Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Clock
Z-value: 1.00
Transcription factors associated with Clock
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Clock
|
ENSMUSG00000029238.12 | Clock |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Clock | mm39_v1_chr5_-_76452365_76452398 | -0.05 | 6.5e-01 | Click! |
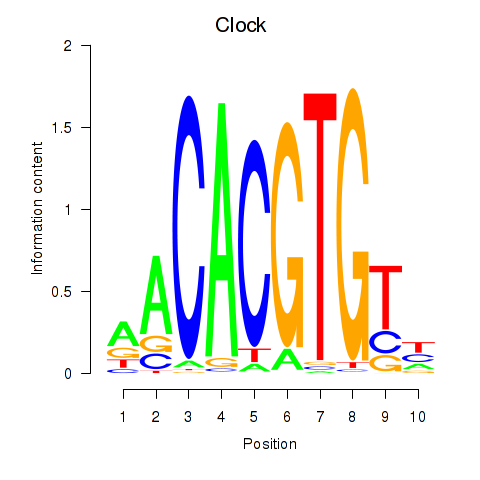
Activity profile of Clock motif
Sorted Z-values of Clock motif
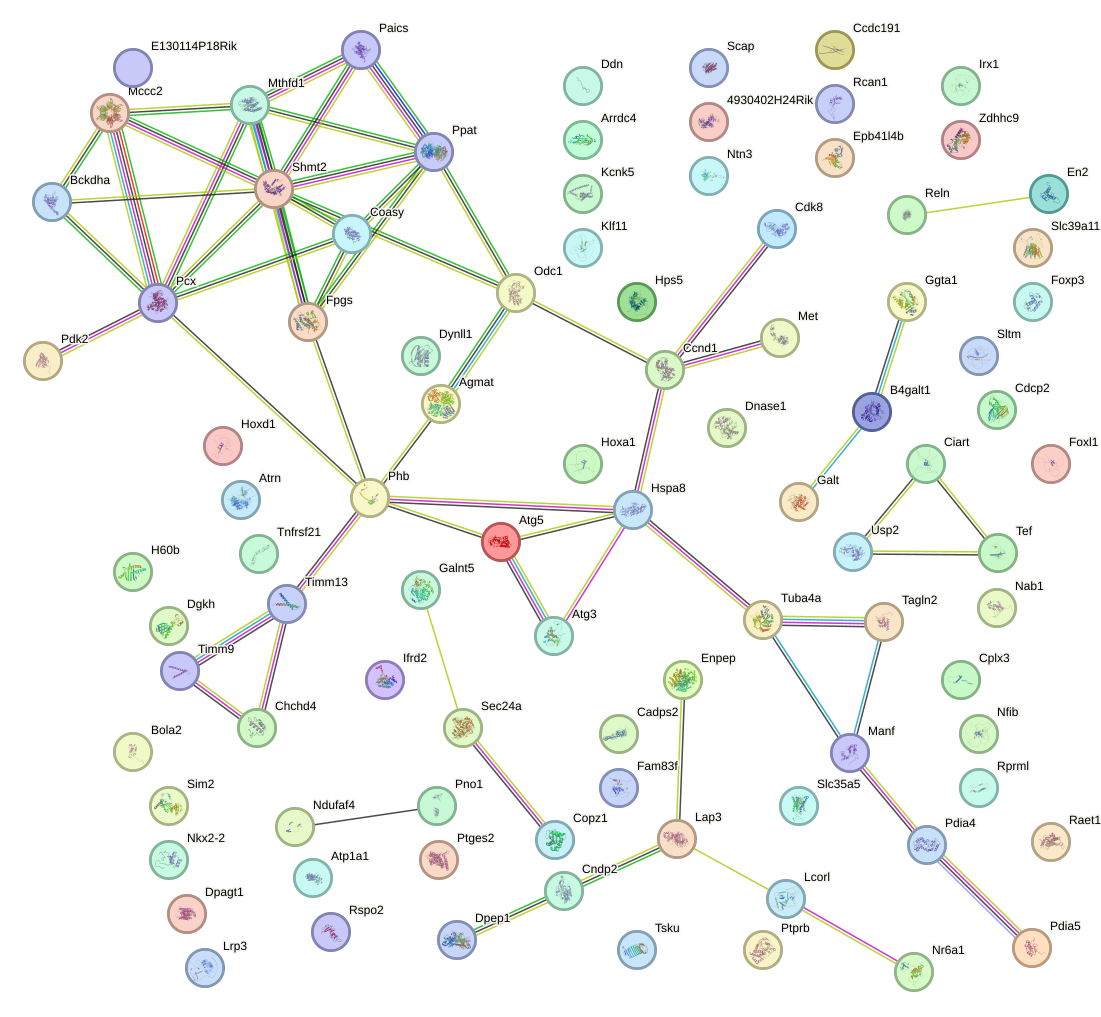
Network of associatons between targets according to the STRING database.
First level regulatory network of Clock
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_57143437 | 7.91 |
ENSMUST00000095076.10
ENSMUST00000030142.4 |
Epb41l4b
|
erythrocyte membrane protein band 4.1 like 4b |
| chr3_-_129126362 | 6.87 |
ENSMUST00000029658.14
|
Enpep
|
glutamyl aminopeptidase |
| chr19_+_4560500 | 6.39 |
ENSMUST00000068004.13
ENSMUST00000224726.3 |
Pcx
|
pyruvate carboxylase |
| chr6_+_17463925 | 6.09 |
ENSMUST00000115442.8
|
Met
|
met proto-oncogene |
| chr6_+_17463748 | 5.56 |
ENSMUST00000115443.8
|
Met
|
met proto-oncogene |
| chr14_-_20231871 | 5.46 |
ENSMUST00000024011.10
|
Kcnk5
|
potassium channel, subfamily K, member 5 |
| chr3_-_95789505 | 5.43 |
ENSMUST00000159863.2
ENSMUST00000159739.8 ENSMUST00000036418.10 ENSMUST00000161866.8 |
Ciart
|
circadian associated repressor of transcription |
| chr5_+_45650821 | 5.19 |
ENSMUST00000198534.2
|
Lap3
|
leucine aminopeptidase 3 |
| chr7_-_98010478 | 5.01 |
ENSMUST00000094161.11
ENSMUST00000164726.8 ENSMUST00000206414.2 ENSMUST00000167405.3 |
Tsku
|
tsukushi, small leucine rich proteoglycan |
| chr7_-_144493560 | 4.78 |
ENSMUST00000093962.5
|
Ccnd1
|
cyclin D1 |
| chr4_+_141473983 | 4.68 |
ENSMUST00000038161.5
|
Agmat
|
agmatine ureohydrolase (agmatinase) |
| chr12_+_76302145 | 4.52 |
ENSMUST00000220046.2
|
Mthfd1
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent), methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthase |
| chr12_+_76302064 | 4.43 |
ENSMUST00000021443.7
|
Mthfd1
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent), methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthase |
| chr5_+_45650716 | 4.37 |
ENSMUST00000046122.11
|
Lap3
|
leucine aminopeptidase 3 |
| chr7_-_98010534 | 4.19 |
ENSMUST00000165257.8
|
Tsku
|
tsukushi, small leucine rich proteoglycan |
| chr6_-_23839136 | 4.10 |
ENSMUST00000166458.9
ENSMUST00000142913.9 ENSMUST00000069074.14 ENSMUST00000115361.9 ENSMUST00000018122.14 ENSMUST00000115356.3 |
Cadps2
|
Ca2+-dependent activator protein for secretion 2 |
| chr6_+_17463819 | 4.06 |
ENSMUST00000140070.8
|
Met
|
met proto-oncogene |
| chr15_+_81695615 | 3.97 |
ENSMUST00000023024.8
|
Tef
|
thyrotroph embryonic factor |
| chr9_-_106769069 | 3.91 |
ENSMUST00000160503.4
ENSMUST00000159620.9 |
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr16_+_3854806 | 3.88 |
ENSMUST00000137748.8
ENSMUST00000006136.11 ENSMUST00000157044.8 ENSMUST00000120009.8 |
Dnase1
|
deoxyribonuclease I |
| chr5_-_22549688 | 3.88 |
ENSMUST00000062372.14
ENSMUST00000161356.8 |
Reln
|
reelin |
| chr6_-_47790272 | 3.57 |
ENSMUST00000077290.9
|
Pdia4
|
protein disulfide isomerase associated 4 |
| chr9_-_106769131 | 3.46 |
ENSMUST00000159283.8
|
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr3_-_95789338 | 3.42 |
ENSMUST00000161994.2
|
Ciart
|
circadian associated repressor of transcription |
| chr2_-_32584132 | 3.21 |
ENSMUST00000028148.11
|
Fpgs
|
folylpolyglutamyl synthetase |
| chr16_-_35311243 | 3.16 |
ENSMUST00000023550.9
|
Pdia5
|
protein disulfide isomerase associated 5 |
| chr11_-_113456568 | 3.12 |
ENSMUST00000071539.10
ENSMUST00000106633.10 ENSMUST00000042657.16 ENSMUST00000149034.8 |
Slc39a11
|
solute carrier family 39 (metal ion transporter), member 11 |
| chr10_+_22034460 | 3.11 |
ENSMUST00000181645.8
ENSMUST00000105522.9 |
Raet1e
H60b
|
retinoic acid early transcript 1E histocompatibility 60b |
| chr18_-_84703738 | 3.05 |
ENSMUST00000025546.17
|
Cndp2
|
CNDP dipeptidase 2 (metallopeptidase M20 family) |
| chr17_-_87573294 | 3.02 |
ENSMUST00000145895.8
ENSMUST00000129616.8 ENSMUST00000155904.2 ENSMUST00000151155.8 ENSMUST00000144236.9 ENSMUST00000024963.11 |
Mcfd2
|
multiple coagulation factor deficiency 2 |
| chr3_+_135531548 | 2.97 |
ENSMUST00000167390.8
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8 |
| chr11_-_95966407 | 2.85 |
ENSMUST00000107686.8
ENSMUST00000107684.2 |
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 (subunit 9) |
| chr7_-_25358406 | 2.83 |
ENSMUST00000071329.8
|
Bckdha
|
branched chain ketoacid dehydrogenase E1, alpha polypeptide |
| chr13_-_72111832 | 2.82 |
ENSMUST00000077337.9
|
Irx1
|
Iroquois homeobox 1 |
| chr6_-_23839419 | 2.72 |
ENSMUST00000115358.9
ENSMUST00000163871.9 |
Cadps2
|
Ca2+-dependent activator protein for secretion 2 |
| chrX_-_47297436 | 2.70 |
ENSMUST00000037960.11
|
Zdhhc9
|
zinc finger, DHHC domain containing 9 |
| chr3_+_135531409 | 2.69 |
ENSMUST00000180196.8
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8 |
| chr13_-_100152069 | 2.66 |
ENSMUST00000022148.7
|
Mccc2
|
methylcrotonoyl-Coenzyme A carboxylase 2 (beta) |
| chr3_+_135531834 | 2.56 |
ENSMUST00000029810.6
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8 |
| chr10_-_127358231 | 2.52 |
ENSMUST00000219239.2
|
Shmt2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr10_+_116137277 | 2.50 |
ENSMUST00000092167.7
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr7_-_68398917 | 2.39 |
ENSMUST00000118110.3
|
Arrdc4
|
arrestin domain containing 4 |
| chr10_-_127358300 | 2.32 |
ENSMUST00000026470.6
|
Shmt2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr6_-_91450486 | 2.31 |
ENSMUST00000040835.9
|
Chchd4
|
coiled-coil-helix-coiled-coil-helix domain containing 4 |
| chr3_-_101511971 | 2.19 |
ENSMUST00000036493.8
|
Atp1a1
|
ATPase, Na+/K+ transporting, alpha 1 polypeptide |
| chr11_-_94932158 | 2.13 |
ENSMUST00000038431.8
|
Pdk2
|
pyruvate dehydrogenase kinase, isoenzyme 2 |
| chr17_+_43327412 | 2.06 |
ENSMUST00000024708.6
|
Tnfrsf21
|
tumor necrosis factor receptor superfamily, member 21 |
| chr10_+_69048914 | 2.06 |
ENSMUST00000163497.8
ENSMUST00000164212.8 ENSMUST00000067908.14 |
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr17_-_28705055 | 2.06 |
ENSMUST00000233870.2
|
Fkbp5
|
FK506 binding protein 5 |
| chr15_-_98705791 | 2.05 |
ENSMUST00000075444.8
|
Ddn
|
dendrin |
| chr5_+_77099229 | 2.04 |
ENSMUST00000141687.2
|
Paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoribosylaminoimidazole, succinocarboxamide synthetase |
| chr11_-_51647290 | 2.00 |
ENSMUST00000109097.9
|
Sec24a
|
Sec24 related gene family, member A (S. cerevisiae) |
| chr16_+_44979086 | 1.99 |
ENSMUST00000023343.4
|
Atg3
|
autophagy related 3 |
| chr11_+_95557783 | 1.99 |
ENSMUST00000125172.8
ENSMUST00000036374.6 |
Phb
|
prohibitin |
| chr4_-_82623972 | 1.99 |
ENSMUST00000155821.2
|
Nfib
|
nuclear factor I/B |
| chr16_-_92262969 | 1.99 |
ENSMUST00000232239.2
ENSMUST00000060005.15 |
Rcan1
|
regulator of calcineurin 1 |
| chr11_-_95966477 | 1.98 |
ENSMUST00000090541.12
|
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 (subunit 9) |
| chr17_-_28705082 | 1.97 |
ENSMUST00000079413.11
|
Fkbp5
|
FK506 binding protein 5 |
| chr10_+_69048464 | 1.97 |
ENSMUST00000020101.12
|
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr9_+_107464841 | 1.95 |
ENSMUST00000010192.11
|
Ifrd2
|
interferon-related developmental regulator 2 |
| chr9_+_40712562 | 1.95 |
ENSMUST00000117557.8
|
Hspa8
|
heat shock protein 8 |
| chr7_-_34914675 | 1.94 |
ENSMUST00000118444.3
ENSMUST00000122409.8 |
Lrp3
|
low density lipoprotein receptor-related protein 3 |
| chr2_+_121279842 | 1.92 |
ENSMUST00000110615.8
ENSMUST00000099475.12 |
Serf2
|
small EDRK-rich factor 2 |
| chr9_+_110162345 | 1.91 |
ENSMUST00000198976.5
|
Scap
|
SREBF chaperone |
| chr9_-_106768601 | 1.90 |
ENSMUST00000069036.14
|
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr10_+_69048506 | 1.82 |
ENSMUST00000167384.8
|
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr5_+_28370687 | 1.79 |
ENSMUST00000036177.9
|
En2
|
engrailed 2 |
| chr5_-_77099406 | 1.76 |
ENSMUST00000140076.2
|
Ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr11_-_51647204 | 1.76 |
ENSMUST00000109092.8
ENSMUST00000064297.5 |
Sec24a
|
Sec24 related gene family, member A (S. cerevisiae) |
| chr10_+_44144346 | 1.72 |
ENSMUST00000039286.5
|
Atg5
|
autophagy related 5 |
| chr5_+_77099154 | 1.67 |
ENSMUST00000031160.16
ENSMUST00000120912.8 ENSMUST00000117536.8 |
Paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoribosylaminoimidazole, succinocarboxamide synthetase |
| chr9_+_110162408 | 1.55 |
ENSMUST00000098350.10
|
Scap
|
SREBF chaperone |
| chr9_+_110162470 | 1.53 |
ENSMUST00000198761.5
ENSMUST00000197630.3 |
Scap
|
SREBF chaperone |
| chr5_-_74863514 | 1.51 |
ENSMUST00000117388.8
|
Lnx1
|
ligand of numb-protein X 1 |
| chr15_+_103181311 | 1.51 |
ENSMUST00000100162.5
ENSMUST00000230893.2 |
Copz1
|
coatomer protein complex, subunit zeta 1 |
| chr4_+_24898074 | 1.48 |
ENSMUST00000029925.10
ENSMUST00000151249.2 |
Ndufaf4
|
NADH:ubiquinone oxidoreductase complex assembly factor 4 |
| chr1_-_52539395 | 1.46 |
ENSMUST00000186764.7
|
Nab1
|
Ngfi-A binding protein 1 |
| chr8_+_123912976 | 1.44 |
ENSMUST00000019422.6
|
Dpep1
|
dipeptidase 1 |
| chr15_+_80556023 | 1.43 |
ENSMUST00000023044.7
|
Fam83f
|
family with sequence similarity 83, member F |
| chr4_+_41755208 | 1.42 |
ENSMUST00000084695.11
ENSMUST00000108038.2 |
Galt
|
galactose-1-phosphate uridyl transferase |
| chr7_-_68398989 | 1.42 |
ENSMUST00000048068.15
|
Arrdc4
|
arrestin domain containing 4 |
| chr12_+_17594795 | 1.40 |
ENSMUST00000171737.3
|
Odc1
|
ornithine decarboxylase, structural 1 |
| chr1_-_75195127 | 1.37 |
ENSMUST00000079464.13
|
Tuba4a
|
tubulin, alpha 4A |
| chr16_+_93885775 | 1.34 |
ENSMUST00000072182.9
|
Sim2
|
single-minded family bHLH transcription factor 2 |
| chr7_-_46445305 | 1.31 |
ENSMUST00000107653.8
ENSMUST00000107654.8 ENSMUST00000014562.14 ENSMUST00000152759.8 |
Hps5
|
HPS5, biogenesis of lysosomal organelles complex 2 subunit 2 |
| chr16_-_44978929 | 1.26 |
ENSMUST00000181177.2
|
Slc35a5
|
solute carrier family 35, member A5 |
| chr2_+_57887896 | 1.25 |
ENSMUST00000112616.8
ENSMUST00000166729.2 |
Galnt5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr6_-_52135261 | 1.21 |
ENSMUST00000000964.6
ENSMUST00000120363.2 |
Hoxa1
|
homeobox A1 |
| chr9_+_43978290 | 1.20 |
ENSMUST00000034508.14
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr2_-_38816229 | 1.17 |
ENSMUST00000076275.11
ENSMUST00000142130.2 |
Nr6a1
|
nuclear receptor subfamily 6, group A, member 1 |
| chr2_-_130748259 | 1.16 |
ENSMUST00000128176.2
ENSMUST00000133766.2 ENSMUST00000135110.2 ENSMUST00000146975.2 |
A730017L22Rik
4930402H24Rik
|
RIKEN cDNA A730017L22 gene RIKEN cDNA 4930402H24 gene |
| chr9_-_57513510 | 1.13 |
ENSMUST00000215487.2
ENSMUST00000045068.10 |
Cplx3
|
complexin 3 |
| chr12_+_24701273 | 1.12 |
ENSMUST00000020982.7
|
Klf11
|
Kruppel-like factor 11 |
| chr16_-_44978816 | 1.11 |
ENSMUST00000181750.2
|
Slc35a5
|
solute carrier family 35, member A5 |
| chr5_-_46014809 | 1.09 |
ENSMUST00000190036.7
ENSMUST00000189859.7 ENSMUST00000186633.3 ENSMUST00000016026.14 ENSMUST00000045586.13 ENSMUST00000238522.2 |
Lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr8_+_121854566 | 1.07 |
ENSMUST00000181609.2
|
Foxl1
|
forkhead box L1 |
| chr7_+_46445512 | 1.06 |
ENSMUST00000006774.11
ENSMUST00000165031.8 |
Gtf2h1
|
general transcription factor II H, polypeptide 1 |
| chr2_+_130748380 | 1.05 |
ENSMUST00000028781.9
|
Atrn
|
attractin |
| chr7_-_46445085 | 1.02 |
ENSMUST00000123725.2
|
Hps5
|
HPS5, biogenesis of lysosomal organelles complex 2 subunit 2 |
| chr2_+_32285908 | 1.00 |
ENSMUST00000028162.5
|
Ptges2
|
prostaglandin E synthase 2 |
| chrX_+_7439839 | 0.96 |
ENSMUST00000144719.9
ENSMUST00000234896.2 |
Flicr
Foxp3
|
Foxp3 regulating long intergenic noncoding RNA forkhead box P3 |
| chr5_+_146168020 | 0.96 |
ENSMUST00000161181.8
ENSMUST00000161652.8 ENSMUST00000031640.15 ENSMUST00000159467.2 |
Cdk8
|
cyclin-dependent kinase 8 |
| chr16_+_43710299 | 0.91 |
ENSMUST00000151183.2
|
Ccdc191
|
coiled-coil domain containing 191 |
| chr16_-_44978986 | 0.90 |
ENSMUST00000180636.8
|
Slc35a5
|
solute carrier family 35, member A5 |
| chr1_+_172327812 | 0.88 |
ENSMUST00000192460.2
|
Tagln2
|
transgelin 2 |
| chr14_-_78970160 | 0.88 |
ENSMUST00000226342.3
|
Dgkh
|
diacylglycerol kinase, eta |
| chr9_+_44238089 | 0.85 |
ENSMUST00000054708.5
|
Dpagt1
|
dolichyl-phosphate (UDP-N-acetylglucosamine) acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase) |
| chr2_-_147028309 | 0.85 |
ENSMUST00000067075.7
|
Nkx2-2
|
NK2 homeobox 2 |
| chr16_-_44979013 | 0.83 |
ENSMUST00000023344.10
|
Slc35a5
|
solute carrier family 35, member A5 |
| chr17_-_24428351 | 0.83 |
ENSMUST00000024931.6
|
Ntn3
|
netrin 3 |
| chr8_-_37081091 | 0.81 |
ENSMUST00000033923.14
|
Dlc1
|
deleted in liver cancer 1 |
| chr2_+_74593324 | 0.81 |
ENSMUST00000047793.6
|
Hoxd1
|
homeobox D1 |
| chr12_-_71183371 | 0.80 |
ENSMUST00000221367.2
ENSMUST00000220482.2 ENSMUST00000221892.2 ENSMUST00000221178.2 ENSMUST00000221559.2 ENSMUST00000166120.9 ENSMUST00000021486.10 ENSMUST00000221797.2 ENSMUST00000221815.2 |
Timm9
|
translocase of inner mitochondrial membrane 9 |
| chr1_+_172328768 | 0.79 |
ENSMUST00000111228.2
|
Tagln2
|
transgelin 2 |
| chr15_-_43034205 | 0.79 |
ENSMUST00000063492.8
ENSMUST00000226810.2 |
Rspo2
|
R-spondin 2 |
| chr11_-_17161504 | 0.77 |
ENSMUST00000020317.8
|
Pno1
|
partner of NOB1 homolog |
| chr4_-_97666279 | 0.75 |
ENSMUST00000146447.8
|
E130114P18Rik
|
RIKEN cDNA E130114P18 gene |
| chr11_+_103540391 | 0.75 |
ENSMUST00000057870.4
|
Rprml
|
reprimo-like |
| chr4_+_106954794 | 0.75 |
ENSMUST00000221740.2
|
Cdcp2
|
CUB domain containing protein 2 |
| chr7_+_126295114 | 0.72 |
ENSMUST00000106369.2
|
Bola2
|
bolA-like 2 (E. coli) |
| chr1_-_186437760 | 0.71 |
ENSMUST00000195201.2
|
Tgfb2
|
transforming growth factor, beta 2 |
| chr13_+_20369377 | 0.68 |
ENSMUST00000221067.2
|
Elmo1
|
engulfment and cell motility 1 |
| chr13_-_63712140 | 0.66 |
ENSMUST00000195756.6
|
Ptch1
|
patched 1 |
| chr9_+_43978369 | 0.66 |
ENSMUST00000177054.8
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr3_-_58599812 | 0.66 |
ENSMUST00000070368.8
|
Siah2
|
siah E3 ubiquitin protein ligase 2 |
| chr12_+_102095260 | 0.64 |
ENSMUST00000079020.12
|
Slc24a4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chr3_+_41519289 | 0.64 |
ENSMUST00000168086.7
|
Jade1
|
jade family PHD finger 1 |
| chr2_-_76720097 | 0.62 |
ENSMUST00000148747.8
|
Ttn
|
titin |
| chr17_-_45883421 | 0.60 |
ENSMUST00000130406.2
|
Hsp90ab1
|
heat shock protein 90 alpha (cytosolic), class B member 1 |
| chr8_-_49296915 | 0.58 |
ENSMUST00000211812.2
|
Tenm3
|
teneurin transmembrane protein 3 |
| chr4_-_131802606 | 0.57 |
ENSMUST00000146021.8
|
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr17_+_71511642 | 0.56 |
ENSMUST00000126681.8
|
Lpin2
|
lipin 2 |
| chr18_-_24842405 | 0.52 |
ENSMUST00000067987.2
|
Gm9955
|
predicted gene 9955 |
| chr13_-_63712349 | 0.52 |
ENSMUST00000192155.6
|
Ptch1
|
patched 1 |
| chr4_-_131802561 | 0.51 |
ENSMUST00000105970.8
ENSMUST00000105975.8 |
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr11_+_74540284 | 0.51 |
ENSMUST00000117818.2
ENSMUST00000092915.12 |
Cluh
|
clustered mitochondria (cluA/CLU1) homolog |
| chr11_+_60308077 | 0.50 |
ENSMUST00000070681.7
|
Gid4
|
GID complex subunit 4, VID24 homolog |
| chr5_-_74692327 | 0.46 |
ENSMUST00000072857.13
ENSMUST00000113542.9 ENSMUST00000151474.3 |
Scfd2
|
Sec1 family domain containing 2 |
| chr18_+_58969739 | 0.46 |
ENSMUST00000052907.7
|
Adamts19
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 19 |
| chr16_+_23247883 | 0.45 |
ENSMUST00000038730.7
|
Rtp1
|
receptor transporter protein 1 |
| chr3_+_41519355 | 0.45 |
ENSMUST00000191952.2
|
Jade1
|
jade family PHD finger 1 |
| chr5_+_77122530 | 0.44 |
ENSMUST00000101087.10
ENSMUST00000120550.2 |
Srp72
|
signal recognition particle 72 |
| chr2_+_31777273 | 0.43 |
ENSMUST00000138325.8
ENSMUST00000028187.7 |
Lamc3
|
laminin gamma 3 |
| chr16_-_44978546 | 0.42 |
ENSMUST00000114600.2
|
Slc35a5
|
solute carrier family 35, member A5 |
| chr11_+_94520567 | 0.42 |
ENSMUST00000021239.7
|
Lrrc59
|
leucine rich repeat containing 59 |
| chr1_-_186438177 | 0.41 |
ENSMUST00000045288.14
|
Tgfb2
|
transforming growth factor, beta 2 |
| chr11_+_101207021 | 0.41 |
ENSMUST00000142640.8
ENSMUST00000019470.14 |
Psme3
|
proteaseome (prosome, macropain) activator subunit 3 (PA28 gamma, Ki) |
| chr11_-_106107132 | 0.40 |
ENSMUST00000002043.10
|
Ccdc47
|
coiled-coil domain containing 47 |
| chr17_+_71923210 | 0.38 |
ENSMUST00000047086.10
|
Wdr43
|
WD repeat domain 43 |
| chr2_-_48839218 | 0.38 |
ENSMUST00000090976.10
ENSMUST00000149679.8 |
Orc4
|
origin recognition complex, subunit 4 |
| chr10_+_69369854 | 0.38 |
ENSMUST00000182557.8
|
Ank3
|
ankyrin 3, epithelial |
| chr2_+_160487801 | 0.38 |
ENSMUST00000109468.3
|
Top1
|
topoisomerase (DNA) I |
| chr12_+_59178072 | 0.38 |
ENSMUST00000176464.8
ENSMUST00000170992.9 ENSMUST00000176322.8 |
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr11_+_67586140 | 0.36 |
ENSMUST00000021290.2
|
Rcvrn
|
recoverin |
| chr11_-_101676076 | 0.36 |
ENSMUST00000164750.8
ENSMUST00000107176.8 ENSMUST00000017868.7 |
Etv4
|
ets variant 4 |
| chrX_+_5959507 | 0.35 |
ENSMUST00000103007.4
|
Nudt11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11 |
| chr11_-_70873773 | 0.33 |
ENSMUST00000078528.7
|
C1qbp
|
complement component 1, q subcomponent binding protein |
| chr14_+_4230658 | 0.31 |
ENSMUST00000225491.2
|
Nr1d2
|
nuclear receptor subfamily 1, group D, member 2 |
| chr2_-_48839276 | 0.31 |
ENSMUST00000028098.11
|
Orc4
|
origin recognition complex, subunit 4 |
| chr11_-_100650566 | 0.30 |
ENSMUST00000107361.9
|
Kcnh4
|
potassium voltage-gated channel, subfamily H (eag-related), member 4 |
| chr9_-_121324744 | 0.28 |
ENSMUST00000035120.6
|
Cck
|
cholecystokinin |
| chr9_+_58041818 | 0.28 |
ENSMUST00000128378.8
ENSMUST00000150820.8 ENSMUST00000167479.8 |
Stra6
|
stimulated by retinoic acid gene 6 |
| chr17_-_45884179 | 0.26 |
ENSMUST00000165127.8
ENSMUST00000166469.8 ENSMUST00000024739.14 |
Hsp90ab1
|
heat shock protein 90 alpha (cytosolic), class B member 1 |
| chr15_-_44651411 | 0.26 |
ENSMUST00000090057.6
ENSMUST00000110269.8 ENSMUST00000228639.2 |
Sybu
|
syntabulin (syntaxin-interacting) |
| chr1_-_75195889 | 0.25 |
ENSMUST00000186213.7
|
Tuba4a
|
tubulin, alpha 4A |
| chr9_-_103243039 | 0.24 |
ENSMUST00000035484.11
|
Cdv3
|
carnitine deficiency-associated gene expressed in ventricle 3 |
| chr4_-_97472844 | 0.22 |
ENSMUST00000107067.8
ENSMUST00000107068.9 |
E130114P18Rik
|
RIKEN cDNA E130114P18 gene |
| chr2_+_48839505 | 0.22 |
ENSMUST00000112745.8
ENSMUST00000112754.8 |
Mbd5
|
methyl-CpG binding domain protein 5 |
| chr18_+_76374651 | 0.22 |
ENSMUST00000168423.9
ENSMUST00000091831.13 |
Smad2
|
SMAD family member 2 |
| chr9_-_121324705 | 0.20 |
ENSMUST00000216138.2
|
Cck
|
cholecystokinin |
| chr4_+_148025316 | 0.20 |
ENSMUST00000103232.2
|
2510039O18Rik
|
RIKEN cDNA 2510039O18 gene |
| chr11_-_42073737 | 0.20 |
ENSMUST00000206085.2
ENSMUST00000020707.12 ENSMUST00000132971.3 |
Gabra1
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1 |
| chr7_+_87233554 | 0.19 |
ENSMUST00000125009.9
|
Grm5
|
glutamate receptor, metabotropic 5 |
| chrX_-_36127891 | 0.19 |
ENSMUST00000115249.10
ENSMUST00000115248.10 |
C330007P06Rik
|
RIKEN cDNA C330007P06 gene |
| chr17_-_90763300 | 0.17 |
ENSMUST00000159778.8
ENSMUST00000174337.8 ENSMUST00000172466.8 |
Nrxn1
|
neurexin I |
| chr12_+_59178258 | 0.16 |
ENSMUST00000177162.8
|
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr11_+_78069528 | 0.16 |
ENSMUST00000108338.2
|
Tlcd1
|
TLC domain containing 1 |
| chr3_+_51131868 | 0.15 |
ENSMUST00000023849.15
ENSMUST00000167780.2 |
Noct
|
nocturnin |
| chr16_-_33787399 | 0.15 |
ENSMUST00000023510.7
|
Umps
|
uridine monophosphate synthetase |
| chr10_+_17598961 | 0.14 |
ENSMUST00000038107.9
ENSMUST00000219558.2 ENSMUST00000218370.2 |
Cited2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
| chr12_+_69418886 | 0.13 |
ENSMUST00000050063.9
|
Arf6
|
ADP-ribosylation factor 6 |
| chr4_+_148675939 | 0.12 |
ENSMUST00000006611.9
|
Srm
|
spermidine synthase |
| chr9_-_48391838 | 0.11 |
ENSMUST00000216470.2
ENSMUST00000217037.2 ENSMUST00000034524.5 ENSMUST00000213895.2 |
Rexo2
|
RNA exonuclease 2 |
| chr11_-_98980122 | 0.09 |
ENSMUST00000017751.3
|
Tns4
|
tensin 4 |
| chr14_-_52628228 | 0.07 |
ENSMUST00000078171.2
|
Olfr1511
|
olfactory receptor 1511 |
| chr7_-_126294902 | 0.07 |
ENSMUST00000144897.2
|
Slx1b
|
SLX1 structure-specific endonuclease subunit homolog B (S. cerevisiae) |
| chr18_+_67597929 | 0.07 |
ENSMUST00000025411.9
|
Prelid3a
|
PRELI domain containing 3A |
| chr6_+_94477294 | 0.06 |
ENSMUST00000061118.11
|
Slc25a26
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 26 |
| chr1_+_181180183 | 0.03 |
ENSMUST00000161880.8
ENSMUST00000027795.14 |
Cnih3
|
cornichon family AMPA receptor auxiliary protein 3 |
| chrX_+_8137881 | 0.03 |
ENSMUST00000115590.2
|
Slc38a5
|
solute carrier family 38, member 5 |
| chr16_+_20430335 | 0.01 |
ENSMUST00000115522.10
ENSMUST00000119224.8 ENSMUST00000120394.8 ENSMUST00000079600.12 |
Eef1akmt4
Gm49333
|
EEF1A lysine methyltransferase 4 predicted gene, 49333 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 9.3 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 2.2 | 15.7 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 2.2 | 9.0 | GO:0000105 | histidine biosynthetic process(GO:0000105) 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 1.8 | 5.5 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 1.6 | 4.8 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 1.5 | 9.2 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 1.3 | 6.4 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 1.1 | 3.2 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 1.0 | 6.9 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 1.0 | 3.9 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.9 | 6.8 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.8 | 4.8 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.8 | 2.3 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.7 | 8.2 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.7 | 3.7 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.7 | 2.8 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.7 | 4.8 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.7 | 7.9 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.5 | 2.1 | GO:0050812 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.5 | 2.1 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.5 | 2.0 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.5 | 1.4 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) glycolytic process from galactose(GO:0061623) |
| 0.4 | 2.2 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.4 | 1.7 | GO:0061739 | aggrephagy(GO:0035973) protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.4 | 1.2 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.4 | 2.0 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.4 | 1.1 | GO:0070237 | positive regulation of activation-induced cell death of T cells(GO:0070237) negative regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905006) |
| 0.4 | 1.4 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.3 | 1.0 | GO:0002654 | positive regulation of tolerance induction dependent upon immune response(GO:0002654) regulation of peripheral tolerance induction(GO:0002658) positive regulation of peripheral tolerance induction(GO:0002660) regulation of peripheral T cell tolerance induction(GO:0002849) positive regulation of peripheral T cell tolerance induction(GO:0002851) |
| 0.3 | 3.8 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.3 | 1.8 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.3 | 0.9 | GO:0036363 | transforming growth factor beta activation(GO:0036363) |
| 0.3 | 10.3 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.3 | 0.9 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.3 | 1.4 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.3 | 5.5 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.3 | 1.1 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.2 | 1.2 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.2 | 2.7 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.2 | 5.0 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.2 | 0.8 | GO:0072321 | protein import into mitochondrial inner membrane(GO:0045039) chaperone-mediated protein transport(GO:0072321) |
| 0.2 | 0.8 | GO:0060437 | lung growth(GO:0060437) trachea cartilage morphogenesis(GO:0060535) |
| 0.1 | 4.8 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 2.8 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 4.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.6 | GO:0043056 | forward locomotion(GO:0043056) |
| 0.1 | 0.9 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 2.0 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 3.9 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.1 | 3.8 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.1 | 1.8 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.1 | 2.7 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.1 | 2.0 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.1 | 3.1 | GO:0001913 | T cell mediated cytotoxicity(GO:0001913) |
| 0.1 | 1.1 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.1 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.1 | 1.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.1 | 3.1 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.1 | 0.3 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.1 | 0.2 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.1 | 0.4 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.1 | 1.1 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 1.1 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 0.2 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.1 | 0.2 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.1 | 0.4 | GO:1901906 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.4 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 1.0 | GO:0090209 | negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.0 | 0.8 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 4.2 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.4 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.0 | 0.4 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.4 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.9 | GO:0046473 | phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.5 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.0 | 1.5 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 1.5 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0006222 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.6 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.4 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.5 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.2 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 0.0 | 1.5 | GO:0014037 | Schwann cell differentiation(GO:0014037) |
| 0.0 | 0.1 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.0 | 0.6 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.0 | 4.5 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.4 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.0 | 0.5 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.0 | 0.8 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 1.8 | GO:0043473 | pigmentation(GO:0043473) |
| 0.0 | 0.5 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 1.3 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.0 | 0.6 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.8 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 1.1 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.5 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.6 | 2.3 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.6 | 1.7 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.5 | 6.9 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.5 | 2.8 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.4 | 1.9 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.3 | 3.1 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.3 | 4.8 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.3 | 3.0 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.3 | 0.8 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.2 | 3.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 2.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.2 | 3.8 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.2 | 2.7 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.2 | 0.9 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.2 | 16.1 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 2.0 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.1 | 1.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 1.5 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 2.0 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.1 | 2.0 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 0.7 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 1.2 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 12.7 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 7.2 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 0.4 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.4 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 8.5 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.3 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.2 | GO:0032444 | activin responsive factor complex(GO:0032444) SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.5 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.7 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 2.4 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 8.8 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.6 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 3.9 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 9.1 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.5 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 3.4 | GO:0005759 | mitochondrial matrix(GO:0005759) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 15.7 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 2.2 | 9.0 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 1.6 | 4.8 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) |
| 1.6 | 6.4 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 1.2 | 3.7 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 1.1 | 3.2 | GO:0004326 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) |
| 0.9 | 2.8 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.7 | 2.0 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.6 | 3.9 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.5 | 2.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.5 | 1.9 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.4 | 4.7 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.4 | 6.7 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.4 | 4.0 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.3 | 11.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.3 | 17.9 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.3 | 2.7 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.3 | 5.5 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.3 | 4.8 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 1.0 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.2 | 2.0 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 0.9 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.2 | 1.4 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.2 | 4.0 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 3.1 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.2 | 1.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.2 | 2.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) steroid hormone binding(GO:1990239) |
| 0.2 | 4.5 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.2 | 1.2 | GO:0008158 | smoothened binding(GO:0005119) hedgehog receptor activity(GO:0008158) |
| 0.2 | 0.3 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.2 | 3.1 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.1 | 0.4 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.1 | 0.9 | GO:0002135 | CTP binding(GO:0002135) |
| 0.1 | 3.7 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.1 | 2.3 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 8.4 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 4.8 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 2.0 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 0.6 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.4 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.1 | 2.7 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 11.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.0 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.2 | GO:0001639 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.1 | 4.5 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 1.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.9 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.4 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 0.5 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 9.1 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 1.1 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.8 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 1.1 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 1.5 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.9 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.2 | GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.3 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.6 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.9 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 5.5 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 2.5 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.9 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 9.0 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 15.8 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.3 | 4.8 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 5.0 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 14.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 2.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 2.0 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.1 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.0 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.0 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 1.5 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.7 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.3 | 15.7 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.3 | 5.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.3 | 8.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.2 | 4.8 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.2 | 5.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 2.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 12.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 1.5 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 4.8 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 3.1 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 1.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.9 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 2.0 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 3.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 0.7 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 3.0 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 0.9 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 3.1 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.1 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 2.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.7 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 6.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.2 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 1.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.9 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.9 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.9 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.7 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 1.2 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.7 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |