Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Cebpe
Z-value: 2.99
Transcription factors associated with Cebpe
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Cebpe
|
ENSMUSG00000052435.8 | Cebpe |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Cebpe | mm39_v1_chr14_-_54949596_54949631 | 0.32 | 6.4e-03 | Click! |
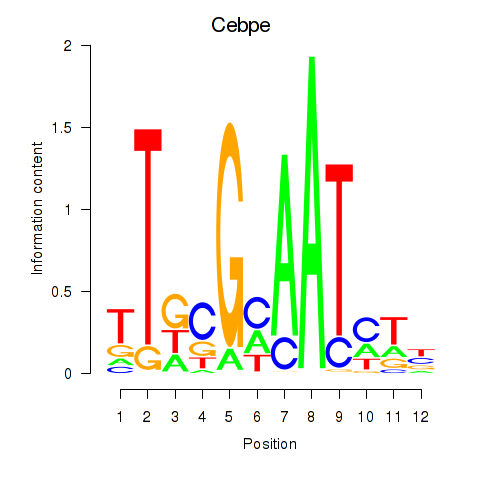
Activity profile of Cebpe motif
Sorted Z-values of Cebpe motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Cebpe
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_87288177 | 50.25 |
ENSMUST00000067790.7
|
Ugt2b5
|
UDP glucuronosyltransferase 2 family, polypeptide B5 |
| chr5_-_87240405 | 45.50 |
ENSMUST00000132667.2
ENSMUST00000145617.8 ENSMUST00000094649.11 |
Ugt2b36
|
UDP glucuronosyltransferase 2 family, polypeptide B36 |
| chr14_+_40827317 | 40.52 |
ENSMUST00000047286.7
|
Mat1a
|
methionine adenosyltransferase I, alpha |
| chr3_+_59939175 | 35.42 |
ENSMUST00000029325.5
|
Aadac
|
arylacetamide deacetylase |
| chr14_+_40826970 | 35.06 |
ENSMUST00000225720.2
|
Mat1a
|
methionine adenosyltransferase I, alpha |
| chr5_-_87485023 | 34.92 |
ENSMUST00000031195.3
|
Ugt2a3
|
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr14_+_40827108 | 34.07 |
ENSMUST00000224514.2
|
Mat1a
|
methionine adenosyltransferase I, alpha |
| chr12_+_8062331 | 30.30 |
ENSMUST00000171239.2
|
Apob
|
apolipoprotein B |
| chr6_-_128503666 | 29.10 |
ENSMUST00000143664.2
ENSMUST00000112132.8 |
Pzp
|
PZP, alpha-2-macroglobulin like |
| chr19_-_39729431 | 27.22 |
ENSMUST00000099472.4
|
Cyp2c68
|
cytochrome P450, family 2, subfamily c, polypeptide 68 |
| chr19_+_39980868 | 27.19 |
ENSMUST00000049178.3
|
Cyp2c37
|
cytochrome P450, family 2. subfamily c, polypeptide 37 |
| chr13_+_4486105 | 26.23 |
ENSMUST00000156277.2
|
Akr1c6
|
aldo-keto reductase family 1, member C6 |
| chr10_+_128089965 | 24.67 |
ENSMUST00000060782.5
ENSMUST00000218722.2 |
Apon
|
apolipoprotein N |
| chr5_-_87572060 | 23.71 |
ENSMUST00000072818.6
|
Ugt2b38
|
UDP glucuronosyltransferase 2 family, polypeptide B38 |
| chr1_-_139708906 | 23.55 |
ENSMUST00000111986.8
ENSMUST00000027612.11 ENSMUST00000111989.9 |
Cfhr4
|
complement factor H-related 4 |
| chr19_-_39637489 | 22.13 |
ENSMUST00000067328.7
|
Cyp2c67
|
cytochrome P450, family 2, subfamily c, polypeptide 67 |
| chr15_+_9335636 | 21.86 |
ENSMUST00000072403.7
|
Ugt3a2
|
UDP glycosyltransferases 3 family, polypeptide A2 |
| chr4_-_60538151 | 21.81 |
ENSMUST00000098047.3
|
Mup10
|
major urinary protein 10 |
| chr4_-_60455331 | 21.49 |
ENSMUST00000135953.2
|
Mup1
|
major urinary protein 1 |
| chr9_-_44714263 | 21.11 |
ENSMUST00000044694.8
|
Ttc36
|
tetratricopeptide repeat domain 36 |
| chr4_-_60377932 | 20.92 |
ENSMUST00000107506.9
ENSMUST00000122381.8 ENSMUST00000118759.8 ENSMUST00000132829.3 |
Mup9
|
major urinary protein 9 |
| chr4_-_60618357 | 20.71 |
ENSMUST00000084544.5
ENSMUST00000098046.10 |
Mup11
|
major urinary protein 11 |
| chr16_+_22710785 | 20.68 |
ENSMUST00000023583.7
ENSMUST00000232098.2 |
Ahsg
|
alpha-2-HS-glycoprotein |
| chr4_-_61592331 | 20.01 |
ENSMUST00000098040.4
|
Mup18
|
major urinary protein 18 |
| chr16_+_22710027 | 20.01 |
ENSMUST00000231848.2
|
Ahsg
|
alpha-2-HS-glycoprotein |
| chr10_+_21253190 | 19.87 |
ENSMUST00000042699.14
|
Aldh8a1
|
aldehyde dehydrogenase 8 family, member A1 |
| chr12_+_104304631 | 19.71 |
ENSMUST00000043058.5
ENSMUST00000101078.12 |
Serpina3k
Serpina3m
|
serine (or cysteine) peptidase inhibitor, clade A, member 3K serine (or cysteine) peptidase inhibitor, clade A, member 3M |
| chr1_-_172722589 | 19.35 |
ENSMUST00000027824.7
|
Apcs
|
serum amyloid P-component |
| chr14_+_75479727 | 19.29 |
ENSMUST00000022576.10
|
Cpb2
|
carboxypeptidase B2 (plasma) |
| chr2_+_43445333 | 18.83 |
ENSMUST00000028223.9
ENSMUST00000112826.8 |
Kynu
|
kynureninase |
| chr15_-_96947963 | 18.53 |
ENSMUST00000230907.2
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr9_-_71075939 | 18.29 |
ENSMUST00000113570.8
|
Aqp9
|
aquaporin 9 |
| chr16_+_37400500 | 17.72 |
ENSMUST00000160847.2
|
Hgd
|
homogentisate 1, 2-dioxygenase |
| chr12_+_8027640 | 17.36 |
ENSMUST00000171271.8
ENSMUST00000037811.13 |
Apob
|
apolipoprotein B |
| chr4_-_107975701 | 17.16 |
ENSMUST00000149106.8
|
Scp2
|
sterol carrier protein 2, liver |
| chr16_+_37400590 | 17.15 |
ENSMUST00000159787.8
|
Hgd
|
homogentisate 1, 2-dioxygenase |
| chr8_-_41668182 | 17.05 |
ENSMUST00000034003.5
|
Fgl1
|
fibrinogen-like protein 1 |
| chr5_-_87074380 | 17.05 |
ENSMUST00000031183.3
|
Ugt2b1
|
UDP glucuronosyltransferase 2 family, polypeptide B1 |
| chr5_-_145946408 | 17.01 |
ENSMUST00000138870.2
ENSMUST00000068317.13 |
Cyp3a25
|
cytochrome P450, family 3, subfamily a, polypeptide 25 |
| chr4_-_60139857 | 16.92 |
ENSMUST00000107490.5
ENSMUST00000074700.9 |
Mup2
|
major urinary protein 2 |
| chr7_+_13467422 | 16.63 |
ENSMUST00000086148.8
|
Sult2a2
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 2 |
| chr19_+_39275518 | 16.55 |
ENSMUST00000003137.15
|
Cyp2c29
|
cytochrome P450, family 2, subfamily c, polypeptide 29 |
| chr1_-_184543367 | 16.54 |
ENSMUST00000048462.13
ENSMUST00000110992.9 |
Mtarc1
|
mitochondrial amidoxime reducing component 1 |
| chr1_+_87998487 | 16.16 |
ENSMUST00000073772.5
|
Ugt1a9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr2_-_69172944 | 15.82 |
ENSMUST00000102709.8
ENSMUST00000102710.10 ENSMUST00000180142.2 |
Abcb11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
| chr19_-_39451509 | 15.72 |
ENSMUST00000035488.3
|
Cyp2c38
|
cytochrome P450, family 2, subfamily c, polypeptide 38 |
| chr16_+_22769822 | 15.67 |
ENSMUST00000023590.9
|
Hrg
|
histidine-rich glycoprotein |
| chr2_+_172994841 | 15.61 |
ENSMUST00000029017.6
|
Pck1
|
phosphoenolpyruvate carboxykinase 1, cytosolic |
| chr10_-_24803336 | 15.60 |
ENSMUST00000020161.10
|
Arg1
|
arginase, liver |
| chr7_-_13571334 | 15.22 |
ENSMUST00000108522.5
|
Sult2a1
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 1 |
| chr5_+_8010445 | 15.17 |
ENSMUST00000115421.3
|
Steap4
|
STEAP family member 4 |
| chr19_+_12577317 | 14.99 |
ENSMUST00000181868.8
ENSMUST00000092931.7 |
Gm4952
|
predicted gene 4952 |
| chr11_+_108286114 | 14.90 |
ENSMUST00000000049.6
|
Apoh
|
apolipoprotein H |
| chr11_-_77784922 | 14.87 |
ENSMUST00000017597.5
|
Pipox
|
pipecolic acid oxidase |
| chr19_-_40175709 | 14.82 |
ENSMUST00000051846.13
|
Cyp2c70
|
cytochrome P450, family 2, subfamily c, polypeptide 70 |
| chr4_-_61259801 | 14.75 |
ENSMUST00000125461.8
|
Mup14
|
major urinary protein 14 |
| chr4_-_60222580 | 14.59 |
ENSMUST00000095058.5
ENSMUST00000163931.8 |
Mup8
|
major urinary protein 8 |
| chr19_-_7779943 | 14.52 |
ENSMUST00000120522.8
|
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr6_-_55152002 | 14.44 |
ENSMUST00000003569.6
|
Inmt
|
indolethylamine N-methyltransferase |
| chr4_-_61259997 | 14.43 |
ENSMUST00000071005.9
ENSMUST00000075206.12 |
Mup14
|
major urinary protein 14 |
| chr19_-_39801188 | 14.31 |
ENSMUST00000162507.2
ENSMUST00000160476.9 ENSMUST00000239028.2 |
Cyp2c40
|
cytochrome P450, family 2, subfamily c, polypeptide 40 |
| chr16_+_22769844 | 13.79 |
ENSMUST00000232422.2
|
Hrg
|
histidine-rich glycoprotein |
| chr2_+_43445359 | 13.73 |
ENSMUST00000050511.7
|
Kynu
|
kynureninase |
| chr3_-_98537568 | 13.63 |
ENSMUST00000044094.6
|
Hsd3b5
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 5 |
| chr16_+_22877000 | 13.62 |
ENSMUST00000039492.14
ENSMUST00000023589.15 ENSMUST00000089902.8 |
Kng1
|
kininogen 1 |
| chr15_+_6474808 | 13.56 |
ENSMUST00000022749.17
ENSMUST00000239466.2 |
C9
|
complement component 9 |
| chr2_-_10135449 | 13.55 |
ENSMUST00000042290.14
|
Itih2
|
inter-alpha trypsin inhibitor, heavy chain 2 |
| chr7_+_25872836 | 13.33 |
ENSMUST00000082214.5
|
Cyp2b9
|
cytochrome P450, family 2, subfamily b, polypeptide 9 |
| chr3_+_20039775 | 13.13 |
ENSMUST00000172860.2
|
Cp
|
ceruloplasmin |
| chr15_+_9279915 | 12.63 |
ENSMUST00000022861.9
|
Ugt3a1
|
UDP glycosyltransferases 3 family, polypeptide A1 |
| chr1_+_172525613 | 12.54 |
ENSMUST00000038495.5
|
Crp
|
C-reactive protein, pentraxin-related |
| chr4_-_60457902 | 12.54 |
ENSMUST00000084548.11
ENSMUST00000103012.10 ENSMUST00000107499.4 |
Mup1
|
major urinary protein 1 |
| chr5_-_87054796 | 12.48 |
ENSMUST00000031181.16
ENSMUST00000113333.2 |
Ugt2b34
|
UDP glucuronosyltransferase 2 family, polypeptide B34 |
| chr12_-_81014849 | 12.32 |
ENSMUST00000095572.5
|
Slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 1 |
| chr11_-_59937302 | 12.27 |
ENSMUST00000000310.14
ENSMUST00000102693.9 ENSMUST00000148512.2 |
Pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr1_-_139786421 | 12.05 |
ENSMUST00000194186.6
ENSMUST00000094489.5 ENSMUST00000239380.2 |
Cfhr2
|
complement factor H-related 2 |
| chr4_-_107975723 | 11.88 |
ENSMUST00000030340.15
|
Scp2
|
sterol carrier protein 2, liver |
| chr5_-_104125270 | 11.81 |
ENSMUST00000112803.3
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr13_-_24098981 | 11.80 |
ENSMUST00000110407.4
|
Slc17a4
|
solute carrier family 17 (sodium phosphate), member 4 |
| chr1_+_21310821 | 11.67 |
ENSMUST00000121676.8
ENSMUST00000124990.3 |
Gsta3
|
glutathione S-transferase, alpha 3 |
| chr1_+_88093726 | 11.64 |
ENSMUST00000097659.5
|
Ugt1a5
|
UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr4_-_96552349 | 11.61 |
ENSMUST00000030299.8
|
Cyp2j5
|
cytochrome P450, family 2, subfamily j, polypeptide 5 |
| chr1_+_21310803 | 11.58 |
ENSMUST00000027067.15
|
Gsta3
|
glutathione S-transferase, alpha 3 |
| chr5_-_104125192 | 11.42 |
ENSMUST00000120320.8
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr11_-_109986763 | 11.23 |
ENSMUST00000046223.14
ENSMUST00000106664.10 ENSMUST00000106662.2 |
Abca8a
|
ATP-binding cassette, sub-family A (ABC1), member 8a |
| chr6_-_141892686 | 11.19 |
ENSMUST00000042119.6
|
Slco1a1
|
solute carrier organic anion transporter family, member 1a1 |
| chr7_+_51530060 | 11.14 |
ENSMUST00000145049.2
|
Gas2
|
growth arrest specific 2 |
| chr19_-_39875192 | 10.85 |
ENSMUST00000168838.3
|
Cyp2c69
|
cytochrome P450, family 2, subfamily c, polypeptide 69 |
| chr13_-_24098951 | 10.83 |
ENSMUST00000021769.16
|
Slc17a4
|
solute carrier family 17 (sodium phosphate), member 4 |
| chr5_-_87402659 | 10.78 |
ENSMUST00000075858.4
|
Ugt2b37
|
UDP glucuronosyltransferase 2 family, polypeptide B37 |
| chr7_+_119125443 | 10.74 |
ENSMUST00000207440.2
|
Acsm5
|
acyl-CoA synthetase medium-chain family member 5 |
| chr7_+_119125426 | 10.70 |
ENSMUST00000066465.3
|
Acsm5
|
acyl-CoA synthetase medium-chain family member 5 |
| chr1_+_139429430 | 10.66 |
ENSMUST00000027615.7
|
F13b
|
coagulation factor XIII, beta subunit |
| chr17_-_35081129 | 10.65 |
ENSMUST00000154526.8
|
Cfb
|
complement factor B |
| chr19_-_7943365 | 10.59 |
ENSMUST00000182102.8
ENSMUST00000075619.5 |
Slc22a27
|
solute carrier family 22, member 27 |
| chr5_+_90708962 | 10.58 |
ENSMUST00000094615.8
ENSMUST00000200765.2 |
Albfm1
|
albumin superfamily member 1 |
| chr12_-_81014755 | 10.54 |
ENSMUST00000218342.2
|
Slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 1 |
| chr17_-_35081456 | 10.52 |
ENSMUST00000025229.11
ENSMUST00000176203.9 ENSMUST00000128767.8 |
Cfb
|
complement factor B |
| chr10_+_23727325 | 10.51 |
ENSMUST00000020190.8
|
Vnn3
|
vanin 3 |
| chr17_-_57535003 | 10.46 |
ENSMUST00000177046.2
ENSMUST00000024988.15 |
C3
|
complement component 3 |
| chr3_-_137945419 | 10.18 |
ENSMUST00000199804.3
ENSMUST00000185122.8 ENSMUST00000183783.8 |
4930579F01Rik
0610031O16Rik
|
RIKEN cDNA 4930579F01 gene RIKEN cDNA 0610031O16 gene |
| chr10_-_89342493 | 10.17 |
ENSMUST00000058126.15
ENSMUST00000105296.9 |
Nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr6_-_141892517 | 10.03 |
ENSMUST00000168119.8
|
Slco1a1
|
solute carrier organic anion transporter family, member 1a1 |
| chr2_-_60114684 | 9.99 |
ENSMUST00000028356.9
ENSMUST00000074606.11 |
Cd302
|
CD302 antigen |
| chr4_+_141473983 | 9.96 |
ENSMUST00000038161.5
|
Agmat
|
agmatine ureohydrolase (agmatinase) |
| chr5_-_104125226 | 9.94 |
ENSMUST00000048118.15
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr4_-_62069046 | 9.89 |
ENSMUST00000077719.4
|
Mup21
|
major urinary protein 21 |
| chr5_+_87148697 | 9.87 |
ENSMUST00000031186.9
|
Ugt2b35
|
UDP glucuronosyltransferase 2 family, polypeptide B35 |
| chr16_-_30086317 | 9.79 |
ENSMUST00000064856.9
|
Cpn2
|
carboxypeptidase N, polypeptide 2 |
| chr3_-_67422821 | 9.55 |
ENSMUST00000054825.5
|
Rarres1
|
retinoic acid receptor responder (tazarotene induced) 1 |
| chr7_+_119125546 | 9.48 |
ENSMUST00000207387.2
ENSMUST00000207813.2 |
Acsm5
|
acyl-CoA synthetase medium-chain family member 5 |
| chr10_+_87694117 | 9.41 |
ENSMUST00000122386.8
|
Igf1
|
insulin-like growth factor 1 |
| chr7_+_13357892 | 9.21 |
ENSMUST00000108525.4
|
Sult2a5
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 5 |
| chr15_+_4756684 | 9.18 |
ENSMUST00000161997.8
ENSMUST00000022788.15 |
C6
|
complement component 6 |
| chr1_+_171041583 | 9.07 |
ENSMUST00000111328.8
|
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr19_-_8382424 | 8.82 |
ENSMUST00000064507.12
ENSMUST00000120540.2 ENSMUST00000096269.11 |
Slc22a30
|
solute carrier family 22, member 30 |
| chr4_+_115420817 | 8.79 |
ENSMUST00000141033.8
ENSMUST00000030486.15 |
Cyp4a31
|
cytochrome P450, family 4, subfamily a, polypeptide 31 |
| chr6_-_115569504 | 8.76 |
ENSMUST00000112957.2
|
Mkrn2os
|
makorin, ring finger protein 2, opposite strand |
| chr7_-_26638802 | 8.63 |
ENSMUST00000170227.3
|
Cyp2a22
|
cytochrome P450, family 2, subfamily a, polypeptide 22 |
| chr15_+_4756657 | 8.62 |
ENSMUST00000162585.8
|
C6
|
complement component 6 |
| chr17_+_80434874 | 8.53 |
ENSMUST00000039205.11
|
Galm
|
galactose mutarotase |
| chr5_+_90666791 | 8.51 |
ENSMUST00000113179.9
ENSMUST00000128740.2 |
Afm
|
afamin |
| chr7_+_119773070 | 8.50 |
ENSMUST00000033201.7
|
Anks4b
|
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr19_-_8109346 | 8.47 |
ENSMUST00000065651.5
|
Slc22a28
|
solute carrier family 22, member 28 |
| chr8_+_35842872 | 8.46 |
ENSMUST00000210337.2
ENSMUST00000070481.8 ENSMUST00000211648.2 |
Ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr11_-_109986804 | 8.39 |
ENSMUST00000100287.9
|
Abca8a
|
ATP-binding cassette, sub-family A (ABC1), member 8a |
| chr7_-_126873219 | 8.34 |
ENSMUST00000082428.6
|
Sephs2
|
selenophosphate synthetase 2 |
| chr1_-_119576347 | 8.28 |
ENSMUST00000027632.14
ENSMUST00000187194.2 |
Epb41l5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr16_-_22848153 | 8.23 |
ENSMUST00000232459.2
|
Kng2
|
kininogen 2 |
| chr7_+_46401214 | 8.09 |
ENSMUST00000210769.2
ENSMUST00000210272.2 ENSMUST00000075982.4 |
Saa2
|
serum amyloid A 2 |
| chr3_+_20011201 | 8.04 |
ENSMUST00000091309.12
ENSMUST00000108329.8 ENSMUST00000003714.13 |
Cp
|
ceruloplasmin |
| chr8_-_40095661 | 8.00 |
ENSMUST00000026021.14
|
Msr1
|
macrophage scavenger receptor 1 |
| chr6_+_17463748 | 7.95 |
ENSMUST00000115443.8
|
Met
|
met proto-oncogene |
| chr1_-_162812087 | 7.94 |
ENSMUST00000028010.9
|
Fmo3
|
flavin containing monooxygenase 3 |
| chr16_+_22738987 | 7.84 |
ENSMUST00000023587.12
|
Fetub
|
fetuin beta |
| chr11_+_75358866 | 7.81 |
ENSMUST00000043598.14
ENSMUST00000108435.2 |
Tlcd2
|
TLC domain containing 2 |
| chr8_-_72966663 | 7.78 |
ENSMUST00000098630.5
|
Cib3
|
calcium and integrin binding family member 3 |
| chr6_-_141801897 | 7.78 |
ENSMUST00000165990.8
|
Slco1a4
|
solute carrier organic anion transporter family, member 1a4 |
| chr1_+_13738967 | 7.74 |
ENSMUST00000088542.4
|
Xkr9
|
X-linked Kx blood group related 9 |
| chr7_-_25176959 | 7.73 |
ENSMUST00000098668.3
ENSMUST00000206687.2 ENSMUST00000206676.2 ENSMUST00000205308.2 ENSMUST00000098669.8 ENSMUST00000206171.2 ENSMUST00000098666.9 |
Ceacam1
|
carcinoembryonic antigen-related cell adhesion molecule 1 |
| chr6_+_40619913 | 7.64 |
ENSMUST00000238599.2
|
Mgam
|
maltase-glucoamylase |
| chr15_+_54274151 | 7.59 |
ENSMUST00000036737.4
|
Colec10
|
collectin sub-family member 10 |
| chr14_-_34032311 | 7.54 |
ENSMUST00000111917.3
ENSMUST00000228704.2 |
Shld2
|
shieldin complex subunit 2 |
| chr3_+_20011251 | 7.53 |
ENSMUST00000108328.8
|
Cp
|
ceruloplasmin |
| chr19_-_20704896 | 7.47 |
ENSMUST00000025656.4
|
Aldh1a7
|
aldehyde dehydrogenase family 1, subfamily A7 |
| chr9_+_103940575 | 7.45 |
ENSMUST00000120854.8
|
Acad11
|
acyl-Coenzyme A dehydrogenase family, member 11 |
| chr1_+_171041539 | 7.43 |
ENSMUST00000005820.11
ENSMUST00000075469.12 ENSMUST00000155126.8 |
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr11_-_89893707 | 7.41 |
ENSMUST00000020864.9
|
Pctp
|
phosphatidylcholine transfer protein |
| chr9_+_119170360 | 7.37 |
ENSMUST00000039784.12
|
Acaa1a
|
acetyl-Coenzyme A acyltransferase 1A |
| chr12_-_28673311 | 7.34 |
ENSMUST00000036136.9
|
Colec11
|
collectin sub-family member 11 |
| chr14_+_51181956 | 7.24 |
ENSMUST00000178092.2
ENSMUST00000227052.2 |
Pnp
Gm49342
|
purine-nucleoside phosphorylase predicted gene, 49342 |
| chr6_-_141801918 | 7.24 |
ENSMUST00000163678.2
|
Slco1a4
|
solute carrier organic anion transporter family, member 1a4 |
| chr6_+_90442269 | 7.13 |
ENSMUST00000113530.4
|
Klf15
|
Kruppel-like factor 15 |
| chr15_+_7120089 | 7.02 |
ENSMUST00000228723.2
|
Lifr
|
LIF receptor alpha |
| chr9_+_103940879 | 7.01 |
ENSMUST00000047799.13
ENSMUST00000189998.3 |
Acad11
|
acyl-Coenzyme A dehydrogenase family, member 11 |
| chr17_-_59320257 | 6.95 |
ENSMUST00000174122.2
ENSMUST00000025065.12 |
Nudt12
|
nudix (nucleoside diphosphate linked moiety X)-type motif 12 |
| chr5_-_108823435 | 6.94 |
ENSMUST00000051757.14
|
Slc26a1
|
solute carrier family 26 (sulfate transporter), member 1 |
| chr2_-_32314017 | 6.93 |
ENSMUST00000113307.9
|
Slc25a25
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 |
| chr9_+_108216466 | 6.92 |
ENSMUST00000193987.2
|
Gpx1
|
glutathione peroxidase 1 |
| chr7_+_25760922 | 6.84 |
ENSMUST00000005669.9
|
Cyp2b13
|
cytochrome P450, family 2, subfamily b, polypeptide 13 |
| chr16_-_38253507 | 6.82 |
ENSMUST00000002926.8
|
Pla1a
|
phospholipase A1 member A |
| chr8_-_40095706 | 6.77 |
ENSMUST00000210525.2
ENSMUST00000170091.3 |
Msr1
|
macrophage scavenger receptor 1 |
| chr7_-_46365108 | 6.74 |
ENSMUST00000006956.9
ENSMUST00000210913.2 |
Saa3
|
serum amyloid A 3 |
| chr3_+_20011405 | 6.73 |
ENSMUST00000108325.9
|
Cp
|
ceruloplasmin |
| chr9_+_119170486 | 6.72 |
ENSMUST00000175743.8
ENSMUST00000176397.8 |
Acaa1a
|
acetyl-Coenzyme A acyltransferase 1A |
| chr3_-_82811269 | 6.65 |
ENSMUST00000029632.7
|
Lrat
|
lecithin-retinol acyltransferase (phosphatidylcholine-retinol-O-acyltransferase) |
| chr4_-_150087587 | 6.64 |
ENSMUST00000084117.13
|
H6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr12_-_28673259 | 6.64 |
ENSMUST00000220836.2
|
Colec11
|
collectin sub-family member 11 |
| chr11_-_78875689 | 6.57 |
ENSMUST00000108269.10
ENSMUST00000108268.10 |
Lgals9
|
lectin, galactose binding, soluble 9 |
| chrX_+_100419965 | 6.56 |
ENSMUST00000119080.8
|
Gjb1
|
gap junction protein, beta 1 |
| chr17_-_35100980 | 6.55 |
ENSMUST00000152417.8
ENSMUST00000146299.8 |
C2
Gm20547
|
complement component 2 (within H-2S) predicted gene 20547 |
| chr7_-_132178101 | 6.54 |
ENSMUST00000084500.8
|
Oat
|
ornithine aminotransferase |
| chr9_+_108216433 | 6.53 |
ENSMUST00000191997.2
|
Gpx1
|
glutathione peroxidase 1 |
| chr2_+_121978156 | 6.44 |
ENSMUST00000102476.5
|
B2m
|
beta-2 microglobulin |
| chr17_-_13159204 | 6.41 |
ENSMUST00000043923.12
|
Acat3
|
acetyl-Coenzyme A acetyltransferase 3 |
| chr18_+_84869456 | 6.29 |
ENSMUST00000160180.9
|
Cyb5a
|
cytochrome b5 type A (microsomal) |
| chr4_+_115420876 | 6.25 |
ENSMUST00000126645.8
ENSMUST00000030480.4 |
Cyp4a31
|
cytochrome P450, family 4, subfamily a, polypeptide 31 |
| chr9_+_108216233 | 6.02 |
ENSMUST00000082429.8
|
Gpx1
|
glutathione peroxidase 1 |
| chr6_+_121709891 | 6.00 |
ENSMUST00000204124.2
|
Gm7298
|
predicted gene 7298 |
| chr4_-_129472328 | 5.88 |
ENSMUST00000052835.9
|
Fam167b
|
family with sequence similarity 167, member B |
| chr1_+_165596961 | 5.83 |
ENSMUST00000040298.5
|
Creg1
|
cellular repressor of E1A-stimulated genes 1 |
| chr10_-_24712034 | 5.76 |
ENSMUST00000218044.2
ENSMUST00000020169.9 |
Enpp3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr3_-_129126362 | 5.64 |
ENSMUST00000029658.14
|
Enpep
|
glutamyl aminopeptidase |
| chr4_+_115268821 | 5.64 |
ENSMUST00000094887.4
|
Cyp4a12b
|
cytochrome P450, family 4, subfamily a, polypeptide 12B |
| chr1_+_88022776 | 5.63 |
ENSMUST00000150634.8
ENSMUST00000058237.14 |
Ugt1a7c
|
UDP glucuronosyltransferase 1 family, polypeptide A7C |
| chr17_+_56056977 | 5.61 |
ENSMUST00000025004.7
|
Adgre4
|
adhesion G protein-coupled receptor E4 |
| chr7_+_67297152 | 5.60 |
ENSMUST00000032774.16
ENSMUST00000107471.8 |
Ttc23
|
tetratricopeptide repeat domain 23 |
| chr15_+_31224616 | 5.59 |
ENSMUST00000186547.7
|
Dap
|
death-associated protein |
| chr9_+_74860133 | 5.55 |
ENSMUST00000215370.2
|
Fam214a
|
family with sequence similarity 214, member A |
| chr5_-_87630117 | 5.53 |
ENSMUST00000079811.13
ENSMUST00000144144.3 |
Ugt2a2
|
UDP glucuronosyltransferase 2 family, polypeptide A2 |
| chr19_+_39499288 | 5.52 |
ENSMUST00000025968.5
|
Cyp2c39
|
cytochrome P450, family 2, subfamily c, polypeptide 39 |
| chr19_-_34724689 | 5.49 |
ENSMUST00000009522.4
|
Slc16a12
|
solute carrier family 16 (monocarboxylic acid transporters), member 12 |
| chr13_+_4283729 | 5.45 |
ENSMUST00000081326.7
|
Akr1c19
|
aldo-keto reductase family 1, member C19 |
| chr10_-_86843878 | 5.43 |
ENSMUST00000035288.17
|
Stab2
|
stabilin 2 |
| chrX_-_16683578 | 5.42 |
ENSMUST00000040820.13
|
Maob
|
monoamine oxidase B |
| chr4_-_49473904 | 5.42 |
ENSMUST00000135976.2
|
Acnat1
|
acyl-coenzyme A amino acid N-acyltransferase 1 |
| chr13_+_24511387 | 5.40 |
ENSMUST00000224953.2
ENSMUST00000050859.13 ENSMUST00000167746.8 ENSMUST00000224819.2 |
Cmah
|
cytidine monophospho-N-acetylneuraminic acid hydroxylase |
| chr5_-_116560916 | 5.26 |
ENSMUST00000036991.5
|
Hspb8
|
heat shock protein 8 |
| chr1_-_13730732 | 5.23 |
ENSMUST00000027071.7
|
Lactb2
|
lactamase, beta 2 |
| chr3_+_129326004 | 5.21 |
ENSMUST00000199910.5
ENSMUST00000197070.5 ENSMUST00000071402.7 |
Elovl6
|
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 36.5 | 109.6 | GO:0009087 | methionine catabolic process(GO:0009087) |
| 10.9 | 32.6 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 9.8 | 29.5 | GO:0097037 | heme export(GO:0097037) |
| 8.7 | 26.2 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 7.7 | 23.2 | GO:1901377 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) |
| 7.3 | 29.0 | GO:0032382 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) |
| 7.1 | 28.3 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 6.4 | 19.3 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 5.8 | 11.6 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) |
| 5.7 | 17.0 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 5.5 | 181.7 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 5.4 | 16.2 | GO:0052055 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 5.0 | 14.9 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 4.9 | 69.0 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 4.9 | 19.5 | GO:0009608 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) response to selenium ion(GO:0010269) |
| 4.6 | 18.3 | GO:0015851 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 4.5 | 53.7 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 4.2 | 62.4 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 4.1 | 102.4 | GO:0080184 | response to phenylpropanoid(GO:0080184) |
| 3.9 | 15.6 | GO:0006114 | glycerol biosynthetic process(GO:0006114) positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 3.9 | 15.6 | GO:0010958 | regulation of amino acid import(GO:0010958) |
| 3.9 | 34.9 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 3.9 | 50.3 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 3.7 | 11.1 | GO:1904437 | positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
| 3.2 | 15.8 | GO:0046618 | drug export(GO:0046618) |
| 3.0 | 44.4 | GO:0015747 | urate transport(GO:0015747) |
| 2.9 | 43.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 2.8 | 8.3 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 2.7 | 2.7 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 2.6 | 7.7 | GO:0070237 | positive regulation of activation-induced cell death of T cells(GO:0070237) |
| 2.5 | 10.2 | GO:0038183 | histone H3-R17 methylation(GO:0034971) bile acid signaling pathway(GO:0038183) |
| 2.4 | 7.2 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 2.3 | 6.9 | GO:1904736 | negative regulation of electron carrier activity(GO:1904733) regulation of fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:1904735) negative regulation of fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:1904736) |
| 2.3 | 2.3 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 2.2 | 19.9 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 2.2 | 6.6 | GO:0046491 | L-methylmalonyl-CoA metabolic process(GO:0046491) |
| 2.1 | 8.5 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 1.9 | 15.0 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 1.8 | 5.5 | GO:0015881 | creatine transport(GO:0015881) |
| 1.8 | 12.8 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 1.7 | 5.1 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 1.7 | 8.5 | GO:1904970 | brush border assembly(GO:1904970) |
| 1.7 | 11.7 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 1.7 | 10.0 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 1.6 | 12.5 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 1.5 | 15.2 | GO:0033216 | ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 1.5 | 4.5 | GO:0034240 | negative regulation of macrophage fusion(GO:0034240) |
| 1.4 | 9.9 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 1.3 | 9.4 | GO:1904073 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 1.3 | 6.7 | GO:0010877 | lipid transport involved in lipid storage(GO:0010877) |
| 1.3 | 4.0 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 1.3 | 3.9 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 1.3 | 6.6 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) |
| 1.3 | 6.5 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 1.2 | 4.9 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 1.2 | 15.7 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 1.1 | 39.8 | GO:0046852 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 1.1 | 32.6 | GO:0006825 | copper ion transport(GO:0006825) |
| 1.1 | 3.3 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 1.1 | 6.6 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 1.1 | 4.3 | GO:0021502 | neural fold elevation formation(GO:0021502) |
| 1.1 | 5.3 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 1.0 | 4.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 1.0 | 7.0 | GO:0048861 | oncostatin-M-mediated signaling pathway(GO:0038165) leukemia inhibitory factor signaling pathway(GO:0048861) |
| 1.0 | 3.9 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.9 | 5.6 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.9 | 6.1 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.9 | 4.3 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.9 | 7.8 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.8 | 7.6 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.8 | 5.8 | GO:0015862 | uridine transport(GO:0015862) |
| 0.8 | 3.3 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.8 | 6.9 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.7 | 22.0 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.7 | 2.9 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.7 | 15.5 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.7 | 2.7 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.7 | 2.0 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.7 | 4.0 | GO:0032902 | nerve growth factor production(GO:0032902) |
| 0.7 | 15.2 | GO:0051923 | sulfation(GO:0051923) |
| 0.7 | 3.9 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.7 | 1.3 | GO:0001998 | angiotensin mediated vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001998) |
| 0.6 | 3.9 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.6 | 2.5 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.6 | 3.1 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.6 | 4.9 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.6 | 2.4 | GO:0010286 | heat acclimation(GO:0010286) |
| 0.6 | 3.0 | GO:1904720 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.6 | 15.0 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.6 | 1.8 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.6 | 11.9 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.6 | 7.7 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.6 | 1.2 | GO:1900620 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.6 | 1.8 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.6 | 2.2 | GO:0030647 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.6 | 5.6 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.6 | 1.7 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.6 | 8.3 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.5 | 3.7 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.5 | 3.1 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.5 | 1.6 | GO:0071315 | cellular response to morphine(GO:0071315) |
| 0.5 | 2.0 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.5 | 2.5 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.5 | 1.5 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.5 | 5.0 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.5 | 1.5 | GO:2000019 | negative regulation of male gonad development(GO:2000019) |
| 0.5 | 2.0 | GO:1902490 | regulation of sperm capacitation(GO:1902490) |
| 0.5 | 43.6 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.5 | 3.8 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.5 | 4.2 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.5 | 4.5 | GO:0019660 | glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.4 | 1.3 | GO:0009955 | adaxial/abaxial pattern specification(GO:0009955) regulation of adaxial/abaxial pattern formation(GO:2000011) |
| 0.4 | 3.1 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.4 | 1.3 | GO:0002215 | defense response to nematode(GO:0002215) positive regulation of memory T cell differentiation(GO:0043382) |
| 0.4 | 5.2 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.4 | 1.3 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.4 | 3.4 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.4 | 2.9 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.4 | 1.7 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.4 | 1.2 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.4 | 7.3 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.4 | 8.1 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.4 | 2.8 | GO:0060309 | elastin catabolic process(GO:0060309) |
| 0.4 | 11.8 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.4 | 2.8 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.4 | 2.3 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.4 | 5.3 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.4 | 1.9 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.4 | 3.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.4 | 2.9 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.4 | 7.9 | GO:0017144 | drug metabolic process(GO:0017144) |
| 0.4 | 4.2 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.3 | 8.0 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.3 | 0.3 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) |
| 0.3 | 11.5 | GO:0014823 | response to activity(GO:0014823) |
| 0.3 | 2.6 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.3 | 14.1 | GO:0042311 | vasodilation(GO:0042311) |
| 0.3 | 0.9 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.3 | 1.5 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.3 | 1.1 | GO:0002238 | response to molecule of fungal origin(GO:0002238) chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.3 | 3.7 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.3 | 1.7 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.3 | 0.8 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.3 | 10.9 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.3 | 1.4 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) |
| 0.3 | 3.3 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.3 | 4.9 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.3 | 1.4 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.3 | 19.4 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.2 | 2.2 | GO:0008212 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.2 | 7.1 | GO:0072112 | renal filtration cell differentiation(GO:0061318) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.2 | 1.6 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 0.2 | 4.7 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.2 | 3.7 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.2 | 4.8 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.2 | 3.2 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.2 | 2.3 | GO:0060283 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.2 | 0.9 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.2 | 0.9 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 | 0.6 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.2 | 1.3 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.2 | 0.6 | GO:0060268 | negative regulation of respiratory burst(GO:0060268) |
| 0.2 | 1.3 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.2 | 4.4 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.2 | 3.2 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.2 | 3.4 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.2 | 2.6 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.2 | 0.9 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.2 | 2.8 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 | 2.5 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.2 | 1.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.2 | 0.9 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.2 | 8.9 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.2 | 3.5 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.2 | 1.9 | GO:0090343 | positive regulation of cell aging(GO:0090343) |
| 0.2 | 0.5 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.2 | 1.0 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.2 | 0.5 | GO:0003127 | detection of nodal flow(GO:0003127) detection of endogenous stimulus(GO:0009726) |
| 0.2 | 6.2 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) |
| 0.2 | 1.8 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.2 | 1.3 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.2 | 16.7 | GO:0046889 | positive regulation of lipid biosynthetic process(GO:0046889) |
| 0.2 | 1.9 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.2 | 5.6 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.2 | 6.4 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.2 | 2.4 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.1 | 0.6 | GO:0090158 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) endoplasmic reticulum membrane organization(GO:0090158) |
| 0.1 | 6.5 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 1.0 | GO:0045583 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.1 | 0.7 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.1 | 1.9 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 2.5 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.1 | 7.5 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 8.8 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 0.9 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 0.4 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.1 | 3.1 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 2.1 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 0.4 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) |
| 0.1 | 1.5 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 4.7 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 0.5 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.1 | 2.8 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.1 | 1.5 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.1 | 0.3 | GO:0002632 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) |
| 0.1 | 1.3 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.1 | 0.8 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.1 | 1.5 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.6 | GO:1901228 | positive regulation of skeletal muscle tissue growth(GO:0048633) positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.1 | 0.4 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.1 | 1.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 5.6 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.1 | 0.6 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.1 | 3.8 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 | 0.3 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 2.4 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 4.1 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.1 | 0.4 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 0.4 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.1 | 0.3 | GO:0060664 | epithelial cell proliferation involved in salivary gland morphogenesis(GO:0060664) |
| 0.1 | 0.7 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.1 | 0.9 | GO:0010918 | positive regulation of mitochondrial membrane potential(GO:0010918) |
| 0.1 | 0.7 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.8 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.1 | 2.4 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.1 | 1.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 3.7 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 | 2.1 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.1 | 2.5 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.1 | 1.0 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.1 | 4.9 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.1 | 0.4 | GO:0035822 | meiotic gene conversion(GO:0006311) gene conversion(GO:0035822) |
| 0.1 | 3.5 | GO:0014037 | Schwann cell differentiation(GO:0014037) |
| 0.1 | 0.7 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.1 | 1.3 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 0.4 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.1 | 1.4 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.1 | 0.5 | GO:0034116 | positive regulation of heterotypic cell-cell adhesion(GO:0034116) |
| 0.1 | 0.4 | GO:0009597 | detection of virus(GO:0009597) |
| 0.1 | 0.6 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.1 | 6.1 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 0.3 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 1.6 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.1 | 2.1 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.1 | 0.3 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.1 | 0.9 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.1 | 0.7 | GO:0060051 | negative regulation of protein glycosylation(GO:0060051) |
| 0.1 | 0.8 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.1 | 0.5 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 0.7 | GO:0008228 | opsonization(GO:0008228) |
| 0.1 | 1.9 | GO:0033622 | integrin activation(GO:0033622) |
| 0.1 | 0.3 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.1 | 0.7 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 0.6 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 0.2 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.1 | 0.2 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 | 8.0 | GO:0009308 | amine metabolic process(GO:0009308) |
| 0.1 | 0.6 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.1 | 1.2 | GO:0007614 | short-term memory(GO:0007614) |
| 0.1 | 0.9 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 1.6 | GO:0034340 | response to type I interferon(GO:0034340) |
| 0.1 | 0.8 | GO:0015697 | quaternary ammonium group transport(GO:0015697) |
| 0.1 | 4.9 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 1.3 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.0 | 1.1 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.6 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.2 | GO:0009115 | xanthine catabolic process(GO:0009115) xanthine metabolic process(GO:0046110) |
| 0.0 | 2.5 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.0 | 0.8 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 1.7 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.2 | GO:0043056 | forward locomotion(GO:0043056) |
| 0.0 | 0.6 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 | 53.4 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 1.8 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.7 | GO:1902571 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.8 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 0.3 | GO:0006662 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 | 0.3 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 1.0 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.6 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.0 | 1.4 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.2 | GO:0008078 | zygotic determination of anterior/posterior axis, embryo(GO:0007354) mesodermal cell migration(GO:0008078) |
| 0.0 | 0.6 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.7 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.0 | 1.4 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.5 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 1.0 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.0 | 0.3 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.2 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.2 | GO:0044821 | telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.2 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.0 | 1.2 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 1.0 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 1.2 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 | 2.9 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.9 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 | 0.4 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.2 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.5 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.4 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) |
| 0.0 | 1.1 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.0 | GO:0060722 | spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) |
| 0.0 | 0.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.7 | GO:0001756 | somitogenesis(GO:0001756) |
| 0.0 | 0.3 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.5 | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway(GO:2001243) |
| 0.0 | 0.7 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 1.5 | GO:0007568 | aging(GO:0007568) |
| 0.0 | 0.1 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.3 | GO:0060416 | response to growth hormone(GO:0060416) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.9 | 47.7 | GO:0034359 | mature chylomicron(GO:0034359) |
| 9.8 | 29.5 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 3.5 | 31.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 3.3 | 13.3 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 2.8 | 22.6 | GO:0097413 | Lewy body(GO:0097413) |
| 2.4 | 31.3 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.9 | 9.4 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.9 | 7.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.9 | 15.8 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.9 | 11.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.8 | 5.0 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.8 | 204.6 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.8 | 32.0 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.7 | 10.3 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.7 | 2.9 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.7 | 107.5 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.7 | 2.8 | GO:0070722 | Tle3-Aes complex(GO:0070722) |
| 0.7 | 7.7 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.7 | 2.0 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.6 | 3.9 | GO:0071256 | Sec61 translocon complex(GO:0005784) endoplasmic reticulum Sec complex(GO:0031205) translocon complex(GO:0071256) |
| 0.5 | 3.3 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.5 | 3.2 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.5 | 5.6 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.5 | 4.6 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.5 | 1.5 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 0.5 | 1.4 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.4 | 6.9 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.4 | 2.0 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.4 | 1.2 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.4 | 5.6 | GO:0044754 | autolysosome(GO:0044754) |
| 0.3 | 2.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.3 | 1.9 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.3 | 9.8 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.3 | 1.5 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.3 | 2.0 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.3 | 29.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.3 | 3.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.3 | 1.8 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.3 | 2.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.3 | 60.1 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.3 | 2.0 | GO:0042825 | TAP complex(GO:0042825) |
| 0.2 | 1.7 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.2 | 6.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 112.0 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.2 | 0.6 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.2 | 4.7 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.2 | 1.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 1.6 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.2 | 19.7 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.2 | 5.1 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.2 | 2.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.2 | 7.8 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.2 | 13.2 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.2 | 4.0 | GO:0032982 | myosin filament(GO:0032982) |
| 0.2 | 0.5 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.2 | 1.0 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.2 | 1.1 | GO:1990923 | PET complex(GO:1990923) |
| 0.2 | 1.5 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.2 | 0.8 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.2 | 2.0 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.1 | 0.6 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 54.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 2.7 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 0.4 | GO:0036501 | UFD1-NPL4 complex(GO:0036501) |
| 0.1 | 18.4 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 31.0 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 2.4 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 1.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 1.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 1.2 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 0.9 | GO:0048500 | signal recognition particle(GO:0048500) |
| 0.1 | 1.3 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 0.3 | GO:0060473 | cortical granule(GO:0060473) |
| 0.1 | 2.7 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 3.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 0.8 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 167.5 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.1 | 2.1 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 2.0 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 0.2 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 0.2 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 0.9 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 3.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.3 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.1 | 0.3 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 0.6 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 1.7 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 1.6 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.1 | 172.2 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 0.5 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 1.2 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.4 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.6 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.6 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.3 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 26.9 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 29.7 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.0 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 1.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.2 | GO:0042575 | DNA polymerase complex(GO:0042575) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 27.4 | 109.6 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 11.3 | 34.0 | GO:0005009 | insulin-activated receptor activity(GO:0005009) |
| 8.7 | 26.2 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 8.1 | 32.6 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 7.3 | 29.0 | GO:0050632 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 6.8 | 278.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 6.8 | 40.7 | GO:0030294 | receptor signaling protein tyrosine kinase inhibitor activity(GO:0030294) |
| 5.7 | 17.0 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 5.3 | 196.7 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 5.2 | 15.6 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 4.8 | 14.5 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 4.7 | 14.1 | GO:0008775 | acetate CoA-transferase activity(GO:0008775) |
| 4.2 | 33.8 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 4.1 | 24.3 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 3.9 | 11.8 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 3.7 | 15.0 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 3.7 | 7.5 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 3.7 | 14.9 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 3.7 | 29.5 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 3.7 | 47.7 | GO:0035473 | lipase binding(GO:0035473) |
| 3.6 | 17.9 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 3.4 | 6.8 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 3.2 | 12.8 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 2.9 | 22.9 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 2.8 | 42.7 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 2.8 | 8.5 | GO:0008431 | vitamin E binding(GO:0008431) |
| 2.8 | 8.3 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 2.7 | 32.9 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 2.6 | 10.3 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 2.5 | 14.9 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 2.5 | 42.1 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 2.3 | 25.6 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 2.2 | 6.6 | GO:0047936 | glucose 1-dehydrogenase [NAD(P)] activity(GO:0047936) |
| 2.2 | 6.6 | GO:0004493 | methylmalonyl-CoA epimerase activity(GO:0004493) |
| 2.1 | 12.7 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 2.1 | 26.7 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 2.0 | 26.2 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 1.9 | 15.2 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 1.8 | 5.5 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 1.8 | 14.4 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 1.8 | 5.4 | GO:0030338 | CMP-N-acetylneuraminate monooxygenase activity(GO:0030338) |
| 1.7 | 18.9 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 1.7 | 35.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 1.6 | 22.6 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 1.6 | 7.9 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 1.5 | 10.8 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 1.5 | 6.1 | GO:0036468 | aromatic-L-amino-acid decarboxylase activity(GO:0004058) L-dopa decarboxylase activity(GO:0036468) |
| 1.4 | 7.0 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 1.2 | 6.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 1.2 | 12.8 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 1.1 | 3.4 | GO:0042602 | riboflavin reductase (NADPH) activity(GO:0042602) |
| 1.1 | 3.2 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 1.0 | 2.9 | GO:0050479 | glyceryl-ether monooxygenase activity(GO:0050479) |
| 0.9 | 14.8 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.9 | 21.2 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.9 | 5.3 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.8 | 2.5 | GO:0052642 | lysophosphatidic acid phosphatase activity(GO:0052642) |
| 0.8 | 3.3 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.8 | 15.7 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.8 | 3.1 | GO:0033797 | selenate reductase activity(GO:0033797) |
| 0.8 | 4.7 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.8 | 16.5 | GO:0070513 | death domain binding(GO:0070513) |
| 0.8 | 26.7 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.8 | 27.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.7 | 3.0 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.7 | 3.7 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.7 | 6.6 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.7 | 2.8 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.7 | 5.6 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.7 | 6.3 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.7 | 5.6 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.7 | 17.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.7 | 23.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.7 | 2.0 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.7 | 4.0 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.7 | 3.3 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.7 | 7.9 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.7 | 6.5 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.6 | 3.9 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.6 | 4.4 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.6 | 2.4 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.6 | 4.2 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.6 | 1.8 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.6 | 10.5 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.5 | 3.7 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.5 | 3.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.5 | 4.6 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.5 | 3.0 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.5 | 15.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.5 | 8.5 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.5 | 3.5 | GO:0008430 | selenium binding(GO:0008430) |
| 0.5 | 2.5 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.5 | 1.0 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.5 | 2.9 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.5 | 4.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.5 | 2.3 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.4 | 3.9 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.4 | 5.2 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.4 | 2.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.4 | 4.5 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.4 | 8.5 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.4 | 1.2 | GO:0033265 | choline binding(GO:0033265) |
| 0.4 | 7.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.4 | 6.9 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.4 | 2.6 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.4 | 4.5 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.4 | 6.6 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.4 | 4.7 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.4 | 7.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.4 | 1.8 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.3 | 4.8 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.3 | 1.5 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.3 | 30.1 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.3 | 0.9 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.3 | 0.9 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.3 | 1.1 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.3 | 3.9 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.3 | 3.6 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.3 | 4.8 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.3 | 2.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.3 | 3.8 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.3 | 4.0 | GO:0043121 | neurotrophin binding(GO:0043121) nerve growth factor binding(GO:0048406) |
| 0.2 | 2.2 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.2 | 9.6 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.2 | 1.2 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.2 | 0.9 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.2 | 2.8 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.2 | 4.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 2.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.2 | 0.9 | GO:0019778 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.2 | 2.0 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.2 | 1.6 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.2 | 5.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.2 | 3.4 | GO:0048038 | quinone binding(GO:0048038) |
| 0.2 | 1.4 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.2 | 0.6 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.2 | 4.3 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.2 | 5.4 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.2 | 1.8 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.2 | 3.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.2 | 12.9 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.2 | 4.9 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.2 | 1.9 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.2 | 0.6 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.2 | 3.0 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.2 | 6.5 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.2 | 5.6 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.2 | 1.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.2 | 0.9 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.2 | 4.9 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 0.5 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) |
| 0.2 | 5.3 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.2 | 3.2 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.2 | 0.2 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.2 | 1.2 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 4.0 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.1 | 1.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 3.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.8 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 1.3 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 1.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 1.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.4 | GO:0019107 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.1 | 0.5 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 4.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.8 | GO:0005329 | dopamine transmembrane transporter activity(GO:0005329) |
| 0.1 | 0.6 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 0.4 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 2.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 1.0 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 2.9 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 17.3 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.1 | 0.7 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 1.4 | GO:0001054 | RNA polymerase I activity(GO:0001054) RNA polymerase II activity(GO:0001055) |
| 0.1 | 2.7 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.1 | 0.8 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.1 | 2.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 1.0 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.5 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.1 | 3.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 4.7 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 0.8 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 1.1 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 0.3 | GO:0070002 | glutamic-type peptidase activity(GO:0070002) |
| 0.1 | 19.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.3 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.1 | 3.0 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 0.3 | GO:0047874 | dolichyldiphosphatase activity(GO:0047874) |
| 0.1 | 2.2 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 2.1 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 0.7 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 2.1 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 2.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 1.0 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 4.6 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.1 | 10.9 | GO:0015293 | symporter activity(GO:0015293) |
| 0.1 | 2.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 2.5 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 2.3 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 0.9 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 3.2 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 1.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 1.1 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 0.5 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.2 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.1 | 0.9 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 1.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 5.6 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 1.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 57.8 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.2 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.7 | GO:0008235 | metalloexopeptidase activity(GO:0008235) |
| 0.0 | 2.5 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.3 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.0 | 0.1 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.0 | 0.3 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 2.7 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.3 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.0 | 1.6 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.3 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.1 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.7 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 2.6 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.7 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 1.9 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 2.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 2.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.9 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 1.7 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 1.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 3.8 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.4 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.2 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 1.0 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.0 | 0.3 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.4 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.4 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 3.3 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.3 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.6 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.3 | GO:0015026 | coreceptor activity(GO:0015026) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 22.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.8 | 21.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.7 | 50.4 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.6 | 39.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.5 | 4.0 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.4 | 4.5 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.4 | 42.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.3 | 0.6 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.3 | 9.9 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.3 | 17.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 15.8 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.2 | 6.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 12.8 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.2 | 56.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 16.6 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.2 | 12.6 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.2 | 12.9 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.2 | 12.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.2 | 9.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 6.4 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 3.7 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 15.8 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 3.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 8.3 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 1.1 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 17.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 6.5 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 1.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 1.8 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 1.6 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 2.6 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.4 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 1.1 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.2 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.3 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.7 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.2 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.5 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 51.9 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 3.5 | 38.7 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 3.2 | 109.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 2.6 | 28.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 2.6 | 43.9 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 2.3 | 32.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 2.1 | 6.4 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 1.9 | 7.6 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 1.9 | 18.7 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 1.6 | 47.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 1.5 | 29.0 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 1.3 | 17.9 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 1.1 | 15.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 1.1 | 20.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.8 | 10.0 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.8 | 10.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.8 | 34.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.7 | 16.8 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.7 | 30.0 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.7 | 19.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.5 | 8.9 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.5 | 8.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.4 | 5.7 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.4 | 8.9 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.4 | 14.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.4 | 13.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.4 | 8.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.4 | 4.5 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.3 | 5.2 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.3 | 15.2 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.3 | 3.9 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.3 | 56.1 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.3 | 6.6 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.3 | 3.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.3 | 4.7 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.2 | 11.1 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.2 | 15.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.2 | 7.1 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 4.7 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.2 | 10.9 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.2 | 6.1 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 8.6 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 3.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 3.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 4.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.8 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 1.9 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.1 | 3.8 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 2.9 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 6.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 11.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 2.3 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 1.1 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.1 | 4.9 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 0.7 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 2.6 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 1.8 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 6.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.8 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 2.7 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 1.9 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.1 | 3.2 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 2.2 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.1 | 1.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.4 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 1.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 0.8 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 1.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 0.4 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 1.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 1.0 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.6 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 2.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 2.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 2.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.6 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.6 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.7 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 1.1 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |