Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Cenpb
Z-value: 0.60
Transcription factors associated with Cenpb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Cenpb
|
ENSMUSG00000068267.6 | Cenpb |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Cenpb | mm39_v1_chr2_-_131021905_131022005 | 0.21 | 7.7e-02 | Click! |
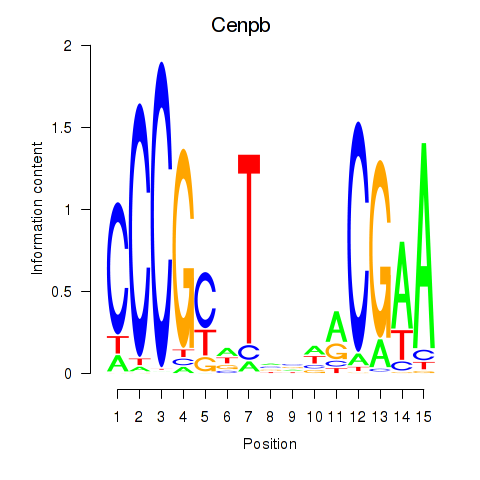
Activity profile of Cenpb motif
Sorted Z-values of Cenpb motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Cenpb
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_48715971 | 2.60 |
ENSMUST00000054368.7
ENSMUST00000140054.3 ENSMUST00000204168.2 ENSMUST00000204408.2 |
Gimap1
Gm28053
|
GTPase, IMAP family member 1 predicted gene, 28053 |
| chr17_+_44445659 | 2.34 |
ENSMUST00000239215.2
|
Clic5
|
chloride intracellular channel 5 |
| chr3_+_106943472 | 2.00 |
ENSMUST00000052718.5
|
Kcna3
|
potassium voltage-gated channel, shaker-related subfamily, member 3 |
| chr5_-_140687995 | 1.66 |
ENSMUST00000135028.5
ENSMUST00000077890.12 ENSMUST00000041783.14 ENSMUST00000142081.6 |
Iqce
|
IQ motif containing E |
| chr8_-_46604742 | 1.57 |
ENSMUST00000041582.15
|
Snx25
|
sorting nexin 25 |
| chr2_+_127178072 | 1.51 |
ENSMUST00000028846.7
|
Dusp2
|
dual specificity phosphatase 2 |
| chrX_-_100312629 | 1.51 |
ENSMUST00000117736.2
|
Gm20489
|
predicted gene 20489 |
| chr8_+_4399588 | 1.49 |
ENSMUST00000110982.8
ENSMUST00000024004.9 |
Ccl25
|
chemokine (C-C motif) ligand 25 |
| chr14_-_44057096 | 1.43 |
ENSMUST00000100691.4
|
Ear1
|
eosinophil-associated, ribonuclease A family, member 1 |
| chr10_+_34173426 | 1.39 |
ENSMUST00000047935.8
|
Tspyl4
|
TSPY-like 4 |
| chr8_+_82069177 | 1.38 |
ENSMUST00000213285.2
ENSMUST00000217122.2 ENSMUST00000215332.2 |
Inpp4b
|
inositol polyphosphate-4-phosphatase, type II |
| chr10_+_67815508 | 1.37 |
ENSMUST00000117086.2
|
Rtkn2
|
rhotekin 2 |
| chr14_-_44112974 | 1.35 |
ENSMUST00000179200.2
|
Ear1
|
eosinophil-associated, ribonuclease A family, member 1 |
| chr10_+_25235748 | 1.29 |
ENSMUST00000218903.2
|
Epb41l2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr11_+_114618209 | 1.20 |
ENSMUST00000069325.14
|
Dnai2
|
dynein axonemal intermediate chain 2 |
| chr1_-_80736165 | 1.17 |
ENSMUST00000077946.12
|
Dock10
|
dedicator of cytokinesis 10 |
| chr6_+_58808733 | 1.16 |
ENSMUST00000126292.8
ENSMUST00000031823.12 |
Herc3
|
hect domain and RLD 3 |
| chr8_-_92039850 | 1.15 |
ENSMUST00000047783.14
|
Rpgrip1l
|
Rpgrip1-like |
| chr10_+_25235696 | 1.10 |
ENSMUST00000053748.16
|
Epb41l2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr6_+_48531710 | 1.10 |
ENSMUST00000114545.8
ENSMUST00000153222.2 ENSMUST00000204071.2 ENSMUST00000101436.3 ENSMUST00000203627.2 |
Lrrc61
|
leucine rich repeat containing 61 |
| chr18_+_90528308 | 1.09 |
ENSMUST00000235634.2
|
Tmx3
|
thioredoxin-related transmembrane protein 3 |
| chr3_-_116388334 | 1.08 |
ENSMUST00000197190.5
ENSMUST00000198454.2 |
Trmt13
|
tRNA methyltransferase 13 |
| chr2_-_117173428 | 1.08 |
ENSMUST00000102534.11
|
Rasgrp1
|
RAS guanyl releasing protein 1 |
| chr14_+_66534539 | 1.06 |
ENSMUST00000121006.2
|
Trim35
|
tripartite motif-containing 35 |
| chr10_+_67815422 | 1.05 |
ENSMUST00000105437.8
|
Rtkn2
|
rhotekin 2 |
| chr14_+_66043281 | 1.05 |
ENSMUST00000022612.10
|
Pbk
|
PDZ binding kinase |
| chr10_+_67815395 | 1.02 |
ENSMUST00000118160.8
|
Rtkn2
|
rhotekin 2 |
| chr6_+_125298372 | 1.00 |
ENSMUST00000176442.8
ENSMUST00000177329.2 |
Scnn1a
|
sodium channel, nonvoltage-gated 1 alpha |
| chr3_-_9898676 | 1.00 |
ENSMUST00000108384.9
|
Pag1
|
phosphoprotein associated with glycosphingolipid microdomains 1 |
| chr3_+_54600268 | 0.97 |
ENSMUST00000199652.5
|
Supt20
|
SPT20 SAGA complex component |
| chr16_+_17577464 | 0.96 |
ENSMUST00000129199.8
|
Klhl22
|
kelch-like 22 |
| chr3_+_116388600 | 0.95 |
ENSMUST00000198386.5
ENSMUST00000198311.5 ENSMUST00000197335.2 |
Sass6
|
SAS-6 centriolar assembly protein |
| chr14_+_66534478 | 0.92 |
ENSMUST00000022623.13
|
Trim35
|
tripartite motif-containing 35 |
| chr3_+_54600196 | 0.88 |
ENSMUST00000197502.5
ENSMUST00000200441.5 ENSMUST00000029315.13 ENSMUST00000199655.5 |
Supt20
|
SPT20 SAGA complex component |
| chr5_-_138169253 | 0.88 |
ENSMUST00000139983.8
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr18_+_10725650 | 0.87 |
ENSMUST00000165555.8
|
Mib1
|
mindbomb E3 ubiquitin protein ligase 1 |
| chr17_+_74111823 | 0.86 |
ENSMUST00000024860.9
|
Ehd3
|
EH-domain containing 3 |
| chr10_+_117977708 | 0.85 |
ENSMUST00000020437.13
ENSMUST00000163238.9 |
Mdm1
|
transformed mouse 3T3 cell double minute 1 |
| chr7_+_125151432 | 0.84 |
ENSMUST00000206217.2
ENSMUST00000205985.2 |
Il4ra
|
interleukin 4 receptor, alpha |
| chr6_-_41012435 | 0.84 |
ENSMUST00000031931.6
|
2210010C04Rik
|
RIKEN cDNA 2210010C04 gene |
| chr3_-_9898713 | 0.83 |
ENSMUST00000161949.8
|
Pag1
|
phosphoprotein associated with glycosphingolipid microdomains 1 |
| chr2_+_180961507 | 0.82 |
ENSMUST00000098971.11
ENSMUST00000054622.15 ENSMUST00000108814.8 ENSMUST00000048608.16 ENSMUST00000108815.8 |
Rtel1
|
regulator of telomere elongation helicase 1 |
| chr16_-_10265204 | 0.82 |
ENSMUST00000051118.7
|
Tvp23a
|
trans-golgi network vesicle protein 23A |
| chr7_-_42962487 | 0.81 |
ENSMUST00000135130.2
ENSMUST00000139061.2 |
Zfp715
|
zinc finger protein 715 |
| chr9_-_105008972 | 0.81 |
ENSMUST00000186925.2
|
Nudt16
|
nudix (nucleoside diphosphate linked moiety X)-type motif 16 |
| chr1_+_87192067 | 0.79 |
ENSMUST00000027472.7
|
Efhd1
|
EF hand domain containing 1 |
| chr3_+_121517158 | 0.77 |
ENSMUST00000029771.13
|
F3
|
coagulation factor III |
| chr5_+_29774437 | 0.77 |
ENSMUST00000199032.2
|
Ube3c
|
ubiquitin protein ligase E3C |
| chr6_-_86710250 | 0.75 |
ENSMUST00000001185.14
|
Gmcl1
|
germ cell-less, spermatogenesis associated 1 |
| chr5_-_124387812 | 0.72 |
ENSMUST00000162812.8
|
Pitpnm2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chr4_-_68872585 | 0.72 |
ENSMUST00000030036.6
|
Brinp1
|
bone morphogenic protein/retinoic acid inducible neural specific 1 |
| chr6_-_86710220 | 0.71 |
ENSMUST00000113679.2
|
Gmcl1
|
germ cell-less, spermatogenesis associated 1 |
| chrX_+_16389108 | 0.71 |
ENSMUST00000115422.2
ENSMUST00000024026.3 |
Cypt1
|
cysteine-rich perinuclear theca 1 |
| chr13_+_38223023 | 0.70 |
ENSMUST00000226110.2
|
Riok1
|
RIO kinase 1 |
| chr3_-_113263974 | 0.70 |
ENSMUST00000098667.5
|
Amy2a2
|
amylase 2a2 |
| chr12_+_113120023 | 0.69 |
ENSMUST00000049271.13
|
Tedc1
|
tubulin epsilon and delta complex 1 |
| chr5_-_138169509 | 0.68 |
ENSMUST00000153867.8
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr19_+_3818112 | 0.67 |
ENSMUST00000005518.16
ENSMUST00000237440.2 ENSMUST00000152935.8 ENSMUST00000176262.8 ENSMUST00000176407.8 ENSMUST00000176926.8 ENSMUST00000176512.8 |
Kmt5b
|
lysine methyltransferase 5B |
| chr19_-_5168251 | 0.67 |
ENSMUST00000113728.8
ENSMUST00000113727.8 ENSMUST00000025798.13 |
Klc2
|
kinesin light chain 2 |
| chr11_+_8998575 | 0.67 |
ENSMUST00000043285.5
|
Gm11992
|
predicted gene 11992 |
| chr2_-_3420102 | 0.66 |
ENSMUST00000115082.10
|
Meig1
|
meiosis expressed gene 1 |
| chr13_+_55300453 | 0.66 |
ENSMUST00000005452.6
|
Fgfr4
|
fibroblast growth factor receptor 4 |
| chr2_-_120801186 | 0.64 |
ENSMUST00000028728.6
|
Ubr1
|
ubiquitin protein ligase E3 component n-recognin 1 |
| chr1_+_64572050 | 0.64 |
ENSMUST00000190348.2
|
Creb1
|
cAMP responsive element binding protein 1 |
| chr9_-_105009023 | 0.63 |
ENSMUST00000035179.9
|
Nudt16
|
nudix (nucleoside diphosphate linked moiety X)-type motif 16 |
| chr17_+_71490540 | 0.62 |
ENSMUST00000156570.8
|
Lpin2
|
lipin 2 |
| chr15_-_81900335 | 0.62 |
ENSMUST00000152227.8
|
Desi1
|
desumoylating isopeptidase 1 |
| chr15_-_83033471 | 0.61 |
ENSMUST00000129372.2
|
Poldip3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr7_+_125151292 | 0.59 |
ENSMUST00000033004.8
|
Il4ra
|
interleukin 4 receptor, alpha |
| chr2_+_180961599 | 0.59 |
ENSMUST00000153112.2
|
Rtel1
|
regulator of telomere elongation helicase 1 |
| chr19_+_3817396 | 0.59 |
ENSMUST00000052699.13
ENSMUST00000113974.11 ENSMUST00000113972.9 ENSMUST00000113973.8 ENSMUST00000113977.9 ENSMUST00000113968.9 |
Kmt5b
|
lysine methyltransferase 5B |
| chr4_-_126954878 | 0.58 |
ENSMUST00000136186.2
ENSMUST00000106099.8 ENSMUST00000106102.9 |
Zmym1
|
zinc finger, MYM domain containing 1 |
| chr6_+_66512401 | 0.57 |
ENSMUST00000101343.2
|
Mad2l1
|
MAD2 mitotic arrest deficient-like 1 |
| chr14_-_54386718 | 0.57 |
ENSMUST00000103685.3
|
Trdv5
|
T cell receptor delta variable 5 |
| chr5_+_138621098 | 0.56 |
ENSMUST00000200521.5
ENSMUST00000032590.14 ENSMUST00000198958.5 ENSMUST00000069862.11 |
A430033K04Rik
|
RIKEN cDNA A430033K04 gene |
| chr11_+_79551358 | 0.55 |
ENSMUST00000155381.2
|
Rab11fip4
|
RAB11 family interacting protein 4 (class II) |
| chr8_-_47070201 | 0.55 |
ENSMUST00000210264.2
ENSMUST00000040468.16 ENSMUST00000209787.2 |
Primpol
|
primase and polymerase (DNA-directed) |
| chr7_-_132454332 | 0.55 |
ENSMUST00000120425.8
ENSMUST00000033257.15 |
Eef1akmt2
|
EEF1A lysine methyltransferase 2 |
| chr16_+_33071784 | 0.55 |
ENSMUST00000023502.6
|
Snx4
|
sorting nexin 4 |
| chr1_-_83016152 | 0.53 |
ENSMUST00000164473.2
ENSMUST00000045560.15 |
Slc19a3
|
solute carrier family 19, member 3 |
| chr17_-_37262495 | 0.53 |
ENSMUST00000040402.14
ENSMUST00000174711.8 |
Ppp1r11
|
protein phosphatase 1, regulatory inhibitor subunit 11 |
| chr11_-_86977576 | 0.53 |
ENSMUST00000020801.14
|
Smg8
|
smg-8 homolog, nonsense mediated mRNA decay factor (C. elegans) |
| chr2_-_3420056 | 0.52 |
ENSMUST00000115084.8
ENSMUST00000115083.8 |
Meig1
|
meiosis expressed gene 1 |
| chr15_+_81900570 | 0.52 |
ENSMUST00000069530.13
ENSMUST00000168581.8 ENSMUST00000164779.2 |
Xrcc6
|
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr11_+_70104929 | 0.52 |
ENSMUST00000094055.10
ENSMUST00000126296.8 ENSMUST00000136328.2 ENSMUST00000153993.3 |
Slc16a11
|
solute carrier family 16 (monocarboxylic acid transporters), member 11 |
| chr13_+_109397184 | 0.51 |
ENSMUST00000153234.8
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr6_-_34887743 | 0.51 |
ENSMUST00000081214.12
|
Wdr91
|
WD repeat domain 91 |
| chr9_-_45923908 | 0.50 |
ENSMUST00000217514.2
|
Pafah1b2
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 |
| chr1_+_134487928 | 0.49 |
ENSMUST00000112198.3
|
Kdm5b
|
lysine (K)-specific demethylase 5B |
| chr4_-_43578823 | 0.49 |
ENSMUST00000030189.14
|
Gba2
|
glucosidase beta 2 |
| chr15_-_89033761 | 0.49 |
ENSMUST00000088823.5
|
Mapk11
|
mitogen-activated protein kinase 11 |
| chr8_+_84728123 | 0.49 |
ENSMUST00000060357.15
ENSMUST00000239176.2 |
1700067K01Rik
|
RIKEN cDNA 1700067K01 gene |
| chr11_+_58868919 | 0.49 |
ENSMUST00000108809.8
ENSMUST00000108810.10 ENSMUST00000093061.7 |
Trim11
|
tripartite motif-containing 11 |
| chr19_+_34169629 | 0.48 |
ENSMUST00000239240.2
ENSMUST00000054956.15 |
Stambpl1
|
STAM binding protein like 1 |
| chr2_-_18001734 | 0.48 |
ENSMUST00000105001.4
|
H2al2a
|
H2A histone family member L2A |
| chr11_+_94881861 | 0.47 |
ENSMUST00000038696.12
|
Ppp1r9b
|
protein phosphatase 1, regulatory subunit 9B |
| chr4_+_132857816 | 0.47 |
ENSMUST00000084241.12
ENSMUST00000138831.2 |
Wasf2
|
WASP family, member 2 |
| chr18_+_90528246 | 0.47 |
ENSMUST00000025515.7
ENSMUST00000235708.2 |
Tmx3
|
thioredoxin-related transmembrane protein 3 |
| chr17_+_6156738 | 0.47 |
ENSMUST00000142030.8
|
Tulp4
|
tubby like protein 4 |
| chr13_-_23882437 | 0.47 |
ENSMUST00000102967.3
|
H4c3
|
H4 clustered histone 3 |
| chr5_+_108777855 | 0.46 |
ENSMUST00000078323.13
ENSMUST00000120327.3 |
Tmem175
|
transmembrane protein 175 |
| chr19_-_59932079 | 0.46 |
ENSMUST00000171986.8
|
Rab11fip2
|
RAB11 family interacting protein 2 (class I) |
| chr7_-_42962640 | 0.45 |
ENSMUST00000012796.14
ENSMUST00000107986.9 |
Zfp715
|
zinc finger protein 715 |
| chr10_+_128247492 | 0.45 |
ENSMUST00000171342.3
|
Rnf41
|
ring finger protein 41 |
| chr1_+_182236728 | 0.45 |
ENSMUST00000117245.2
|
Trp53bp2
|
transformation related protein 53 binding protein 2 |
| chr8_+_92040215 | 0.44 |
ENSMUST00000166548.9
|
Fto
|
fat mass and obesity associated |
| chr19_+_3817953 | 0.44 |
ENSMUST00000113970.8
|
Kmt5b
|
lysine methyltransferase 5B |
| chr14_+_66872699 | 0.44 |
ENSMUST00000159365.8
ENSMUST00000054661.8 ENSMUST00000225182.2 ENSMUST00000159068.2 |
Adra1a
|
adrenergic receptor, alpha 1a |
| chr5_+_108777636 | 0.44 |
ENSMUST00000146207.6
ENSMUST00000063272.13 |
Tmem175
|
transmembrane protein 175 |
| chr3_+_75464837 | 0.44 |
ENSMUST00000161776.8
ENSMUST00000029423.9 |
Serpini1
|
serine (or cysteine) peptidase inhibitor, clade I, member 1 |
| chr8_-_106016496 | 0.43 |
ENSMUST00000014981.8
|
4931428F04Rik
|
RIKEN cDNA 4931428F04 gene |
| chr7_+_12615091 | 0.43 |
ENSMUST00000144578.2
|
Zfp128
|
zinc finger protein 128 |
| chr3_+_66892979 | 0.42 |
ENSMUST00000162362.8
ENSMUST00000065074.14 ENSMUST00000065047.13 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chrX_-_63195706 | 0.42 |
ENSMUST00000114682.4
|
Gm6760
|
predicted gene 6760 |
| chr17_-_25459086 | 0.42 |
ENSMUST00000038973.7
ENSMUST00000115154.11 |
Gnptg
|
N-acetylglucosamine-1-phosphotransferase, gamma subunit |
| chr2_-_3513783 | 0.42 |
ENSMUST00000124331.8
ENSMUST00000140494.2 ENSMUST00000027961.12 |
Gm45902
Hspa14
|
predicted gene 45902 heat shock protein 14 |
| chr7_+_24920840 | 0.41 |
ENSMUST00000055604.6
|
Zfp526
|
zinc finger protein 526 |
| chr10_-_21943978 | 0.41 |
ENSMUST00000092672.6
|
4930444G20Rik
|
RIKEN cDNA 4930444G20 gene |
| chr13_-_12476313 | 0.41 |
ENSMUST00000143693.8
ENSMUST00000144283.2 ENSMUST00000099820.10 ENSMUST00000135166.8 |
Lgals8
|
lectin, galactose binding, soluble 8 |
| chr11_+_62349238 | 0.40 |
ENSMUST00000014389.6
|
Pigl
|
phosphatidylinositol glycan anchor biosynthesis, class L |
| chr10_-_112764879 | 0.40 |
ENSMUST00000099276.4
|
Atxn7l3b
|
ataxin 7-like 3B |
| chr10_+_80591030 | 0.40 |
ENSMUST00000105336.9
|
Dot1l
|
DOT1-like, histone H3 methyltransferase (S. cerevisiae) |
| chr9_-_59432729 | 0.39 |
ENSMUST00000055345.9
|
Tmem202
|
transmembrane protein 202 |
| chr8_+_71358576 | 0.38 |
ENSMUST00000019405.4
ENSMUST00000212511.2 |
Map1s
|
microtubule-associated protein 1S |
| chr7_+_65759198 | 0.38 |
ENSMUST00000036372.8
|
Chsy1
|
chondroitin sulfate synthase 1 |
| chr12_+_3856510 | 0.38 |
ENSMUST00000172719.8
|
Dnmt3a
|
DNA methyltransferase 3A |
| chr3_-_96812610 | 0.37 |
ENSMUST00000029738.14
|
Gpr89
|
G protein-coupled receptor 89 |
| chr9_-_107648144 | 0.37 |
ENSMUST00000183248.3
ENSMUST00000182022.8 ENSMUST00000035199.13 ENSMUST00000182659.8 |
Rbm5
|
RNA binding motif protein 5 |
| chr17_-_36290571 | 0.36 |
ENSMUST00000173724.2
ENSMUST00000172900.8 ENSMUST00000174849.8 |
Prr3
|
proline-rich polypeptide 3 |
| chr10_-_62628008 | 0.36 |
ENSMUST00000217768.2
ENSMUST00000020268.7 ENSMUST00000218946.2 ENSMUST00000219527.2 |
Ccar1
|
cell division cycle and apoptosis regulator 1 |
| chr4_-_117039809 | 0.36 |
ENSMUST00000065896.9
|
Kif2c
|
kinesin family member 2C |
| chr15_-_31601652 | 0.36 |
ENSMUST00000161266.2
|
Cct5
|
chaperonin containing Tcp1, subunit 5 (epsilon) |
| chr7_-_46608193 | 0.34 |
ENSMUST00000094398.13
|
Uevld
|
UEV and lactate/malate dehyrogenase domains |
| chr17_+_80597632 | 0.34 |
ENSMUST00000227729.2
ENSMUST00000061703.10 |
Morn2
|
MORN repeat containing 2 |
| chr16_-_18630722 | 0.34 |
ENSMUST00000000028.14
ENSMUST00000115585.2 |
Cdc45
|
cell division cycle 45 |
| chr17_-_44416665 | 0.34 |
ENSMUST00000024757.14
|
Enpp4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 |
| chr3_+_66893031 | 0.33 |
ENSMUST00000046542.13
ENSMUST00000162693.8 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr1_+_183766572 | 0.33 |
ENSMUST00000048655.8
|
Dusp10
|
dual specificity phosphatase 10 |
| chr11_+_120653613 | 0.33 |
ENSMUST00000105046.4
|
Hmga1b
|
high mobility group AT-hook 1B |
| chr4_+_43578922 | 0.33 |
ENSMUST00000030190.9
|
Rgp1
|
RAB6A GEF compex partner 1 |
| chr16_+_17577493 | 0.32 |
ENSMUST00000165790.9
|
Klhl22
|
kelch-like 22 |
| chr18_+_32970278 | 0.32 |
ENSMUST00000053663.11
|
Wdr36
|
WD repeat domain 36 |
| chr5_-_138169476 | 0.32 |
ENSMUST00000147920.2
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr6_+_66512374 | 0.32 |
ENSMUST00000116605.8
|
Mad2l1
|
MAD2 mitotic arrest deficient-like 1 |
| chr5_+_121535999 | 0.31 |
ENSMUST00000042163.15
|
Naa25
|
N(alpha)-acetyltransferase 25, NatB auxiliary subunit |
| chr8_-_25438784 | 0.30 |
ENSMUST00000119720.8
ENSMUST00000121438.9 |
Adam32
|
a disintegrin and metallopeptidase domain 32 |
| chr18_+_37072232 | 0.30 |
ENSMUST00000115662.9
ENSMUST00000195590.2 |
Pcdha2
|
protocadherin alpha 2 |
| chr10_+_128247598 | 0.29 |
ENSMUST00000096386.13
|
Rnf41
|
ring finger protein 41 |
| chr17_-_66384017 | 0.29 |
ENSMUST00000150766.2
ENSMUST00000038116.13 |
Ankrd12
|
ankyrin repeat domain 12 |
| chr2_-_122199604 | 0.29 |
ENSMUST00000151130.8
|
Shf
|
Src homology 2 domain containing F |
| chr3_+_89979948 | 0.28 |
ENSMUST00000121503.8
ENSMUST00000119570.8 |
Tpm3
|
tropomyosin 3, gamma |
| chr11_-_113641980 | 0.28 |
ENSMUST00000153453.2
|
Cdc42ep4
|
CDC42 effector protein (Rho GTPase binding) 4 |
| chr19_-_32689687 | 0.28 |
ENSMUST00000237752.2
ENSMUST00000235412.2 |
Atad1
|
ATPase family, AAA domain containing 1 |
| chr1_+_85949657 | 0.28 |
ENSMUST00000052854.13
ENSMUST00000125083.2 ENSMUST00000152501.8 ENSMUST00000113344.8 ENSMUST00000130504.8 ENSMUST00000153247.3 |
Spata3
|
spermatogenesis associated 3 |
| chr5_-_35732390 | 0.28 |
ENSMUST00000030980.12
|
Trmt44
|
tRNA methyltransferase 44 |
| chr4_+_12906838 | 0.28 |
ENSMUST00000143186.8
ENSMUST00000183345.2 |
Triqk
|
triple QxxK/R motif containing |
| chr12_+_74011255 | 0.27 |
ENSMUST00000021532.6
|
Snapc1
|
small nuclear RNA activating complex, polypeptide 1 |
| chr3_-_108747767 | 0.27 |
ENSMUST00000196679.5
|
Stxbp3
|
syntaxin binding protein 3 |
| chr2_-_122199643 | 0.27 |
ENSMUST00000125826.8
|
Shf
|
Src homology 2 domain containing F |
| chr17_+_26028059 | 0.27 |
ENSMUST00000045692.9
|
Fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chrX_+_93768175 | 0.27 |
ENSMUST00000101388.4
|
Zxdb
|
zinc finger, X-linked, duplicated B |
| chr3_-_33136153 | 0.26 |
ENSMUST00000108225.10
|
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr16_+_10652910 | 0.26 |
ENSMUST00000037913.9
|
Rmi2
|
RecQ mediated genome instability 2 |
| chr17_+_24022153 | 0.26 |
ENSMUST00000190686.7
ENSMUST00000088621.11 ENSMUST00000233636.2 |
Srrm2
|
serine/arginine repetitive matrix 2 |
| chr7_+_12568647 | 0.26 |
ENSMUST00000004614.15
|
Zfp110
|
zinc finger protein 110 |
| chr15_+_36179676 | 0.25 |
ENSMUST00000171205.3
|
Spag1
|
sperm associated antigen 1 |
| chr6_-_135231168 | 0.25 |
ENSMUST00000111909.8
|
Gsg1
|
germ cell associated 1 |
| chr17_+_43878989 | 0.25 |
ENSMUST00000167214.8
ENSMUST00000024706.12 |
Pla2g7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr8_+_92040188 | 0.25 |
ENSMUST00000136802.8
|
Fto
|
fat mass and obesity associated |
| chr13_+_81859460 | 0.25 |
ENSMUST00000057598.7
ENSMUST00000224299.2 |
Mblac2
|
metallo-beta-lactamase domain containing 2 |
| chr6_-_99005835 | 0.25 |
ENSMUST00000154163.9
|
Foxp1
|
forkhead box P1 |
| chr8_+_84874654 | 0.24 |
ENSMUST00000143833.8
ENSMUST00000118856.8 |
Brme1
|
break repair meiotic recombinase recruitment factor 1 |
| chr1_-_55401851 | 0.23 |
ENSMUST00000114423.7
|
Boll
|
boule homolog, RNA binding protein |
| chr13_-_95661726 | 0.22 |
ENSMUST00000022185.10
|
F2rl1
|
coagulation factor II (thrombin) receptor-like 1 |
| chr8_-_106706035 | 0.22 |
ENSMUST00000034371.9
|
Dpep3
|
dipeptidase 3 |
| chr11_-_113642135 | 0.22 |
ENSMUST00000106616.2
|
Cdc42ep4
|
CDC42 effector protein (Rho GTPase binding) 4 |
| chr4_+_152423075 | 0.21 |
ENSMUST00000030775.12
ENSMUST00000164662.8 |
Chd5
|
chromodomain helicase DNA binding protein 5 |
| chr4_-_8239034 | 0.21 |
ENSMUST00000066674.8
|
Car8
|
carbonic anhydrase 8 |
| chr4_-_140787852 | 0.21 |
ENSMUST00000144196.2
ENSMUST00000097816.9 |
Crocc
|
ciliary rootlet coiled-coil, rootletin |
| chrX_-_20955370 | 0.20 |
ENSMUST00000040667.13
|
Zfp300
|
zinc finger protein 300 |
| chr8_-_34575946 | 0.20 |
ENSMUST00000033913.11
|
Dctn6
|
dynactin 6 |
| chr6_-_88852017 | 0.20 |
ENSMUST00000145944.3
|
Podxl2
|
podocalyxin-like 2 |
| chr17_+_25459138 | 0.20 |
ENSMUST00000063574.7
|
Tsr3
|
TSR3 20S rRNA accumulation |
| chr5_-_4154681 | 0.20 |
ENSMUST00000001507.5
|
Cyp51
|
cytochrome P450, family 51 |
| chr12_+_69771761 | 0.19 |
ENSMUST00000222950.2
|
Dmac2l
|
distal membrane arm assembly complex 2 like |
| chr2_+_105054657 | 0.19 |
ENSMUST00000068813.3
|
Them7
|
thioesterase superfamily member 7 |
| chr3_+_69629318 | 0.19 |
ENSMUST00000029358.15
|
Nmd3
|
NMD3 ribosome export adaptor |
| chr1_+_85949681 | 0.18 |
ENSMUST00000159876.8
ENSMUST00000135440.2 |
Spata3
|
spermatogenesis associated 3 |
| chr16_-_44153498 | 0.18 |
ENSMUST00000047446.13
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr4_-_111759951 | 0.18 |
ENSMUST00000102719.8
ENSMUST00000102721.8 |
Slc5a9
|
solute carrier family 5 (sodium/glucose cotransporter), member 9 |
| chr5_+_147797425 | 0.18 |
ENSMUST00000201376.4
ENSMUST00000201120.2 |
Pomp
|
proteasome maturation protein |
| chr11_+_101137231 | 0.18 |
ENSMUST00000122006.8
ENSMUST00000151830.2 |
Ramp2
|
receptor (calcitonin) activity modifying protein 2 |
| chr13_+_120151982 | 0.18 |
ENSMUST00000179869.3
ENSMUST00000224188.2 |
Hmgcs1
|
3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 |
| chr1_+_63312420 | 0.18 |
ENSMUST00000239483.2
ENSMUST00000114132.8 ENSMUST00000126932.2 |
Zdbf2
|
zinc finger, DBF-type containing 2 |
| chr17_-_24005020 | 0.17 |
ENSMUST00000024704.10
|
Flywch2
|
FLYWCH family member 2 |
| chr17_+_66418525 | 0.17 |
ENSMUST00000072383.14
|
Washc1
|
WASH complex subunit 1 |
| chr14_-_30973164 | 0.17 |
ENSMUST00000226565.2
ENSMUST00000022459.5 |
Phf7
|
PHD finger protein 7 |
| chr2_-_157121440 | 0.17 |
ENSMUST00000143663.2
|
Mroh8
|
maestro heat-like repeat family member 8 |
| chr5_+_4242367 | 0.17 |
ENSMUST00000177258.2
|
Mterf1b
|
mitochondrial transcription termination factor 1b |
| chr1_-_64995891 | 0.16 |
ENSMUST00000123225.2
|
Plekhm3
|
pleckstrin homology domain containing, family M, member 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.4 | 1.4 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) |
| 0.4 | 1.4 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) negative regulation of t-circle formation(GO:1904430) |
| 0.3 | 1.4 | GO:0046709 | IDP metabolic process(GO:0046707) IDP catabolic process(GO:0046709) |
| 0.3 | 0.8 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.2 | 1.9 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.2 | 1.6 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.2 | 0.7 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.2 | 0.6 | GO:1904550 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.2 | 0.7 | GO:0042245 | RNA repair(GO:0042245) |
| 0.2 | 0.6 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.2 | 2.9 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.2 | 1.9 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.2 | 1.8 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.1 | 0.4 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.1 | 0.4 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.1 | 0.5 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.1 | 0.5 | GO:0035938 | estradiol secretion(GO:0035938) regulation of estradiol secretion(GO:2000864) |
| 0.1 | 0.3 | GO:0071163 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) DNA replication preinitiation complex assembly(GO:0071163) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 2.3 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.1 | 0.2 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.1 | 0.9 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 0.5 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.1 | 2.5 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.1 | 0.4 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.1 | 0.9 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.1 | 0.5 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.1 | 1.6 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.4 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.1 | 0.7 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 0.9 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 1.2 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 1.1 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.1 | 0.5 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.1 | 0.6 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.1 | 0.7 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.1 | 1.4 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 0.4 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 0.7 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 0.6 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.1 | 0.2 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) termination of mitochondrial transcription(GO:0006393) |
| 0.1 | 0.5 | GO:0002349 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.1 | 1.2 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.1 | 0.2 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.2 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.0 | 0.3 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 | 0.7 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.5 | GO:0022615 | protein to membrane docking(GO:0022615) |
| 0.0 | 0.3 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 1.4 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 1.3 | GO:0007094 | mitotic spindle assembly checkpoint(GO:0007094) |
| 0.0 | 0.2 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.1 | GO:0000239 | pachytene(GO:0000239) |
| 0.0 | 0.5 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.3 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.4 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.3 | GO:1990009 | retinal cell apoptotic process(GO:1990009) |
| 0.0 | 0.6 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.4 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.2 | GO:1903566 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.4 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.2 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.0 | 1.0 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 1.5 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 0.7 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.7 | GO:0007614 | short-term memory(GO:0007614) |
| 0.0 | 0.2 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.4 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.1 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.4 | GO:1904871 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 0.1 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 1.0 | GO:0032435 | negative regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032435) |
| 0.0 | 0.5 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.5 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 1.0 | GO:0050891 | multicellular organismal water homeostasis(GO:0050891) |
| 0.0 | 0.0 | GO:1900104 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.5 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 1.4 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 2.6 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.2 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.3 | GO:0006152 | purine nucleoside catabolic process(GO:0006152) purine ribonucleoside catabolic process(GO:0046130) |
| 0.0 | 0.1 | GO:0060434 | bronchus morphogenesis(GO:0060434) |
| 0.0 | 0.3 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 1.9 | GO:0050868 | negative regulation of T cell activation(GO:0050868) |
| 0.0 | 0.2 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.8 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.5 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.3 | 0.9 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.2 | 1.3 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.2 | 0.6 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 0.4 | GO:0060205 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.1 | 1.0 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 1.9 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.3 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.1 | 1.9 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 0.3 | GO:0034066 | RIC1-RGP1 guanyl-nucleotide exchange factor complex(GO:0034066) |
| 0.1 | 0.5 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.1 | 0.3 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.1 | 0.7 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 1.7 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.5 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 2.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.5 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.3 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 0.7 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.9 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.3 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 1.0 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 2.3 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.0 | 0.7 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.1 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.3 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.0 | 0.1 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 1.9 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 1.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.2 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 1.4 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0035870 | dITP diphosphatase activity(GO:0035870) |
| 0.4 | 1.5 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.3 | 1.4 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.2 | 0.7 | GO:0005135 | interleukin-3 receptor binding(GO:0005135) |
| 0.2 | 1.7 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.2 | 0.7 | GO:1990931 | oxidative DNA demethylase activity(GO:0035516) RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.2 | 0.6 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.2 | 2.4 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.2 | 0.5 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.2 | 0.6 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.1 | 0.5 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.1 | 0.5 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.1 | 0.4 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 0.5 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 1.8 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.4 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 1.8 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.6 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 0.4 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 0.7 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 2.0 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 1.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 0.6 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.1 | 1.6 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.1 | 3.8 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 0.3 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.1 | 1.0 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 0.5 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.1 | 0.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.7 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.3 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.1 | 0.2 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 0.6 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 0.3 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.1 | 0.2 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.1 | 0.9 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.3 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.3 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.1 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.0 | 0.2 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.3 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.3 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.4 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 1.1 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.2 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.7 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.4 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 2.4 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 0.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 2.2 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.0 | 0.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 2.0 | GO:0005253 | anion channel activity(GO:0005253) |
| 0.0 | 0.9 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.9 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 1.1 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.6 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.0 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.7 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 1.8 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 1.5 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.9 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.5 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 1.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.6 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 1.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.5 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 1.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.6 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.9 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.8 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |