Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
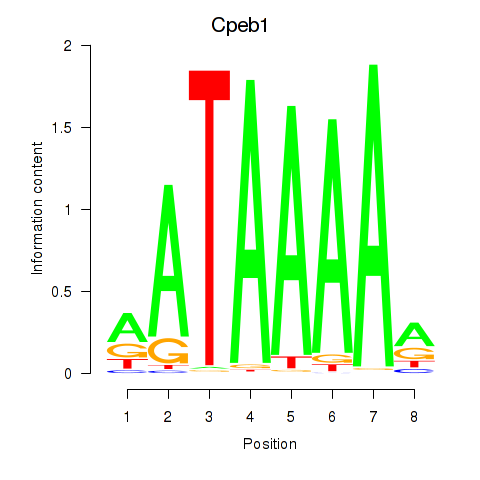
Results for Cpeb1
Z-value: 2.17
Transcription factors associated with Cpeb1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Cpeb1
|
ENSMUSG00000025586.18 | Cpeb1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Cpeb1 | mm39_v1_chr7_-_81104423_81104512 | 0.51 | 4.6e-06 | Click! |
Activity profile of Cpeb1 motif
Sorted Z-values of Cpeb1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Cpeb1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_55825033 | 13.14 |
ENSMUST00000114772.9
ENSMUST00000114768.10 ENSMUST00000155882.8 |
Fhl1
|
four and a half LIM domains 1 |
| chr3_+_106020545 | 13.13 |
ENSMUST00000079132.12
ENSMUST00000139086.2 |
Chia1
|
chitinase, acidic 1 |
| chrX_+_55824797 | 12.70 |
ENSMUST00000114773.10
|
Fhl1
|
four and a half LIM domains 1 |
| chr8_+_94537910 | 12.35 |
ENSMUST00000138659.9
|
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chr3_-_92616380 | 11.61 |
ENSMUST00000090867.2
|
Lce1e
|
late cornified envelope 1E |
| chr3_+_92899521 | 11.35 |
ENSMUST00000090863.5
|
Lce3f
|
late cornified envelope 3F |
| chr3_-_92922976 | 11.21 |
ENSMUST00000107301.2
ENSMUST00000029521.5 |
Crct1
|
cysteine-rich C-terminal 1 |
| chr3_+_68491487 | 11.10 |
ENSMUST00000182997.3
|
Schip1
|
schwannomin interacting protein 1 |
| chr18_-_15284476 | 11.03 |
ENSMUST00000025992.7
|
Kctd1
|
potassium channel tetramerisation domain containing 1 |
| chr3_-_92564232 | 10.94 |
ENSMUST00000029531.4
|
Lce1b
|
late cornified envelope 1B |
| chr7_+_30475819 | 10.60 |
ENSMUST00000041703.10
|
Dmkn
|
dermokine |
| chr14_-_57371041 | 10.39 |
ENSMUST00000039380.9
|
Gjb6
|
gap junction protein, beta 6 |
| chr2_+_109522781 | 10.15 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr3_+_87855973 | 10.10 |
ENSMUST00000005019.6
|
Crabp2
|
cellular retinoic acid binding protein II |
| chr18_+_69726431 | 9.90 |
ENSMUST00000201091.4
ENSMUST00000201037.4 ENSMUST00000114977.5 |
Tcf4
|
transcription factor 4 |
| chr3_-_92833537 | 9.87 |
ENSMUST00000067318.6
|
Lce3a
|
late cornified envelope 3A |
| chr3_-_92715195 | 9.77 |
ENSMUST00000179917.2
|
Lce1k
|
late cornified envelope 1K |
| chr3_-_92594516 | 9.46 |
ENSMUST00000029524.4
|
Lce1d
|
late cornified envelope 1D |
| chr3_-_92577621 | 9.45 |
ENSMUST00000029530.6
|
Lce1a2
|
late cornified envelope 1A2 |
| chr1_-_171023798 | 9.42 |
ENSMUST00000111332.2
|
Pcp4l1
|
Purkinje cell protein 4-like 1 |
| chr19_+_39049442 | 9.35 |
ENSMUST00000087236.5
|
Cyp2c65
|
cytochrome P450, family 2, subfamily c, polypeptide 65 |
| chr5_+_76988444 | 9.32 |
ENSMUST00000120639.9
ENSMUST00000163347.8 ENSMUST00000121851.2 |
Cracd
|
capping protein inhibiting regulator of actin |
| chr3_+_92586546 | 9.23 |
ENSMUST00000047055.4
|
Lce1c
|
late cornified envelope 1C |
| chr3_-_92777521 | 9.14 |
ENSMUST00000163439.3
|
2310050C09Rik
|
RIKEN cDNA 2310050C09 gene |
| chr1_-_79838897 | 8.97 |
ENSMUST00000190724.2
|
Serpine2
|
serine (or cysteine) peptidase inhibitor, clade E, member 2 |
| chr11_-_42073737 | 8.96 |
ENSMUST00000206085.2
ENSMUST00000020707.12 ENSMUST00000132971.3 |
Gabra1
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1 |
| chr3_-_92686198 | 8.94 |
ENSMUST00000090866.2
|
Lce1i
|
late cornified envelope 1I |
| chr10_-_127025851 | 8.91 |
ENSMUST00000222006.2
ENSMUST00000019611.15 ENSMUST00000219245.2 |
Arhgef25
|
Rho guanine nucleotide exchange factor (GEF) 25 |
| chr3_-_92366022 | 8.64 |
ENSMUST00000058142.4
|
Sprr3
|
small proline-rich protein 3 |
| chr19_+_17114108 | 8.46 |
ENSMUST00000223920.2
ENSMUST00000225351.2 |
Prune2
|
prune homolog 2 |
| chr3_-_92659644 | 8.46 |
ENSMUST00000029527.6
|
Lce1g
|
late cornified envelope 1G |
| chr5_+_107645626 | 8.31 |
ENSMUST00000152474.8
ENSMUST00000060553.8 |
Btbd8
|
BTB (POZ) domain containing 8 |
| chr3_-_92627651 | 8.29 |
ENSMUST00000047153.4
|
Lce1f
|
late cornified envelope 1F |
| chr11_-_79394904 | 8.24 |
ENSMUST00000164465.3
|
Omg
|
oligodendrocyte myelin glycoprotein |
| chr16_+_25620652 | 8.15 |
ENSMUST00000115304.8
ENSMUST00000115305.2 ENSMUST00000040231.13 ENSMUST00000115306.8 |
Trp63
|
transformation related protein 63 |
| chr3_+_92851790 | 8.11 |
ENSMUST00000055375.6
|
Lce3c
|
late cornified envelope 3C |
| chr18_+_20443795 | 8.07 |
ENSMUST00000077146.4
|
Dsg1a
|
desmoglein 1 alpha |
| chr1_-_169575203 | 8.02 |
ENSMUST00000027991.12
ENSMUST00000111357.2 |
Rgs4
|
regulator of G-protein signaling 4 |
| chr5_+_149335214 | 7.64 |
ENSMUST00000093110.12
|
Medag
|
mesenteric estrogen dependent adipogenesis |
| chr3_+_93227047 | 7.61 |
ENSMUST00000090856.10
ENSMUST00000093774.4 |
Hrnr
|
hornerin |
| chr3_-_63758672 | 7.59 |
ENSMUST00000162269.9
ENSMUST00000159676.9 ENSMUST00000175947.8 |
Plch1
|
phospholipase C, eta 1 |
| chr10_+_29087658 | 7.58 |
ENSMUST00000213489.2
|
9330159F19Rik
|
RIKEN cDNA 9330159F19 gene |
| chr3_-_92697821 | 7.43 |
ENSMUST00000186525.2
|
Lce1j
|
late cornified envelope 1J |
| chr10_+_5589210 | 7.34 |
ENSMUST00000019906.6
|
Vip
|
vasoactive intestinal polypeptide |
| chr10_+_29087602 | 7.19 |
ENSMUST00000092627.6
|
9330159F19Rik
|
RIKEN cDNA 9330159F19 gene |
| chr11_+_67477347 | 7.19 |
ENSMUST00000108682.9
|
Gas7
|
growth arrest specific 7 |
| chr3_-_92672367 | 7.06 |
ENSMUST00000051521.5
|
Lce1h
|
late cornified envelope 1H |
| chr2_-_110781268 | 6.95 |
ENSMUST00000099623.10
|
Ano3
|
anoctamin 3 |
| chr19_-_5399368 | 6.94 |
ENSMUST00000238111.2
|
Cst6
|
cystatin E/M |
| chr3_+_93184854 | 6.89 |
ENSMUST00000180308.3
|
Flg
|
filaggrin |
| chr11_+_67477501 | 6.87 |
ENSMUST00000108680.2
|
Gas7
|
growth arrest specific 7 |
| chrX_-_94209913 | 6.86 |
ENSMUST00000113873.9
ENSMUST00000113876.9 ENSMUST00000199920.5 ENSMUST00000113885.8 ENSMUST00000113883.8 ENSMUST00000196012.2 ENSMUST00000182001.8 ENSMUST00000113878.8 ENSMUST00000113882.8 ENSMUST00000182562.2 |
Arhgef9
|
CDC42 guanine nucleotide exchange factor (GEF) 9 |
| chr11_-_42072990 | 6.84 |
ENSMUST00000205546.2
|
Gabra1
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1 |
| chr6_-_136852792 | 6.72 |
ENSMUST00000032342.3
|
Mgp
|
matrix Gla protein |
| chr10_+_70276604 | 6.72 |
ENSMUST00000173042.9
ENSMUST00000062883.7 |
Fam13c
|
family with sequence similarity 13, member C |
| chr11_+_104467791 | 6.70 |
ENSMUST00000106957.8
|
Myl4
|
myosin, light polypeptide 4 |
| chr15_-_101862711 | 6.66 |
ENSMUST00000164932.3
|
Krt78
|
keratin 78 |
| chr11_-_118246566 | 6.66 |
ENSMUST00000155707.3
|
Timp2
|
tissue inhibitor of metalloproteinase 2 |
| chr19_+_29902506 | 6.46 |
ENSMUST00000120388.9
ENSMUST00000144528.8 ENSMUST00000177518.8 |
Il33
|
interleukin 33 |
| chr11_+_42312150 | 6.36 |
ENSMUST00000192403.2
|
Gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 2 |
| chr8_+_40807344 | 6.21 |
ENSMUST00000136835.2
|
Micu3
|
mitochondrial calcium uptake family, member 3 |
| chr19_+_34078333 | 6.13 |
ENSMUST00000025685.8
|
Lipm
|
lipase, family member M |
| chrX_+_133618693 | 6.07 |
ENSMUST00000113201.8
ENSMUST00000051256.10 ENSMUST00000113199.8 ENSMUST00000035748.14 ENSMUST00000113198.8 ENSMUST00000113197.2 |
Armcx1
|
armadillo repeat containing, X-linked 1 |
| chr3_+_92874366 | 6.05 |
ENSMUST00000098886.5
|
Lce3e
|
late cornified envelope 3E |
| chr2_+_83642910 | 5.86 |
ENSMUST00000051454.4
|
Fam171b
|
family with sequence similarity 171, member B |
| chr11_+_100902572 | 5.84 |
ENSMUST00000092663.4
|
Atp6v0a1
|
ATPase, H+ transporting, lysosomal V0 subunit A1 |
| chr1_-_22031718 | 5.76 |
ENSMUST00000029667.13
ENSMUST00000173058.8 ENSMUST00000173404.2 |
Kcnq5
|
potassium voltage-gated channel, subfamily Q, member 5 |
| chr3_+_92864693 | 5.75 |
ENSMUST00000059053.11
|
Lce3d
|
late cornified envelope 3D |
| chr8_+_94537460 | 5.73 |
ENSMUST00000034198.15
ENSMUST00000125716.8 |
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chr11_+_104468107 | 5.72 |
ENSMUST00000106956.10
|
Myl4
|
myosin, light polypeptide 4 |
| chr18_+_69726654 | 5.60 |
ENSMUST00000200921.4
|
Tcf4
|
transcription factor 4 |
| chr3_+_113824181 | 5.52 |
ENSMUST00000123619.8
ENSMUST00000092155.12 |
Col11a1
|
collagen, type XI, alpha 1 |
| chr12_-_90705212 | 5.52 |
ENSMUST00000082432.6
|
Dio2
|
deiodinase, iodothyronine, type II |
| chr3_+_102641822 | 5.43 |
ENSMUST00000029451.12
|
Tspan2
|
tetraspanin 2 |
| chr14_-_28691423 | 5.42 |
ENSMUST00000225985.2
|
Cacna2d3
|
calcium channel, voltage-dependent, alpha2/delta subunit 3 |
| chr18_+_69652837 | 5.41 |
ENSMUST00000201410.4
ENSMUST00000202937.4 |
Tcf4
|
transcription factor 4 |
| chr3_-_59170245 | 5.39 |
ENSMUST00000050360.14
ENSMUST00000199609.2 |
P2ry12
|
purinergic receptor P2Y, G-protein coupled 12 |
| chr16_+_96001865 | 5.38 |
ENSMUST00000171181.9
ENSMUST00000233818.2 ENSMUST00000233945.2 ENSMUST00000166952.8 |
Sh3bgr
|
SH3-binding domain glutamic acid-rich protein |
| chr18_+_44237577 | 5.37 |
ENSMUST00000239465.2
|
Spink12
|
serine peptidase inhibitor, Kazal type 12 |
| chr11_-_42072920 | 5.36 |
ENSMUST00000207274.2
|
Gabra1
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1 |
| chr13_+_93441447 | 5.31 |
ENSMUST00000109497.8
ENSMUST00000109498.8 ENSMUST00000060490.11 ENSMUST00000109492.9 ENSMUST00000109496.8 ENSMUST00000109495.8 |
Homer1
|
homer scaffolding protein 1 |
| chr3_-_92926364 | 5.30 |
ENSMUST00000193944.2
ENSMUST00000029520.9 |
Lce1m
|
late cornified envelope 1M |
| chr3_-_92393193 | 5.29 |
ENSMUST00000054599.8
|
Sprr1a
|
small proline-rich protein 1A |
| chr3_-_106126794 | 5.24 |
ENSMUST00000082219.6
|
Chil4
|
chitinase-like 4 |
| chr17_+_72088281 | 5.24 |
ENSMUST00000229874.2
|
Clip4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr13_-_78347876 | 5.23 |
ENSMUST00000091458.13
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr10_-_49659355 | 5.21 |
ENSMUST00000105484.10
ENSMUST00000218598.2 ENSMUST00000079751.9 ENSMUST00000218441.2 |
Grik2
|
glutamate receptor, ionotropic, kainate 2 (beta 2) |
| chr18_+_44096300 | 5.20 |
ENSMUST00000069245.8
|
Spink5
|
serine peptidase inhibitor, Kazal type 5 |
| chr7_-_109380745 | 5.17 |
ENSMUST00000207400.2
ENSMUST00000033331.7 |
Nrip3
|
nuclear receptor interacting protein 3 |
| chr11_-_118246332 | 5.16 |
ENSMUST00000017610.10
|
Timp2
|
tissue inhibitor of metalloproteinase 2 |
| chr13_+_83652352 | 5.14 |
ENSMUST00000198916.5
ENSMUST00000200123.5 ENSMUST00000005722.14 ENSMUST00000163888.8 |
Mef2c
|
myocyte enhancer factor 2C |
| chr3_+_92840279 | 5.11 |
ENSMUST00000046234.5
|
Lce3b
|
late cornified envelope 3B |
| chr10_+_45453907 | 5.09 |
ENSMUST00000037044.13
|
Hace1
|
HECT domain and ankyrin repeat containing, E3 ubiquitin protein ligase 1 |
| chr1_-_45542442 | 5.03 |
ENSMUST00000086430.5
|
Col5a2
|
collagen, type V, alpha 2 |
| chr10_+_70276473 | 5.02 |
ENSMUST00000105436.9
|
Fam13c
|
family with sequence similarity 13, member C |
| chr8_+_94879235 | 4.86 |
ENSMUST00000034211.10
ENSMUST00000211930.2 ENSMUST00000211915.2 |
Mt3
|
metallothionein 3 |
| chr18_-_25886750 | 4.81 |
ENSMUST00000224553.2
ENSMUST00000025117.14 |
Celf4
|
CUGBP, Elav-like family member 4 |
| chrX_+_140258381 | 4.81 |
ENSMUST00000112931.8
ENSMUST00000112930.8 |
Col4a5
|
collagen, type IV, alpha 5 |
| chr2_-_165997551 | 4.80 |
ENSMUST00000109249.9
ENSMUST00000146497.9 |
Sulf2
|
sulfatase 2 |
| chr2_+_51928017 | 4.80 |
ENSMUST00000065927.6
|
Tnfaip6
|
tumor necrosis factor alpha induced protein 6 |
| chr11_-_81859241 | 4.79 |
ENSMUST00000066197.7
|
Asic2
|
acid-sensing (proton-gated) ion channel 2 |
| chr18_-_43032359 | 4.78 |
ENSMUST00000117687.8
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr2_+_14393127 | 4.78 |
ENSMUST00000114731.8
ENSMUST00000082290.8 |
Slc39a12
|
solute carrier family 39 (zinc transporter), member 12 |
| chr3_-_141637245 | 4.77 |
ENSMUST00000106232.8
|
Bmpr1b
|
bone morphogenetic protein receptor, type 1B |
| chr1_-_22551594 | 4.77 |
ENSMUST00000239255.2
|
Rims1
|
regulating synaptic membrane exocytosis 1 |
| chr1_-_168259465 | 4.75 |
ENSMUST00000176540.8
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr10_-_43050516 | 4.64 |
ENSMUST00000040275.9
|
Sobp
|
sine oculis binding protein |
| chr12_+_38830081 | 4.56 |
ENSMUST00000095767.11
|
Etv1
|
ets variant 1 |
| chr3_+_109481223 | 4.52 |
ENSMUST00000106576.3
|
Vav3
|
vav 3 oncogene |
| chr15_+_36179676 | 4.51 |
ENSMUST00000171205.3
|
Spag1
|
sperm associated antigen 1 |
| chr18_-_43032535 | 4.49 |
ENSMUST00000120632.2
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr16_+_56298228 | 4.42 |
ENSMUST00000231832.2
ENSMUST00000096013.11 ENSMUST00000048471.15 ENSMUST00000231870.2 ENSMUST00000171000.3 ENSMUST00000231781.2 ENSMUST00000096012.11 |
Abi3bp
|
ABI family member 3 binding protein |
| chr6_-_7693110 | 4.41 |
ENSMUST00000126303.8
|
Asns
|
asparagine synthetase |
| chr2_+_65760477 | 4.33 |
ENSMUST00000176109.8
|
Csrnp3
|
cysteine-serine-rich nuclear protein 3 |
| chrX_+_158623460 | 4.31 |
ENSMUST00000112451.8
ENSMUST00000112453.9 |
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr5_+_92540444 | 4.31 |
ENSMUST00000126281.8
|
Art3
|
ADP-ribosyltransferase 3 |
| chr11_-_42070517 | 4.27 |
ENSMUST00000206105.2
ENSMUST00000153147.3 |
Gabra1
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1 |
| chr1_-_168259839 | 4.21 |
ENSMUST00000188912.7
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr2_-_103133524 | 4.19 |
ENSMUST00000090475.10
|
Ehf
|
ets homologous factor |
| chr1_+_106908709 | 4.18 |
ENSMUST00000027564.8
|
Serpinb13
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 13 |
| chr16_-_56652241 | 4.17 |
ENSMUST00000135672.2
|
Tmem45a
|
transmembrane protein 45a |
| chr7_+_87233554 | 4.17 |
ENSMUST00000125009.9
|
Grm5
|
glutamate receptor, metabotropic 5 |
| chr10_+_115653152 | 4.17 |
ENSMUST00000080630.11
ENSMUST00000179196.3 ENSMUST00000035563.15 |
Tspan8
|
tetraspanin 8 |
| chr2_-_52448552 | 4.17 |
ENSMUST00000102760.10
ENSMUST00000102761.9 |
Cacnb4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr4_-_14621805 | 4.17 |
ENSMUST00000042221.14
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr17_+_14087827 | 4.16 |
ENSMUST00000239324.2
|
Afdn
|
afadin, adherens junction formation factor |
| chr17_-_52133594 | 4.16 |
ENSMUST00000129667.8
ENSMUST00000169480.8 ENSMUST00000148559.2 |
Satb1
|
special AT-rich sequence binding protein 1 |
| chr16_-_21814190 | 4.16 |
ENSMUST00000231766.2
ENSMUST00000074230.12 |
Liph
|
lipase, member H |
| chr3_+_67799510 | 4.14 |
ENSMUST00000063263.5
ENSMUST00000182006.4 |
Iqcj
Iqschfp
|
IQ motif containing J Iqcj and Schip1 fusion protein |
| chr1_+_179928709 | 4.13 |
ENSMUST00000133890.8
|
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr2_+_14393245 | 4.10 |
ENSMUST00000133258.2
|
Slc39a12
|
solute carrier family 39 (zinc transporter), member 12 |
| chr7_-_30750856 | 4.10 |
ENSMUST00000073892.6
|
Fxyd7
|
FXYD domain-containing ion transport regulator 7 |
| chr16_+_96001650 | 4.09 |
ENSMUST00000048770.16
|
Sh3bgr
|
SH3-binding domain glutamic acid-rich protein |
| chr5_-_73805063 | 4.09 |
ENSMUST00000081170.9
|
Sgcb
|
sarcoglycan, beta (dystrophin-associated glycoprotein) |
| chr13_+_93441307 | 4.05 |
ENSMUST00000080127.12
|
Homer1
|
homer scaffolding protein 1 |
| chr3_+_96577447 | 4.05 |
ENSMUST00000048427.9
|
Ankrd35
|
ankyrin repeat domain 35 |
| chr13_-_78344492 | 4.04 |
ENSMUST00000125176.3
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr6_-_112923715 | 4.04 |
ENSMUST00000113169.9
|
Srgap3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr13_-_113182891 | 4.03 |
ENSMUST00000231962.2
ENSMUST00000022282.6 |
Gpx8
|
glutathione peroxidase 8 (putative) |
| chr1_+_107289659 | 4.00 |
ENSMUST00000027566.3
|
Serpinb11
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 11 |
| chr2_+_16361582 | 3.99 |
ENSMUST00000114703.10
|
Plxdc2
|
plexin domain containing 2 |
| chr18_+_36431732 | 3.99 |
ENSMUST00000210490.3
|
Igip
|
IgA inducing protein |
| chr7_+_43339842 | 3.97 |
ENSMUST00000056329.7
|
Klk14
|
kallikrein related-peptidase 14 |
| chr11_-_100036792 | 3.96 |
ENSMUST00000007317.8
|
Krt19
|
keratin 19 |
| chr2_+_131751848 | 3.95 |
ENSMUST00000091288.13
ENSMUST00000124100.8 ENSMUST00000136783.2 |
Prnp
Prn
|
prion protein prion protein readthrough transcript |
| chr4_-_68872585 | 3.95 |
ENSMUST00000030036.6
|
Brinp1
|
bone morphogenic protein/retinoic acid inducible neural specific 1 |
| chr18_-_12438695 | 3.93 |
ENSMUST00000122408.8
ENSMUST00000234183.2 ENSMUST00000142066.2 ENSMUST00000118525.9 |
Ankrd29
|
ankyrin repeat domain 29 |
| chr9_+_122752116 | 3.91 |
ENSMUST00000051667.14
|
Zfp105
|
zinc finger protein 105 |
| chr18_+_69726055 | 3.91 |
ENSMUST00000114978.9
|
Tcf4
|
transcription factor 4 |
| chr4_-_87951565 | 3.90 |
ENSMUST00000078090.12
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr5_+_13449276 | 3.90 |
ENSMUST00000030714.9
ENSMUST00000141968.2 |
Sema3a
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr13_+_83723743 | 3.83 |
ENSMUST00000198217.5
ENSMUST00000199210.5 |
Mef2c
|
myocyte enhancer factor 2C |
| chr4_-_154721288 | 3.79 |
ENSMUST00000030902.13
ENSMUST00000105637.8 ENSMUST00000070313.14 ENSMUST00000105636.8 ENSMUST00000105638.9 ENSMUST00000097759.9 ENSMUST00000124771.2 |
Prdm16
|
PR domain containing 16 |
| chr10_-_63926044 | 3.79 |
ENSMUST00000105439.2
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr11_+_81992662 | 3.78 |
ENSMUST00000000194.4
|
Ccl12
|
chemokine (C-C motif) ligand 12 |
| chr11_+_29413734 | 3.78 |
ENSMUST00000155854.8
|
Ccdc88a
|
coiled coil domain containing 88A |
| chr17_-_82045800 | 3.77 |
ENSMUST00000235015.2
ENSMUST00000163123.3 |
Slc8a1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr7_+_4122523 | 3.75 |
ENSMUST00000119661.8
ENSMUST00000129423.8 |
Ttyh1
|
tweety family member 1 |
| chr16_+_94225942 | 3.74 |
ENSMUST00000141176.2
|
Ttc3
|
tetratricopeptide repeat domain 3 |
| chr3_-_26386609 | 3.70 |
ENSMUST00000193603.6
|
Nlgn1
|
neuroligin 1 |
| chr4_+_101365144 | 3.68 |
ENSMUST00000149047.8
ENSMUST00000106929.10 |
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr16_-_21814289 | 3.64 |
ENSMUST00000060673.8
|
Liph
|
lipase, member H |
| chr6_-_112924205 | 3.62 |
ENSMUST00000088373.11
|
Srgap3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr6_+_8520006 | 3.61 |
ENSMUST00000162567.8
ENSMUST00000161217.8 |
Glcci1
|
glucocorticoid induced transcript 1 |
| chr2_-_103133503 | 3.59 |
ENSMUST00000111176.9
|
Ehf
|
ets homologous factor |
| chr1_-_37535202 | 3.59 |
ENSMUST00000143636.8
|
Mgat4a
|
mannoside acetylglucosaminyltransferase 4, isoenzyme A |
| chr18_+_20509771 | 3.59 |
ENSMUST00000076737.7
|
Dsg1b
|
desmoglein 1 beta |
| chr19_+_8828132 | 3.57 |
ENSMUST00000235683.2
ENSMUST00000096257.3 |
Lrrn4cl
|
LRRN4 C-terminal like |
| chr10_-_30647836 | 3.57 |
ENSMUST00000215926.2
ENSMUST00000213836.2 |
Ncoa7
|
nuclear receptor coactivator 7 |
| chr11_-_99134885 | 3.55 |
ENSMUST00000103132.10
ENSMUST00000038214.7 |
Krt222
|
keratin 222 |
| chr12_+_38833501 | 3.53 |
ENSMUST00000159334.8
|
Etv1
|
ets variant 1 |
| chrX_-_58211440 | 3.53 |
ENSMUST00000119306.2
|
Fgf13
|
fibroblast growth factor 13 |
| chr13_+_42454922 | 3.50 |
ENSMUST00000021796.9
|
Edn1
|
endothelin 1 |
| chrX_+_136552469 | 3.50 |
ENSMUST00000075471.4
|
Il1rapl2
|
interleukin 1 receptor accessory protein-like 2 |
| chr18_+_37858753 | 3.48 |
ENSMUST00000066149.9
|
Pcdhga8
|
protocadherin gamma subfamily A, 8 |
| chr10_+_102376109 | 3.47 |
ENSMUST00000055355.6
|
Rassf9
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
| chr19_+_39102342 | 3.45 |
ENSMUST00000087234.3
|
Cyp2c66
|
cytochrome P450, family 2, subfamily c, polypeptide 66 |
| chr1_-_168259264 | 3.44 |
ENSMUST00000176790.8
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr8_-_34237752 | 3.39 |
ENSMUST00000179364.3
|
Smim18
|
small integral membrane protein 18 |
| chr1_+_12762501 | 3.37 |
ENSMUST00000177608.8
ENSMUST00000180062.8 |
Sulf1
|
sulfatase 1 |
| chr10_-_30531832 | 3.37 |
ENSMUST00000217138.2
ENSMUST00000217644.2 ENSMUST00000216172.2 |
Ncoa7
|
nuclear receptor coactivator 7 |
| chrX_+_9751861 | 3.36 |
ENSMUST00000067529.9
ENSMUST00000086165.4 |
Sytl5
|
synaptotagmin-like 5 |
| chr17_+_24026892 | 3.30 |
ENSMUST00000191385.3
|
Srrm2
|
serine/arginine repetitive matrix 2 |
| chr7_+_142052569 | 3.28 |
ENSMUST00000078497.15
ENSMUST00000105953.10 ENSMUST00000179658.8 ENSMUST00000105954.10 ENSMUST00000105952.10 ENSMUST00000105955.8 ENSMUST00000074187.13 ENSMUST00000169299.9 ENSMUST00000105957.10 ENSMUST00000180152.8 ENSMUST00000105950.11 ENSMUST00000105958.10 ENSMUST00000105949.8 |
Tnnt3
|
troponin T3, skeletal, fast |
| chr9_-_37058590 | 3.28 |
ENSMUST00000080754.12
ENSMUST00000188057.7 ENSMUST00000039674.13 |
Pknox2
|
Pbx/knotted 1 homeobox 2 |
| chr3_-_33039225 | 3.26 |
ENSMUST00000108221.2
|
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr7_+_4122555 | 3.25 |
ENSMUST00000079415.12
|
Ttyh1
|
tweety family member 1 |
| chr6_-_53797748 | 3.25 |
ENSMUST00000127748.5
|
Tril
|
TLR4 interactor with leucine-rich repeats |
| chr9_+_54606798 | 3.21 |
ENSMUST00000154690.8
|
Dnaja4
|
DnaJ heat shock protein family (Hsp40) member A4 |
| chr7_-_30750828 | 3.19 |
ENSMUST00000206341.2
|
Fxyd7
|
FXYD domain-containing ion transport regulator 7 |
| chr6_-_87958611 | 3.18 |
ENSMUST00000056403.7
|
H1f10
|
H1.10 linker histone |
| chr6_-_138398376 | 3.17 |
ENSMUST00000163065.8
|
Lmo3
|
LIM domain only 3 |
| chr12_-_36092475 | 3.17 |
ENSMUST00000020896.17
|
Tspan13
|
tetraspanin 13 |
| chrX_+_13498321 | 3.16 |
ENSMUST00000041708.10
|
Gpr34
|
G protein-coupled receptor 34 |
| chr4_-_91264670 | 3.12 |
ENSMUST00000107109.9
ENSMUST00000107111.9 ENSMUST00000107120.8 |
Elavl2
|
ELAV like RNA binding protein 1 |
| chr5_+_92534738 | 3.11 |
ENSMUST00000128246.8
|
Art3
|
ADP-ribosyltransferase 3 |
| chr15_+_16728842 | 3.11 |
ENSMUST00000228307.2
|
Cdh9
|
cadherin 9 |
| chr1_-_157084252 | 3.11 |
ENSMUST00000134543.8
|
Rasal2
|
RAS protein activator like 2 |
| chr14_-_24227857 | 3.10 |
ENSMUST00000042009.13
|
Dlg5
|
discs large MAGUK scaffold protein 5 |
| chr18_+_37637317 | 3.10 |
ENSMUST00000052179.8
|
Pcdhb20
|
protocadherin beta 20 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 11.8 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 3.4 | 10.1 | GO:0061193 | taste bud development(GO:0061193) |
| 3.0 | 9.0 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 2.7 | 8.2 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) cloacal septation(GO:0060197) |
| 2.7 | 8.1 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 2.6 | 10.6 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 2.5 | 178.1 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 2.2 | 19.9 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 1.9 | 11.2 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 1.9 | 5.6 | GO:0070194 | synaptonemal complex disassembly(GO:0070194) |
| 1.8 | 5.4 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 1.7 | 10.2 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 1.7 | 5.1 | GO:1903465 | motogenic signaling involved in postnatal olfactory bulb interneuron migration(GO:0021837) positive regulation of mitotic cell cycle DNA replication(GO:1903465) |
| 1.6 | 4.9 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 1.6 | 18.7 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 1.4 | 31.8 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 1.4 | 5.5 | GO:0035989 | tendon development(GO:0035989) |
| 1.4 | 5.5 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) |
| 1.3 | 3.9 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 1.3 | 5.0 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 1.2 | 3.7 | GO:0098942 | cytoskeletal matrix organization at active zone(GO:0048789) neurexin clustering involved in presynaptic membrane assembly(GO:0097115) retrograde trans-synaptic signaling by trans-synaptic protein complex(GO:0098942) |
| 1.2 | 3.5 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 1.1 | 12.6 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 1.1 | 5.6 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 1.1 | 4.4 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 1.0 | 5.2 | GO:0002784 | regulation of antimicrobial peptide production(GO:0002784) regulation of antibacterial peptide production(GO:0002786) |
| 1.0 | 4.1 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 1.0 | 6.9 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 1.0 | 4.8 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 1.0 | 4.8 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.9 | 6.5 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.9 | 2.8 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.9 | 11.8 | GO:0010579 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.9 | 2.7 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.9 | 1.8 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.9 | 6.0 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.8 | 2.4 | GO:0021627 | olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 0.8 | 4.8 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.8 | 21.4 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.8 | 2.3 | GO:0048936 | neurofilament bundle assembly(GO:0033693) peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.8 | 3.1 | GO:0045186 | zonula adherens assembly(GO:0045186) |
| 0.7 | 11.8 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.7 | 3.7 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.7 | 2.1 | GO:0070904 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.7 | 4.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.7 | 5.4 | GO:0014004 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.7 | 4.7 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.7 | 12.0 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.6 | 10.1 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.6 | 3.8 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.6 | 2.5 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.6 | 0.6 | GO:0099545 | trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.6 | 8.1 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.6 | 1.1 | GO:0021856 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) facioacoustic ganglion development(GO:1903375) |
| 0.6 | 1.7 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.5 | 1.6 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.5 | 25.8 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.5 | 1.0 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) |
| 0.5 | 3.9 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.5 | 8.2 | GO:0048670 | regulation of collateral sprouting(GO:0048670) |
| 0.5 | 9.4 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.5 | 8.9 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.5 | 2.3 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.5 | 2.7 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 0.4 | 3.1 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.4 | 1.3 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.4 | 2.6 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.4 | 15.1 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.4 | 2.5 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.4 | 3.8 | GO:0071313 | cellular response to caffeine(GO:0071313) positive regulation of the force of heart contraction(GO:0098735) |
| 0.4 | 6.2 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.4 | 11.1 | GO:0001553 | luteinization(GO:0001553) |
| 0.4 | 3.3 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.4 | 1.6 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.4 | 1.6 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.4 | 1.6 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.4 | 2.3 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.4 | 1.5 | GO:0035566 | regulation of metanephros size(GO:0035566) |
| 0.4 | 6.4 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.4 | 9.3 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.4 | 4.1 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.4 | 2.9 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.4 | 3.2 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.3 | 3.1 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.3 | 6.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.3 | 1.4 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.3 | 1.0 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.3 | 5.3 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.3 | 2.6 | GO:0009099 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.3 | 6.6 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.3 | 7.6 | GO:0031424 | keratinization(GO:0031424) |
| 0.3 | 3.0 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.3 | 2.9 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.3 | 7.7 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.3 | 2.5 | GO:0060313 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.3 | 12.4 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.3 | 4.0 | GO:0007320 | insemination(GO:0007320) |
| 0.3 | 1.2 | GO:0080163 | regulation of protein serine/threonine phosphatase activity(GO:0080163) |
| 0.3 | 4.2 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.3 | 3.9 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.3 | 0.9 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.3 | 1.7 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.3 | 0.8 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.3 | 1.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.3 | 4.2 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.3 | 2.8 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.3 | 1.7 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.3 | 6.1 | GO:0007614 | short-term memory(GO:0007614) |
| 0.3 | 16.2 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.3 | 1.3 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.3 | 1.0 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.3 | 2.3 | GO:2000510 | positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.2 | 4.5 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.2 | 5.5 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.2 | 0.5 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.2 | 1.4 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.2 | 6.0 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.2 | 0.9 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.2 | 1.4 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 0.5 | GO:0060434 | regulation of tolerance induction to self antigen(GO:0002649) bronchus morphogenesis(GO:0060434) |
| 0.2 | 2.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.2 | 0.5 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.2 | 4.2 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.2 | 2.0 | GO:1902172 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.2 | 4.0 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.2 | 4.8 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.2 | 1.7 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.2 | 1.6 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.2 | 2.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.2 | 1.0 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.2 | 2.0 | GO:1902101 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.2 | 1.0 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.2 | 2.3 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.2 | 5.8 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.2 | 1.3 | GO:0060023 | soft palate development(GO:0060023) |
| 0.2 | 13.6 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.2 | 1.9 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.2 | 0.7 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.2 | 0.9 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.2 | 2.9 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 | 2.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 7.2 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.2 | 1.0 | GO:1903758 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.2 | 0.2 | GO:0035910 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.2 | 0.8 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.2 | 7.4 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.2 | 9.4 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.2 | 0.9 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.1 | 1.0 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.3 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 1.2 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.1 | 1.6 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 | 2.0 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 | 1.2 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 1.1 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.1 | 0.4 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.1 | 1.7 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 4.3 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 1.0 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.1 | 2.5 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.9 | GO:0035509 | negative regulation of myosin-light-chain-phosphatase activity(GO:0035509) |
| 0.1 | 1.1 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.1 | 0.4 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.1 | 3.5 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.1 | 0.1 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.1 | 0.6 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.1 | 0.8 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 4.0 | GO:0060706 | cell differentiation involved in embryonic placenta development(GO:0060706) |
| 0.1 | 0.3 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.1 | 0.7 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 0.9 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 8.1 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 1.3 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 3.0 | GO:0060285 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.1 | 1.0 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 | 14.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.8 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.1 | 0.4 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.1 | 3.6 | GO:0090102 | cochlea development(GO:0090102) |
| 0.1 | 0.6 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) |
| 0.1 | 3.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.4 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.1 | 0.4 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.1 | 0.5 | GO:0009624 | response to nematode(GO:0009624) |
| 0.1 | 0.4 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.1 | 0.4 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 0.5 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.1 | 1.7 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.1 | 6.9 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.1 | 1.4 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 1.6 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 1.3 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 3.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 4.2 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 0.5 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.1 | 2.9 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 1.2 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.1 | 7.8 | GO:0035308 | negative regulation of protein dephosphorylation(GO:0035308) |
| 0.1 | 0.6 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 0.5 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 1.7 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 1.2 | GO:0006027 | aminoglycan catabolic process(GO:0006026) glycosaminoglycan catabolic process(GO:0006027) chondroitin sulfate metabolic process(GO:0030204) |
| 0.1 | 1.1 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 0.8 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 1.7 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.1 | 3.0 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.1 | 3.4 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 0.7 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.1 | 1.7 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 2.4 | GO:0034142 | toll-like receptor 4 signaling pathway(GO:0034142) |
| 0.1 | 1.3 | GO:0072606 | interleukin-8 secretion(GO:0072606) |
| 0.1 | 1.4 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.1 | 2.1 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.1 | 11.5 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.1 | 1.6 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.1 | 0.8 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.1 | 8.6 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 1.0 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 3.7 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 1.2 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.4 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 3.7 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 6.3 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.8 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 0.4 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 1.3 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.3 | GO:0009313 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 4.7 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 0.8 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.0 | 0.5 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 5.5 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 3.8 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.4 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.9 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.6 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 1.8 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 1.3 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.9 | GO:0007099 | centriole replication(GO:0007099) centriole assembly(GO:0098534) |
| 0.0 | 0.6 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.9 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.0 | 1.9 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 3.8 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 2.1 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.0 | 4.3 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 1.0 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.5 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.1 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 6.0 | GO:0046578 | regulation of Ras protein signal transduction(GO:0046578) |
| 0.0 | 4.2 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 1.5 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.0 | 0.4 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.0 | 0.1 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.2 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.4 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.6 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 2.3 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.4 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 1.2 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.2 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.3 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) |
| 0.0 | 2.7 | GO:0016197 | endosomal transport(GO:0016197) |
| 0.0 | 0.8 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.9 | GO:0051225 | spindle assembly(GO:0051225) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 200.2 | GO:0001533 | cornified envelope(GO:0001533) |
| 1.7 | 5.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 1.1 | 31.8 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 1.0 | 5.2 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.8 | 4.8 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.8 | 5.4 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.7 | 5.8 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.7 | 9.0 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.7 | 4.8 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.7 | 5.2 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.6 | 5.5 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.5 | 3.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.5 | 10.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.5 | 3.5 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.5 | 9.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.5 | 1.4 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.5 | 5.6 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.5 | 4.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.4 | 4.0 | GO:1990357 | terminal web(GO:1990357) |
| 0.4 | 10.7 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.4 | 3.3 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.4 | 5.6 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.4 | 13.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.4 | 10.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.3 | 2.8 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.3 | 1.0 | GO:1990844 | subsarcolemmal mitochondrion(GO:1990843) interfibrillar mitochondrion(GO:1990844) |
| 0.3 | 10.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.3 | 5.3 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.3 | 2.5 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.3 | 4.7 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.3 | 18.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.3 | 3.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.2 | 14.3 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 3.9 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 0.9 | GO:0071942 | XPC complex(GO:0071942) |
| 0.2 | 0.9 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.2 | 1.7 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.2 | 1.4 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.2 | 14.4 | GO:0031672 | A band(GO:0031672) |
| 0.2 | 3.0 | GO:0000243 | commitment complex(GO:0000243) |
| 0.2 | 0.8 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.2 | 0.9 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.2 | 2.3 | GO:0005883 | neurofilament(GO:0005883) |
| 0.2 | 2.5 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.2 | 4.2 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.2 | 1.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 3.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 0.4 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.1 | 1.9 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 1.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 10.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 3.0 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 2.2 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.6 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 14.4 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 30.0 | GO:0030016 | myofibril(GO:0030016) |
| 0.1 | 1.0 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.1 | 1.0 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 10.6 | GO:0042641 | actomyosin(GO:0042641) |
| 0.1 | 5.6 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 0.9 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.4 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 18.2 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.1 | 2.9 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 20.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 10.7 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 4.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 3.4 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 0.6 | GO:0005851 | eukaryotic translation initiation factor 2 complex(GO:0005850) eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 14.0 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 3.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 3.8 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 5.4 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 3.9 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 0.6 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 1.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 1.6 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 0.4 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 6.4 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 1.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 2.0 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 3.0 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 2.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 1.3 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 15.7 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 7.2 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 3.0 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.5 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 1.2 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 1.7 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.5 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 18.7 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.1 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.0 | 6.5 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 0.6 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.9 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 3.6 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 4.0 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 2.9 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.4 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 2.3 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 5.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.4 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.3 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.4 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 2.7 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.6 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 1.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.5 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 3.3 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 3.0 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.9 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.1 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.3 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 4.8 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 7.3 | GO:0036477 | somatodendritic compartment(GO:0036477) |
| 0.0 | 3.5 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.6 | GO:0044297 | cell body(GO:0044297) |
| 0.0 | 0.4 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 12.9 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 10.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 3.2 | 25.4 | GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 2.6 | 12.8 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 2.3 | 18.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 2.2 | 19.9 | GO:0004568 | chitinase activity(GO:0004568) |
| 2.2 | 12.9 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 1.8 | 29.4 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 1.6 | 11.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 1.5 | 5.8 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 1.4 | 4.2 | GO:0099530 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 1.4 | 5.5 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 1.3 | 3.8 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 1.1 | 4.4 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 1.0 | 13.3 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.9 | 5.6 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.9 | 5.4 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.9 | 3.5 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.9 | 3.5 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.9 | 5.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.9 | 7.7 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.8 | 3.3 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.7 | 3.0 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.7 | 2.1 | GO:0015229 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.7 | 17.9 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.6 | 5.8 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.6 | 3.9 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.6 | 11.5 | GO:0019841 | retinol binding(GO:0019841) |
| 0.6 | 1.2 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.6 | 6.8 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.6 | 2.9 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.6 | 5.8 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.6 | 4.6 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.5 | 3.3 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.5 | 11.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.5 | 7.4 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.5 | 3.3 | GO:0031013 | troponin C binding(GO:0030172) calcium-dependent ATPase activity(GO:0030899) troponin I binding(GO:0031013) |
| 0.5 | 5.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.5 | 7.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.5 | 1.4 | GO:0004420 | hydroxymethylglutaryl-CoA reductase (NADPH) activity(GO:0004420) hydroxymethylglutaryl-CoA reductase activity(GO:0042282) |
| 0.5 | 2.7 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.5 | 2.3 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.4 | 1.3 | GO:0008061 | chitin binding(GO:0008061) |
| 0.4 | 4.8 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.4 | 4.8 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.4 | 2.8 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.4 | 2.7 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.4 | 10.2 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.4 | 2.6 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.4 | 2.2 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.4 | 2.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.4 | 3.9 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.3 | 7.0 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.3 | 1.0 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.3 | 2.6 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.3 | 1.6 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.3 | 5.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.3 | 1.3 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.3 | 16.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.3 | 4.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.3 | 2.5 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.3 | 2.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.3 | 1.9 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.3 | 7.7 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.3 | 3.0 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.3 | 1.5 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.3 | 0.9 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.3 | 2.1 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.3 | 0.6 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.3 | 7.6 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.3 | 1.7 | GO:0052723 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.3 | 5.6 | GO:0051861 | glycolipid binding(GO:0051861) |
| 0.3 | 4.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.3 | 2.2 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.3 | 2.5 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.3 | 2.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.3 | 2.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.2 | 3.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 6.5 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.2 | 0.5 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.2 | 11.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 1.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.2 | 2.9 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.2 | 177.5 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.2 | 1.1 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) |
| 0.2 | 8.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 3.2 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.2 | 13.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.2 | 14.8 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.2 | 1.0 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.2 | 4.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.2 | 1.4 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.2 | 3.8 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.2 | 3.2 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.2 | 4.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 5.8 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.2 | 2.9 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 14.1 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.2 | 37.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.2 | 1.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.2 | 1.2 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.2 | 0.7 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.2 | 2.6 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.2 | 18.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.2 | 0.5 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.1 | 12.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 7.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 1.6 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 1.5 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 0.9 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.1 | 7.6 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 3.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 4.0 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 3.5 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 0.5 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.1 | 2.5 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 6.0 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 2.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 2.3 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 3.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 3.6 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 0.9 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.1 | 1.0 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 1.6 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 2.6 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 1.0 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 16.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 0.4 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 14.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.8 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 1.0 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 2.8 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 2.6 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 2.9 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 2.0 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 1.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.8 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 12.9 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 0.8 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 1.9 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 5.9 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 1.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 0.3 | GO:0052795 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 3.4 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 1.0 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.4 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.1 | GO:0071820 | N-box binding(GO:0071820) |
| 0.0 | 14.1 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.5 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 3.1 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 3.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.8 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 1.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.8 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.8 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 2.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 3.2 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.7 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 4.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.6 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 2.1 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 2.2 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.0 | 0.5 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.3 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 1.4 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 4.6 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 4.3 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 1.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 3.8 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.6 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 1.3 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 3.6 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 4.1 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 4.9 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.7 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.0 | 0.6 | GO:0035064 | methylated histone binding(GO:0035064) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 18.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.5 | 13.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.3 | 24.8 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.3 | 20.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.2 | 12.0 | PID ATR PATHWAY | ATR signaling pathway |
| 0.2 | 31.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.2 | 13.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 5.8 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.2 | 0.6 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.2 | 2.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.2 | 19.6 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.2 | 8.2 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.2 | 1.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 6.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 2.0 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 1.0 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 1.7 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 4.5 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 4.2 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 5.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 30.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 5.6 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 4.3 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 10.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 1.9 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 10.0 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 2.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 1.2 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 7.2 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 1.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 0.4 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.1 | 1.6 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 2.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 3.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 2.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 2.2 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 3.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 1.6 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 2.0 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 2.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 2.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 2.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.3 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 2.4 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 1.1 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 3.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.6 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 38.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.7 | 46.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.5 | 8.8 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.5 | 10.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.4 | 0.4 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.4 | 27.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.4 | 18.1 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.4 | 10.2 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.3 | 4.8 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.3 | 5.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.3 | 9.8 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.3 | 18.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.3 | 5.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.3 | 6.8 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.2 | 5.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 10.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 8.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.2 | 2.5 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 4.1 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 3.8 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.2 | 4.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 1.7 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 4.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 3.5 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.1 | 0.9 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 5.9 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 1.6 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.1 | 18.2 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 1.0 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 1.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 1.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 3.7 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 1.4 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 4.3 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 2.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 2.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 5.8 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 0.9 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 1.6 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 2.0 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.1 | 2.0 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 2.9 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 2.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.8 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.1 | 0.9 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 0.3 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 3.7 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.1 | 1.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 4.4 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 5.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 3.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 0.6 | REACTOME GAP JUNCTION TRAFFICKING | Genes involved in Gap junction trafficking |
| 0.0 | 1.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 1.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 6.1 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.6 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 2.3 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 5.4 | REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 0.0 | 0.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.3 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 1.0 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.5 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.6 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.6 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 2.7 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |