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.1 | 1.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 0.7 | GO:2000667 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.1 | 3.1 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.1 | 2.5 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 0.4 | GO:1904579 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 0.1 | 1.7 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 3.4 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 0.6 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.1 | 6.5 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 1.5 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 1.4 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.1 | 1.0 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 1.0 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 2.6 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.1 | 1.1 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.1 | 3.5 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.1 | 0.6 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.1 | 0.4 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 0.4 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 0.8 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.1 | 0.6 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 | 0.6 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.1 | 0.6 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 0.8 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 2.2 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 1.5 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 2.9 | GO:0061098 | positive regulation of protein tyrosine kinase activity(GO:0061098) |
| 0.0 | 1.7 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 2.7 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 2.4 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 2.2 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.2 | GO:0034552 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.8 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 1.6 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 1.1 | GO:0060285 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.8 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 12.2 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 1.7 | GO:0071385 | cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 | 2.2 | GO:0035307 | positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 0.7 | GO:0072711 | cellular response to hydroxyurea(GO:0072711) |
| 0.0 | 2.6 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 1.3 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.3 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.3 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.5 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 2.1 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.7 | GO:0060259 | regulation of feeding behavior(GO:0060259) |
| 0.0 | 2.2 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.7 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 4.2 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.5 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 1.3 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.4 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 0.0 | 0.6 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 1.5 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.2 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.0 | 0.9 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 3.9 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.0 | 0.2 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.2 | GO:0046037 | GMP metabolic process(GO:0046037) |
| 0.0 | 0.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 16.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 1.5 | 4.4 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 1.1 | 5.3 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.6 | 4.7 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.6 | 2.2 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.5 | 6.5 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.5 | 1.6 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) cyclin K-CDK13 complex(GO:0002945) |
| 0.4 | 7.4 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.4 | 2.8 | GO:0008091 | spectrin(GO:0008091) |
| 0.3 | 1.7 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.3 | 4.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.3 | 1.8 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.2 | 1.1 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.2 | 6.1 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 2.2 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.2 | 2.7 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 3.9 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 3.5 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 0.7 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 4.0 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 2.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 0.4 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 0.4 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 0.6 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 1.7 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.1 | 3.7 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 1.5 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 3.3 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 0.7 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.5 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 1.0 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.7 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 0.7 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 2.2 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 3.5 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 2.1 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 1.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.5 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.4 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 1.2 | GO:0044215 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 1.7 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 3.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.5 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 2.8 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 3.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.6 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 2.6 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.7 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 1.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 1.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 1.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 5.1 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 2.6 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 1.4 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 2.3 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.1 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 2.4 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.4 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 1.9 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 16.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 1.7 | 6.7 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 1.4 | 4.1 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 1.0 | 3.1 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.9 | 2.7 | GO:0015056 | corticotrophin-releasing factor receptor activity(GO:0015056) corticotropin-releasing hormone receptor activity(GO:0043404) |
| 0.9 | 4.4 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.9 | 2.6 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.8 | 3.4 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.8 | 3.9 | GO:0055056 | dehydroascorbic acid transporter activity(GO:0033300) D-glucose transmembrane transporter activity(GO:0055056) |
| 0.7 | 2.2 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.6 | 2.5 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.5 | 1.6 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.5 | 4.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.5 | 4.4 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.5 | 5.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.5 | 2.3 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.4 | 1.3 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.4 | 4.0 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.3 | 2.7 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.3 | 1.4 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.3 | 3.4 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.3 | 3.3 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.3 | 1.5 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.2 | 2.9 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.2 | 4.6 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.2 | 6.1 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.2 | 1.7 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.2 | 2.8 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.6 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.1 | 1.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 5.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 1.0 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 0.3 | GO:0070039 | rRNA (guanosine-2'-O-)-methyltransferase activity(GO:0070039) |
| 0.1 | 4.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 2.7 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 4.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 1.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 1.1 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 6.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 3.0 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 1.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 1.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 1.7 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 3.2 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 2.2 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 2.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 2.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 3.3 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 3.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 3.1 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 0.8 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 1.6 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 2.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 1.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 1.1 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.6 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 1.7 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.1 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 5.0 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.1 | 3.2 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.9 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 1.0 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 2.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.8 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 6.3 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.3 | GO:0070404 | NADH binding(GO:0070404) |
| 0.0 | 3.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.5 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 1.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.7 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 2.5 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.7 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.7 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 1.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 1.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 2.5 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.2 | GO:0004114 | 3',5'-cyclic-nucleotide phosphodiesterase activity(GO:0004114) |
| 0.0 | 3.1 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.3 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.2 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 1.6 | GO:0051219 | phosphoprotein binding(GO:0051219) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 11.1 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 2.4 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 6.7 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.9 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 1.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 2.2 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 4.0 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 1.7 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 2.4 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.1 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.4 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 3.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.2 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.7 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.3 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.4 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.3 | 3.4 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.3 | 6.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 3.9 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.2 | 6.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 2.7 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 4.0 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 6.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 4.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 6.1 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 1.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 1.9 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 6.2 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 2.8 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 2.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 2.6 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.1 | 3.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 0.6 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 7.3 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 1.8 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 7.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.8 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.7 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.8 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.6 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF CYCLIN B | Genes involved in APC/C:Cdc20 mediated degradation of Cyclin B |
| 0.0 | 1.1 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 2.4 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |