Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
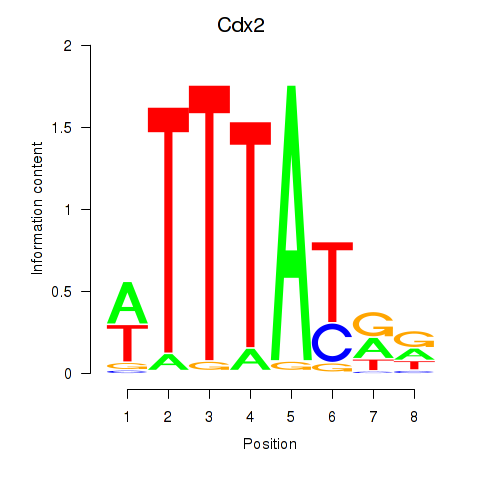
Results for Cdx2
Z-value: 1.04
Transcription factors associated with Cdx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Cdx2
|
ENSMUSG00000029646.4 | Cdx2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Cdx2 | mm39_v1_chr5_-_147244074_147244091 | 0.67 | 1.5e-10 | Click! |
Activity profile of Cdx2 motif
Sorted Z-values of Cdx2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Cdx2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_130633776 | 17.88 |
ENSMUST00000084509.7
ENSMUST00000213064.3 ENSMUST00000208311.4 |
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr17_-_31348576 | 15.86 |
ENSMUST00000024827.5
|
Tff3
|
trefoil factor 3, intestinal |
| chr6_+_71176811 | 13.76 |
ENSMUST00000067492.8
|
Fabp1
|
fatty acid binding protein 1, liver |
| chr11_-_99328969 | 10.91 |
ENSMUST00000017743.3
|
Krt20
|
keratin 20 |
| chr16_-_48232770 | 10.40 |
ENSMUST00000212197.2
|
Gm5485
|
predicted gene 5485 |
| chr3_+_57332735 | 9.27 |
ENSMUST00000029377.8
|
Tm4sf4
|
transmembrane 4 superfamily member 4 |
| chr3_-_14843512 | 8.36 |
ENSMUST00000094365.11
|
Car1
|
carbonic anhydrase 1 |
| chr3_+_3699205 | 8.26 |
ENSMUST00000108394.3
|
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr9_-_65330231 | 8.12 |
ENSMUST00000065894.7
|
Slc51b
|
solute carrier family 51, beta subunit |
| chr1_-_45542442 | 7.04 |
ENSMUST00000086430.5
|
Col5a2
|
collagen, type V, alpha 2 |
| chr3_-_10273628 | 6.62 |
ENSMUST00000029041.6
|
Fabp4
|
fatty acid binding protein 4, adipocyte |
| chr11_+_96085118 | 6.47 |
ENSMUST00000062709.4
|
Hoxb13
|
homeobox B13 |
| chrX_+_21581135 | 5.72 |
ENSMUST00000033414.8
|
Slc6a14
|
solute carrier family 6 (neurotransmitter transporter), member 14 |
| chrX_+_95498965 | 5.61 |
ENSMUST00000033553.14
|
Heph
|
hephaestin |
| chr10_+_97400990 | 5.56 |
ENSMUST00000038160.6
|
Lum
|
lumican |
| chr11_+_67128843 | 5.38 |
ENSMUST00000018632.11
|
Myh4
|
myosin, heavy polypeptide 4, skeletal muscle |
| chr11_+_94102255 | 5.08 |
ENSMUST00000041589.6
|
Tob1
|
transducer of ErbB-2.1 |
| chr6_+_17491231 | 4.88 |
ENSMUST00000080469.12
|
Met
|
met proto-oncogene |
| chr11_+_96214078 | 4.63 |
ENSMUST00000093944.10
|
Hoxb3
|
homeobox B3 |
| chr6_-_52190299 | 4.59 |
ENSMUST00000128102.4
|
Gm28308
|
predicted gene 28308 |
| chr2_+_74552322 | 4.15 |
ENSMUST00000047904.4
|
Hoxd4
|
homeobox D4 |
| chrX_+_138585728 | 4.01 |
ENSMUST00000096313.4
|
Tbc1d8b
|
TBC1 domain family, member 8B |
| chr7_+_140343652 | 3.91 |
ENSMUST00000026552.9
ENSMUST00000209253.2 ENSMUST00000210235.2 |
Cyp2e1
|
cytochrome P450, family 2, subfamily e, polypeptide 1 |
| chr8_-_34333573 | 3.65 |
ENSMUST00000183062.2
|
Rbpms
|
RNA binding protein gene with multiple splicing |
| chr8_-_5155347 | 3.64 |
ENSMUST00000023835.3
|
Slc10a2
|
solute carrier family 10, member 2 |
| chrX_+_162923474 | 3.56 |
ENSMUST00000073973.11
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr10_+_52267702 | 3.53 |
ENSMUST00000067085.7
|
Nepn
|
nephrocan |
| chr4_-_63965161 | 3.27 |
ENSMUST00000107377.10
|
Tnc
|
tenascin C |
| chr3_-_92393193 | 3.13 |
ENSMUST00000054599.8
|
Sprr1a
|
small proline-rich protein 1A |
| chr8_+_15107646 | 2.95 |
ENSMUST00000033842.4
|
Myom2
|
myomesin 2 |
| chr8_+_55003359 | 2.31 |
ENSMUST00000033918.4
|
Asb5
|
ankyrin repeat and SOCs box-containing 5 |
| chr5_-_135773047 | 2.30 |
ENSMUST00000153399.2
|
Tmem120a
|
transmembrane protein 120A |
| chr14_+_54923655 | 2.19 |
ENSMUST00000038539.8
ENSMUST00000228027.2 |
1700123O20Rik
|
RIKEN cDNA 1700123O20 gene |
| chr1_-_144124871 | 2.15 |
ENSMUST00000189061.7
|
Rgs1
|
regulator of G-protein signaling 1 |
| chr17_-_71305003 | 2.15 |
ENSMUST00000024846.13
ENSMUST00000232766.2 |
Myl12a
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr15_+_3300249 | 2.13 |
ENSMUST00000082424.12
ENSMUST00000159158.9 ENSMUST00000159216.10 ENSMUST00000160311.3 |
Selenop
|
selenoprotein P |
| chr14_+_80237691 | 2.08 |
ENSMUST00000228749.2
ENSMUST00000088735.4 |
Olfm4
|
olfactomedin 4 |
| chr4_-_9643636 | 2.02 |
ENSMUST00000108333.8
ENSMUST00000108334.8 ENSMUST00000108335.8 ENSMUST00000152526.8 ENSMUST00000103004.10 |
Asph
|
aspartate-beta-hydroxylase |
| chr3_+_81904229 | 1.92 |
ENSMUST00000029641.10
|
Asic5
|
acid-sensing (proton-gated) ion channel family member 5 |
| chrX_-_50294867 | 1.71 |
ENSMUST00000114876.9
|
Mbnl3
|
muscleblind like splicing factor 3 |
| chr1_+_139429430 | 1.66 |
ENSMUST00000027615.7
|
F13b
|
coagulation factor XIII, beta subunit |
| chrM_+_8603 | 1.64 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr16_-_43836681 | 1.60 |
ENSMUST00000036174.10
|
Gramd1c
|
GRAM domain containing 1C |
| chr15_-_101441254 | 1.53 |
ENSMUST00000023720.8
|
Krt84
|
keratin 84 |
| chr4_-_109059414 | 1.51 |
ENSMUST00000160774.8
ENSMUST00000194478.6 ENSMUST00000030288.14 ENSMUST00000162787.9 |
Osbpl9
|
oxysterol binding protein-like 9 |
| chr17_-_71153283 | 1.50 |
ENSMUST00000156484.2
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr11_-_99501015 | 1.48 |
ENSMUST00000076478.2
|
Gm11937
|
predicted gene 11937 |
| chr2_-_51039112 | 1.46 |
ENSMUST00000154545.2
ENSMUST00000017288.9 |
Rnd3
|
Rho family GTPase 3 |
| chr11_-_99996452 | 1.44 |
ENSMUST00000107416.3
|
Krt36
|
keratin 36 |
| chr8_+_95113066 | 1.36 |
ENSMUST00000161576.8
ENSMUST00000034220.8 |
Herpud1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr1_-_144124787 | 1.36 |
ENSMUST00000185714.2
|
Rgs1
|
regulator of G-protein signaling 1 |
| chr12_-_102844537 | 1.08 |
ENSMUST00000045652.8
ENSMUST00000223554.2 |
Btbd7
|
BTB (POZ) domain containing 7 |
| chr14_-_50425655 | 0.98 |
ENSMUST00000205837.3
|
Olfr730
|
olfactory receptor 730 |
| chr2_-_69542805 | 0.97 |
ENSMUST00000102706.4
ENSMUST00000073152.13 |
Fastkd1
|
FAST kinase domains 1 |
| chr15_-_101482320 | 0.96 |
ENSMUST00000042957.6
|
Krt75
|
keratin 75 |
| chr3_+_93052089 | 0.94 |
ENSMUST00000107300.7
ENSMUST00000195515.2 |
Crnn
|
cornulin |
| chr13_-_24118139 | 0.90 |
ENSMUST00000052776.4
|
H2bc1
|
H2B clustered histone 1 |
| chr10_-_33920289 | 0.87 |
ENSMUST00000218239.2
|
Calhm4
|
calcium homeostasis modulator family member 4 |
| chr9_+_95519654 | 0.86 |
ENSMUST00000015498.9
|
Pcolce2
|
procollagen C-endopeptidase enhancer 2 |
| chr2_+_153742294 | 0.86 |
ENSMUST00000088955.12
ENSMUST00000135501.3 |
Bpifb6
|
BPI fold containing family B, member 6 |
| chrX_+_162873183 | 0.83 |
ENSMUST00000015545.10
|
Cltrn
|
collectrin, amino acid transport regulator |
| chr8_-_106660470 | 0.81 |
ENSMUST00000034368.8
|
Ctrl
|
chymotrypsin-like |
| chr2_+_142920386 | 0.74 |
ENSMUST00000028902.3
|
Otor
|
otoraplin |
| chr18_-_79152504 | 0.74 |
ENSMUST00000025430.11
|
Setbp1
|
SET binding protein 1 |
| chr1_-_37996838 | 0.67 |
ENSMUST00000027254.10
ENSMUST00000114894.2 |
Lyg1
|
lysozyme G-like 1 |
| chr16_-_88609108 | 0.59 |
ENSMUST00000232664.2
|
Gm20741
|
predicted gene, 20741 |
| chr11_-_99869879 | 0.59 |
ENSMUST00000105051.2
|
Krtap29-1
|
keratin associated protein 29-1 |
| chr3_+_126391046 | 0.50 |
ENSMUST00000106401.8
|
Camk2d
|
calcium/calmodulin-dependent protein kinase II, delta |
| chr10_-_33920300 | 0.48 |
ENSMUST00000048052.7
|
Calhm4
|
calcium homeostasis modulator family member 4 |
| chr14_-_4506874 | 0.45 |
ENSMUST00000224934.2
|
Thrb
|
thyroid hormone receptor beta |
| chr3_+_126390951 | 0.45 |
ENSMUST00000171289.8
|
Camk2d
|
calcium/calmodulin-dependent protein kinase II, delta |
| chr11_-_99932373 | 0.43 |
ENSMUST00000056362.3
|
Krt34
|
keratin 34 |
| chr1_-_186437760 | 0.39 |
ENSMUST00000195201.2
|
Tgfb2
|
transforming growth factor, beta 2 |
| chr15_-_50746202 | 0.38 |
ENSMUST00000184885.8
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chrM_+_7758 | 0.37 |
ENSMUST00000082407.1
|
mt-Atp8
|
mitochondrially encoded ATP synthase 8 |
| chr19_-_34856853 | 0.37 |
ENSMUST00000036584.13
|
Pank1
|
pantothenate kinase 1 |
| chr14_-_63221950 | 0.37 |
ENSMUST00000100493.3
|
Defb48
|
defensin beta 48 |
| chr19_+_8828132 | 0.33 |
ENSMUST00000235683.2
ENSMUST00000096257.3 |
Lrrn4cl
|
LRRN4 C-terminal like |
| chrM_+_7779 | 0.31 |
ENSMUST00000082408.1
|
mt-Atp6
|
mitochondrially encoded ATP synthase 6 |
| chr19_-_47680528 | 0.30 |
ENSMUST00000026045.14
ENSMUST00000086923.6 |
Col17a1
|
collagen, type XVII, alpha 1 |
| chr14_-_54923517 | 0.29 |
ENSMUST00000125265.2
|
Acin1
|
apoptotic chromatin condensation inducer 1 |
| chr13_+_23765546 | 0.29 |
ENSMUST00000102968.3
|
H4c4
|
H4 clustered histone 4 |
| chr17_-_65143066 | 0.27 |
ENSMUST00000024896.4
|
4930583I09Rik
|
RIKEN cDNA 4930583I09 gene |
| chr11_-_99884818 | 0.27 |
ENSMUST00000105049.2
|
Krtap17-1
|
keratin associated protein 17-1 |
| chr14_+_64229914 | 0.27 |
ENSMUST00000058229.6
|
Rp1l1
|
retinitis pigmentosa 1 homolog like 1 |
| chr2_-_25982160 | 0.26 |
ENSMUST00000114159.9
|
Nacc2
|
nucleus accumbens associated 2, BEN and BTB (POZ) domain containing |
| chr13_-_22219738 | 0.25 |
ENSMUST00000091742.6
|
H2ac12
|
H2A clustered histone 12 |
| chr10_+_118040729 | 0.25 |
ENSMUST00000096691.5
|
Il22
|
interleukin 22 |
| chr14_+_54453748 | 0.19 |
ENSMUST00000103736.2
|
Traj4
|
T cell receptor alpha joining 4 |
| chr6_+_42263644 | 0.15 |
ENSMUST00000163936.8
|
Clcn1
|
chloride channel, voltage-sensitive 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.0 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 1.6 | 6.5 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.9 | 3.6 | GO:0015801 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 0.9 | 13.8 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.8 | 3.3 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.8 | 4.6 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.7 | 4.9 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.7 | 5.6 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.7 | 17.9 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.7 | 2.0 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.6 | 8.1 | GO:0070863 | positive regulation of protein glycosylation(GO:0060050) positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.4 | 5.1 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) |
| 0.3 | 6.6 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.3 | 10.9 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.3 | 1.4 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.3 | 2.1 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.2 | 2.9 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.2 | 1.0 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.1 | 8.4 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 3.6 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.1 | 5.4 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 5.1 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 3.9 | GO:0017144 | drug metabolic process(GO:0017144) |
| 0.1 | 3.6 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 15.9 | GO:0007586 | digestion(GO:0007586) |
| 0.1 | 0.3 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 0.1 | 0.7 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.1 | 3.1 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 1.5 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 1.1 | GO:0060693 | regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.1 | 0.5 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.1 | 2.9 | GO:0045616 | regulation of keratinocyte differentiation(GO:0045616) |
| 0.0 | 1.7 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 4.0 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 3.5 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) |
| 0.0 | 0.3 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 2.1 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 8.3 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.0 | 0.7 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 5.7 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 4.1 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 1.6 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.4 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.7 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.4 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 1.9 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 1.0 | GO:0045333 | cellular respiration(GO:0045333) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 7.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 1.4 | 17.9 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 1.3 | 13.8 | GO:0045179 | apical cortex(GO:0045179) |
| 0.6 | 5.6 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.3 | 1.4 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.2 | 2.9 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.2 | 2.1 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.2 | 6.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 5.4 | GO:0032982 | myosin filament(GO:0032982) |
| 0.2 | 2.0 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.2 | 5.1 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 3.6 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 2.1 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 3.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 6.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 3.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 4.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 5.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.7 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 3.6 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.3 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 13.6 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 1.0 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 15.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 5.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 3.9 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 3.5 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.3 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.0 | 17.9 | GO:0035375 | zymogen binding(GO:0035375) |
| 1.5 | 13.8 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 1.2 | 4.9 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 1.1 | 5.7 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 1.0 | 6.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.8 | 8.4 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.7 | 3.6 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.6 | 3.6 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.5 | 3.6 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.4 | 5.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.3 | 3.3 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.2 | 1.9 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.1 | 7.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 2.9 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 2.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 3.9 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 8.7 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 1.0 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 0.4 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 6.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 1.5 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 1.6 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 3.4 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 5.4 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 5.1 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.7 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 4.6 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 2.0 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 2.1 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.0 | 3.8 | GO:0017137 | Rab GTPase binding(GO:0017137) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 12.2 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 15.9 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 5.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 6.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 3.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 6.6 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 12.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 3.5 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 1.0 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 1.5 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.4 | 5.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.3 | 3.6 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 5.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.2 | 8.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 6.6 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 5.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 8.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 4.9 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.1 | 1.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 13.8 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 1.4 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 3.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.5 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 3.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.0 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |