Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
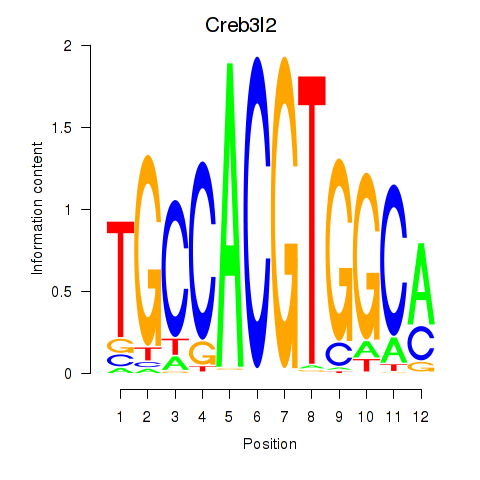
Results for Creb3l2
Z-value: 1.27
Transcription factors associated with Creb3l2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Creb3l2
|
ENSMUSG00000038648.6 | Creb3l2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Creb3l2 | mm39_v1_chr6_-_37419030_37419117 | 0.32 | 5.5e-03 | Click! |
Activity profile of Creb3l2 motif
Sorted Z-values of Creb3l2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Creb3l2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_26417982 | 13.92 |
ENSMUST00000142410.2
ENSMUST00000120333.8 ENSMUST00000039113.14 |
Pdia2
|
protein disulfide isomerase associated 2 |
| chr5_-_31102829 | 10.88 |
ENSMUST00000031051.8
|
Cgref1
|
cell growth regulator with EF hand domain 1 |
| chr2_-_92201342 | 6.42 |
ENSMUST00000176810.8
ENSMUST00000090582.11 ENSMUST00000068586.13 |
Large2
|
LARGE xylosyl- and glucuronyltransferase 2 |
| chrX_+_138464065 | 6.36 |
ENSMUST00000113027.8
|
Rnf128
|
ring finger protein 128 |
| chr5_+_63969706 | 5.74 |
ENSMUST00000081747.8
ENSMUST00000196575.5 |
0610040J01Rik
|
RIKEN cDNA 0610040J01 gene |
| chr1_+_72863641 | 5.46 |
ENSMUST00000047328.11
|
Igfbp2
|
insulin-like growth factor binding protein 2 |
| chr16_-_20972750 | 4.90 |
ENSMUST00000170665.3
|
Teddm3
|
transmembrane epididymal family member 3 |
| chr4_+_133280680 | 4.82 |
ENSMUST00000042706.3
|
Nr0b2
|
nuclear receptor subfamily 0, group B, member 2 |
| chr2_-_179976458 | 4.75 |
ENSMUST00000015771.3
|
Gata5
|
GATA binding protein 5 |
| chr9_-_67739607 | 4.69 |
ENSMUST00000054500.7
|
C2cd4a
|
C2 calcium-dependent domain containing 4A |
| chr10_+_128745214 | 4.60 |
ENSMUST00000220308.2
|
Cd63
|
CD63 antigen |
| chr5_-_139799953 | 4.44 |
ENSMUST00000044002.10
|
Tmem184a
|
transmembrane protein 184a |
| chr8_-_106863521 | 4.40 |
ENSMUST00000115979.9
|
Esrp2
|
epithelial splicing regulatory protein 2 |
| chrX_+_8137372 | 3.99 |
ENSMUST00000127103.8
ENSMUST00000115591.8 |
Slc38a5
|
solute carrier family 38, member 5 |
| chr10_+_125802084 | 3.97 |
ENSMUST00000074807.8
|
Lrig3
|
leucine-rich repeats and immunoglobulin-like domains 3 |
| chr5_-_115109118 | 3.97 |
ENSMUST00000031535.12
|
Hnf1a
|
HNF1 homeobox A |
| chr8_-_106863423 | 3.96 |
ENSMUST00000146940.2
|
Esrp2
|
epithelial splicing regulatory protein 2 |
| chr2_-_92201311 | 3.94 |
ENSMUST00000176774.8
|
Large2
|
LARGE xylosyl- and glucuronyltransferase 2 |
| chrX_+_8137620 | 3.82 |
ENSMUST00000033512.11
|
Slc38a5
|
solute carrier family 38, member 5 |
| chrX_+_8137881 | 3.68 |
ENSMUST00000115590.2
|
Slc38a5
|
solute carrier family 38, member 5 |
| chr12_-_25147139 | 3.25 |
ENSMUST00000221761.2
|
Id2
|
inhibitor of DNA binding 2 |
| chr2_+_160722562 | 3.03 |
ENSMUST00000109456.9
|
Lpin3
|
lipin 3 |
| chr18_+_65933586 | 3.01 |
ENSMUST00000025394.14
ENSMUST00000236847.2 ENSMUST00000153193.3 |
Sec11c
|
SEC11 homolog C, signal peptidase complex subunit |
| chr19_+_46561801 | 3.00 |
ENSMUST00000026011.8
|
Sfxn2
|
sideroflexin 2 |
| chr6_-_22356098 | 2.97 |
ENSMUST00000165576.8
|
Fam3c
|
family with sequence similarity 3, member C |
| chr15_-_83234372 | 2.90 |
ENSMUST00000226124.2
ENSMUST00000067215.9 |
Arfgap3
|
ADP-ribosylation factor GTPase activating protein 3 |
| chr2_+_180367056 | 2.90 |
ENSMUST00000094218.4
|
Slc17a9
|
solute carrier family 17, member 9 |
| chr6_-_83294526 | 2.84 |
ENSMUST00000005810.9
|
Mthfd2
|
methylenetetrahydrofolate dehydrogenase (NAD+ dependent), methenyltetrahydrofolate cyclohydrolase |
| chr18_+_75500600 | 2.82 |
ENSMUST00000026999.10
|
Smad7
|
SMAD family member 7 |
| chr2_-_143853122 | 2.76 |
ENSMUST00000016072.12
ENSMUST00000037875.6 |
Rrbp1
|
ribosome binding protein 1 |
| chr19_+_59447950 | 2.74 |
ENSMUST00000174353.2
|
Emx2
|
empty spiracles homeobox 2 |
| chr6_-_22356067 | 2.68 |
ENSMUST00000163963.8
|
Fam3c
|
family with sequence similarity 3, member C |
| chr17_-_56583715 | 2.61 |
ENSMUST00000058136.9
|
Ticam1
|
toll-like receptor adaptor molecule 1 |
| chr2_+_68691778 | 2.58 |
ENSMUST00000028426.9
|
Cers6
|
ceramide synthase 6 |
| chr5_-_46014809 | 2.55 |
ENSMUST00000190036.7
ENSMUST00000189859.7 ENSMUST00000186633.3 ENSMUST00000016026.14 ENSMUST00000045586.13 ENSMUST00000238522.2 |
Lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr5_+_143389573 | 2.51 |
ENSMUST00000110731.4
|
Kdelr2
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 |
| chr6_-_22356175 | 2.49 |
ENSMUST00000081288.14
|
Fam3c
|
family with sequence similarity 3, member C |
| chr4_+_148675939 | 2.48 |
ENSMUST00000006611.9
|
Srm
|
spermidine synthase |
| chr2_-_32584132 | 2.41 |
ENSMUST00000028148.11
|
Fpgs
|
folylpolyglutamyl synthetase |
| chr6_-_8778439 | 2.39 |
ENSMUST00000115520.8
ENSMUST00000038403.12 ENSMUST00000115518.8 |
Ica1
|
islet cell autoantigen 1 |
| chr1_+_63216281 | 2.26 |
ENSMUST00000188524.2
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr3_+_123061094 | 2.24 |
ENSMUST00000047923.12
ENSMUST00000200333.2 |
Sec24d
|
Sec24 related gene family, member D (S. cerevisiae) |
| chr3_-_63872189 | 2.22 |
ENSMUST00000029402.15
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr19_+_34899752 | 2.20 |
ENSMUST00000087341.7
|
Kif20b
|
kinesin family member 20B |
| chr1_+_63215976 | 2.18 |
ENSMUST00000129339.8
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr19_+_34899778 | 2.18 |
ENSMUST00000223907.2
|
Kif20b
|
kinesin family member 20B |
| chr14_+_26359390 | 2.17 |
ENSMUST00000112318.10
|
Arf4
|
ADP-ribosylation factor 4 |
| chr11_+_20151406 | 2.16 |
ENSMUST00000020358.12
ENSMUST00000109602.8 ENSMUST00000109601.8 ENSMUST00000163483.2 |
Rab1a
|
RAB1A, member RAS oncogene family |
| chr6_-_8778106 | 2.16 |
ENSMUST00000151758.2
ENSMUST00000115519.8 ENSMUST00000153390.8 |
Ica1
|
islet cell autoantigen 1 |
| chr11_-_120534469 | 2.12 |
ENSMUST00000141254.8
ENSMUST00000170556.8 ENSMUST00000151876.8 ENSMUST00000026133.15 ENSMUST00000139706.2 |
Pycr1
|
pyrroline-5-carboxylate reductase 1 |
| chr10_+_61484331 | 2.08 |
ENSMUST00000020286.7
|
Ppa1
|
pyrophosphatase (inorganic) 1 |
| chr4_+_116578117 | 2.08 |
ENSMUST00000045542.13
ENSMUST00000106459.8 |
Tesk2
|
testis-specific kinase 2 |
| chr14_-_56499690 | 2.06 |
ENSMUST00000015581.6
|
Gzmb
|
granzyme B |
| chr3_-_63872079 | 2.03 |
ENSMUST00000161659.8
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr14_-_55880980 | 1.99 |
ENSMUST00000132338.8
|
Tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr5_-_139799780 | 1.97 |
ENSMUST00000146780.3
|
Tmem184a
|
transmembrane protein 184a |
| chr14_+_26359191 | 1.88 |
ENSMUST00000022429.9
|
Arf4
|
ADP-ribosylation factor 4 |
| chr4_-_119279551 | 1.86 |
ENSMUST00000106316.2
ENSMUST00000030385.13 |
Ppcs
|
phosphopantothenoylcysteine synthetase |
| chr9_+_108216466 | 1.80 |
ENSMUST00000193987.2
|
Gpx1
|
glutathione peroxidase 1 |
| chr6_+_129489501 | 1.79 |
ENSMUST00000032263.7
|
Tmem52b
|
transmembrane protein 52B |
| chr18_-_40352372 | 1.77 |
ENSMUST00000025364.6
|
Yipf5
|
Yip1 domain family, member 5 |
| chr19_+_6135013 | 1.76 |
ENSMUST00000025704.3
|
Cdca5
|
cell division cycle associated 5 |
| chr3_-_101511971 | 1.72 |
ENSMUST00000036493.8
|
Atp1a1
|
ATPase, Na+/K+ transporting, alpha 1 polypeptide |
| chr2_+_164675697 | 1.72 |
ENSMUST00000143780.9
|
Ctsa
|
cathepsin A |
| chr14_-_55880708 | 1.70 |
ENSMUST00000120041.8
ENSMUST00000121937.8 ENSMUST00000133707.2 ENSMUST00000002391.15 ENSMUST00000121791.8 |
Tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr7_+_78563513 | 1.69 |
ENSMUST00000038142.15
|
Isg20
|
interferon-stimulated protein |
| chr9_+_108216433 | 1.66 |
ENSMUST00000191997.2
|
Gpx1
|
glutathione peroxidase 1 |
| chr11_-_97673203 | 1.66 |
ENSMUST00000128801.2
ENSMUST00000103146.5 |
Rpl23
|
ribosomal protein L23 |
| chr2_+_68691902 | 1.59 |
ENSMUST00000176018.2
|
Cers6
|
ceramide synthase 6 |
| chr11_-_60702081 | 1.56 |
ENSMUST00000018744.15
|
Shmt1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr7_+_78563964 | 1.54 |
ENSMUST00000120331.4
|
Isg20
|
interferon-stimulated protein |
| chr14_-_55881177 | 1.53 |
ENSMUST00000138085.2
|
Tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr11_+_100960838 | 1.50 |
ENSMUST00000001802.10
|
Naglu
|
alpha-N-acetylglucosaminidase (Sanfilippo disease IIIB) |
| chr14_-_55881257 | 1.48 |
ENSMUST00000122358.8
|
Tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr16_-_95928804 | 1.48 |
ENSMUST00000233292.2
ENSMUST00000050884.16 |
Hmgn1
|
high mobility group nucleosomal binding domain 1 |
| chr11_-_94544748 | 1.47 |
ENSMUST00000039949.5
|
Eme1
|
essential meiotic structure-specific endonuclease 1 |
| chr6_+_56809044 | 1.44 |
ENSMUST00000031795.8
|
Fkbp9
|
FK506 binding protein 9 |
| chr19_-_40576782 | 1.40 |
ENSMUST00000176939.8
|
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr19_-_40576897 | 1.38 |
ENSMUST00000025979.13
|
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr19_-_40576817 | 1.36 |
ENSMUST00000175932.2
ENSMUST00000176955.8 ENSMUST00000149476.3 |
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chrX_-_8011952 | 1.35 |
ENSMUST00000115615.9
ENSMUST00000115616.8 ENSMUST00000115621.9 |
Rbm3
|
RNA binding motif (RNP1, RRM) protein 3 |
| chr12_+_17594795 | 1.31 |
ENSMUST00000171737.3
|
Odc1
|
ornithine decarboxylase, structural 1 |
| chr10_+_12966532 | 1.31 |
ENSMUST00000121646.8
ENSMUST00000121325.8 ENSMUST00000121766.8 |
Plagl1
|
pleiomorphic adenoma gene-like 1 |
| chr6_+_70821481 | 1.30 |
ENSMUST00000034093.15
ENSMUST00000162950.2 |
Eif2ak3
|
eukaryotic translation initiation factor 2 alpha kinase 3 |
| chr11_+_100306523 | 1.26 |
ENSMUST00000001595.10
ENSMUST00000107400.3 |
Fkbp10
|
FK506 binding protein 10 |
| chr19_+_34899811 | 1.26 |
ENSMUST00000223776.2
|
Kif20b
|
kinesin family member 20B |
| chr4_-_106657446 | 1.26 |
ENSMUST00000148688.2
|
Acot11
|
acyl-CoA thioesterase 11 |
| chr17_-_86452564 | 1.24 |
ENSMUST00000095187.4
|
Srbd1
|
S1 RNA binding domain 1 |
| chr18_+_31892933 | 1.24 |
ENSMUST00000115808.4
|
Ammecr1l
|
AMME chromosomal region gene 1-like |
| chr11_+_109253598 | 1.23 |
ENSMUST00000106702.4
ENSMUST00000020930.14 |
Gna13
|
guanine nucleotide binding protein, alpha 13 |
| chrX_+_73372664 | 1.21 |
ENSMUST00000004326.4
|
Plxna3
|
plexin A3 |
| chr15_+_84807582 | 1.14 |
ENSMUST00000165443.4
|
Nup50
|
nucleoporin 50 |
| chrX_-_8011918 | 1.10 |
ENSMUST00000115619.8
ENSMUST00000115617.10 ENSMUST00000040010.10 |
Rbm3
|
RNA binding motif (RNP1, RRM) protein 3 |
| chrX_+_49930311 | 1.09 |
ENSMUST00000114887.9
|
Stk26
|
serine/threonine kinase 26 |
| chr4_-_155737841 | 1.08 |
ENSMUST00000030937.2
|
Mmp23
|
matrix metallopeptidase 23 |
| chr7_-_24950145 | 1.07 |
ENSMUST00000116343.3
ENSMUST00000045847.15 |
Erf
|
Ets2 repressor factor |
| chr2_+_74498551 | 1.05 |
ENSMUST00000001872.5
|
Hoxd13
|
homeobox D13 |
| chr16_-_78173645 | 1.03 |
ENSMUST00000023570.14
|
Btg3
|
BTG anti-proliferation factor 3 |
| chr6_+_87864796 | 1.01 |
ENSMUST00000113607.10
ENSMUST00000049966.6 |
Copg1
|
coatomer protein complex, subunit gamma 1 |
| chr9_-_87137515 | 1.01 |
ENSMUST00000093802.6
|
Cep162
|
centrosomal protein 162 |
| chr15_-_34495329 | 1.01 |
ENSMUST00000022946.6
|
Rida
|
reactive intermediate imine deaminase A homolog |
| chr7_+_78564062 | 0.99 |
ENSMUST00000205981.2
|
Isg20
|
interferon-stimulated protein |
| chr17_-_24382638 | 0.96 |
ENSMUST00000129523.3
ENSMUST00000138685.3 ENSMUST00000040735.12 |
Amdhd2
|
amidohydrolase domain containing 2 |
| chr7_+_79042052 | 0.92 |
ENSMUST00000118959.8
ENSMUST00000036865.13 |
Fanci
|
Fanconi anemia, complementation group I |
| chr19_+_46044972 | 0.91 |
ENSMUST00000111899.8
ENSMUST00000099392.10 ENSMUST00000062322.11 |
Pprc1
|
peroxisome proliferative activated receptor, gamma, coactivator-related 1 |
| chr9_-_118917275 | 0.90 |
ENSMUST00000214470.2
|
Plcd1
|
phospholipase C, delta 1 |
| chr6_-_38276157 | 0.89 |
ENSMUST00000058524.3
|
Zc3hav1l
|
zinc finger CCCH-type, antiviral 1-like |
| chr1_+_87254729 | 0.89 |
ENSMUST00000172794.8
ENSMUST00000164992.9 ENSMUST00000173173.8 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr15_+_84807640 | 0.83 |
ENSMUST00000230411.2
|
Nup50
|
nucleoporin 50 |
| chr4_-_108263873 | 0.82 |
ENSMUST00000184609.2
|
Gpx7
|
glutathione peroxidase 7 |
| chr5_-_25047577 | 0.80 |
ENSMUST00000030787.9
|
Rheb
|
Ras homolog enriched in brain |
| chr4_+_128548479 | 0.76 |
ENSMUST00000030588.13
ENSMUST00000136377.8 |
Phc2
|
polyhomeotic 2 |
| chr17_+_29251602 | 0.71 |
ENSMUST00000130216.3
|
Srsf3
|
serine and arginine-rich splicing factor 3 |
| chr10_+_20024203 | 0.69 |
ENSMUST00000020173.16
|
Map7
|
microtubule-associated protein 7 |
| chr3_+_96604415 | 0.68 |
ENSMUST00000107077.4
|
Pias3
|
protein inhibitor of activated STAT 3 |
| chr10_-_119075910 | 0.68 |
ENSMUST00000020315.13
|
Cand1
|
cullin associated and neddylation disassociated 1 |
| chr15_+_77617804 | 0.67 |
ENSMUST00000230377.2
|
Apol9b
|
apolipoprotein L 9b |
| chrX_-_37653396 | 0.66 |
ENSMUST00000016681.15
|
Cul4b
|
cullin 4B |
| chr4_-_131802606 | 0.66 |
ENSMUST00000146021.8
|
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr3_-_37778470 | 0.64 |
ENSMUST00000108105.2
ENSMUST00000079755.5 ENSMUST00000099128.2 |
Gm5148
|
predicted gene 5148 |
| chr3_+_96604390 | 0.63 |
ENSMUST00000162778.3
ENSMUST00000064900.16 |
Pias3
|
protein inhibitor of activated STAT 3 |
| chr7_-_126859790 | 0.61 |
ENSMUST00000035276.5
|
Dctpp1
|
dCTP pyrophosphatase 1 |
| chr5_+_31212110 | 0.58 |
ENSMUST00000013773.12
ENSMUST00000201838.4 |
Cad
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr10_+_20024724 | 0.57 |
ENSMUST00000116259.5
ENSMUST00000214231.2 |
Map7
|
microtubule-associated protein 7 |
| chr16_-_5021843 | 0.57 |
ENSMUST00000147567.2
ENSMUST00000023911.11 |
Nagpa
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase |
| chr3_+_89680867 | 0.55 |
ENSMUST00000038356.13
|
Ube2q1
|
ubiquitin-conjugating enzyme E2Q family member 1 |
| chr6_+_129489551 | 0.54 |
ENSMUST00000204741.3
|
Tmem52b
|
transmembrane protein 52B |
| chr6_-_13871475 | 0.53 |
ENSMUST00000139231.2
|
2610001J05Rik
|
RIKEN cDNA 2610001J05 gene |
| chr5_+_31212165 | 0.52 |
ENSMUST00000202795.4
ENSMUST00000201182.4 ENSMUST00000200953.4 |
Cad
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr1_+_75119419 | 0.51 |
ENSMUST00000097694.11
ENSMUST00000190240.7 |
Retreg2
|
reticulophagy regulator family member 2 |
| chr5_+_108416763 | 0.49 |
ENSMUST00000031190.5
|
Dr1
|
down-regulator of transcription 1 |
| chr16_-_4867703 | 0.49 |
ENSMUST00000115844.3
ENSMUST00000023189.15 |
Glyr1
|
glyoxylate reductase 1 homolog (Arabidopsis) |
| chr13_-_103911092 | 0.47 |
ENSMUST00000074616.7
|
Srek1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr8_-_105981732 | 0.47 |
ENSMUST00000093217.9
ENSMUST00000161745.3 ENSMUST00000136822.3 |
B3gnt9
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 9 |
| chr11_-_5049223 | 0.47 |
ENSMUST00000079949.13
|
Ewsr1
|
Ewing sarcoma breakpoint region 1 |
| chr9_-_107166543 | 0.46 |
ENSMUST00000192054.2
|
Mapkapk3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr4_+_99544536 | 0.46 |
ENSMUST00000087285.5
|
Foxd3
|
forkhead box D3 |
| chr2_-_126718129 | 0.45 |
ENSMUST00000103224.10
|
Trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr10_+_80008768 | 0.41 |
ENSMUST00000041882.7
|
Fam174c
|
family with sequence similarity 174, member C |
| chr2_+_150628655 | 0.39 |
ENSMUST00000045441.8
|
Pygb
|
brain glycogen phosphorylase |
| chr11_+_100436433 | 0.38 |
ENSMUST00000092684.12
ENSMUST00000006976.8 |
Odad4
|
outer dynein arm complex subunit 4 |
| chr17_-_12988492 | 0.38 |
ENSMUST00000024599.14
|
Igf2r
|
insulin-like growth factor 2 receptor |
| chr6_-_13871454 | 0.37 |
ENSMUST00000185219.2
ENSMUST00000185361.2 ENSMUST00000155856.2 |
2610001J05Rik
|
RIKEN cDNA 2610001J05 gene |
| chr7_+_16609227 | 0.37 |
ENSMUST00000108493.3
|
Dact3
|
dishevelled-binding antagonist of beta-catenin 3 |
| chr6_-_40917431 | 0.35 |
ENSMUST00000122181.8
ENSMUST00000031935.10 |
1700074P13Rik
|
RIKEN cDNA 1700074P13 gene |
| chr4_+_148123554 | 0.34 |
ENSMUST00000141283.8
|
Mthfr
|
methylenetetrahydrofolate reductase |
| chr15_+_34495441 | 0.32 |
ENSMUST00000052290.14
ENSMUST00000079028.6 |
Pop1
|
processing of precursor 1, ribonuclease P/MRP family, (S. cerevisiae) |
| chr14_-_70588803 | 0.32 |
ENSMUST00000143153.2
ENSMUST00000127000.2 ENSMUST00000068044.14 ENSMUST00000022688.10 |
Slc39a14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr2_-_162929732 | 0.32 |
ENSMUST00000094653.6
|
Gtsf1l
|
gametocyte specific factor 1-like |
| chr5_-_137608726 | 0.32 |
ENSMUST00000197912.5
|
Pcolce
|
procollagen C-endopeptidase enhancer protein |
| chr5_-_137608886 | 0.31 |
ENSMUST00000142675.8
|
Pcolce
|
procollagen C-endopeptidase enhancer protein |
| chr16_+_4867876 | 0.25 |
ENSMUST00000230703.2
ENSMUST00000052449.6 |
Ubn1
|
ubinuclein 1 |
| chr13_+_90237824 | 0.25 |
ENSMUST00000012566.9
|
Tmem167
|
transmembrane protein 167 |
| chr7_-_45116316 | 0.24 |
ENSMUST00000033093.10
|
Bax
|
BCL2-associated X protein |
| chr13_-_24390265 | 0.23 |
ENSMUST00000123076.8
|
Carmil1
|
capping protein regulator and myosin 1 linker 1 |
| chr7_-_105289515 | 0.20 |
ENSMUST00000133519.8
ENSMUST00000209550.2 ENSMUST00000210911.2 ENSMUST00000084782.10 ENSMUST00000131446.8 |
Arfip2
|
ADP-ribosylation factor interacting protein 2 |
| chr2_-_126718037 | 0.20 |
ENSMUST00000028843.12
|
Trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr15_+_79575046 | 0.19 |
ENSMUST00000046463.10
|
Gtpbp1
|
GTP binding protein 1 |
| chr4_-_131802561 | 0.19 |
ENSMUST00000105970.8
ENSMUST00000105975.8 |
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr4_+_44756608 | 0.18 |
ENSMUST00000143385.2
|
Zcchc7
|
zinc finger, CCHC domain containing 7 |
| chr6_-_145021816 | 0.17 |
ENSMUST00000111742.8
ENSMUST00000048252.11 |
Bcat1
|
branched chain aminotransferase 1, cytosolic |
| chr13_-_94422337 | 0.17 |
ENSMUST00000022197.15
ENSMUST00000152555.8 |
Scamp1
|
secretory carrier membrane protein 1 |
| chr18_+_67597929 | 0.17 |
ENSMUST00000025411.9
|
Prelid3a
|
PRELI domain containing 3A |
| chr8_-_125390107 | 0.15 |
ENSMUST00000034464.8
|
2310022B05Rik
|
RIKEN cDNA 2310022B05 gene |
| chr19_-_6817955 | 0.13 |
ENSMUST00000239527.1
|
RPS6KA4
|
ribosomal protein S6 kinase, polypeptide 4 |
| chr4_+_136038243 | 0.12 |
ENSMUST00000131671.8
|
Hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr9_+_21867043 | 0.11 |
ENSMUST00000053583.7
|
Swsap1
|
SWIM type zinc finger 7 associated protein 1 |
| chr8_-_95328348 | 0.08 |
ENSMUST00000212547.2
ENSMUST00000212507.2 ENSMUST00000034226.8 |
Psme3ip1
|
proteasome activator subunit 3 interacting protein 1 |
| chr2_+_70491901 | 0.07 |
ENSMUST00000112201.8
ENSMUST00000028509.11 ENSMUST00000133432.8 ENSMUST00000112205.2 |
Gorasp2
|
golgi reassembly stacking protein 2 |
| chr5_+_31026967 | 0.05 |
ENSMUST00000114716.4
|
Tmem214
|
transmembrane protein 214 |
| chr17_-_29566774 | 0.04 |
ENSMUST00000095427.12
ENSMUST00000118366.9 |
Mtch1
|
mitochondrial carrier 1 |
| chr4_+_136038301 | 0.03 |
ENSMUST00000084219.12
|
Hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr6_-_93769426 | 0.02 |
ENSMUST00000204788.2
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr8_-_95328260 | 0.02 |
ENSMUST00000212765.2
ENSMUST00000213008.2 ENSMUST00000212159.2 ENSMUST00000213022.2 ENSMUST00000212258.2 ENSMUST00000212788.2 ENSMUST00000212791.2 |
Psme3ip1
|
proteasome activator subunit 3 interacting protein 1 |
| chr11_+_82671934 | 0.01 |
ENSMUST00000092849.12
ENSMUST00000021039.12 ENSMUST00000080461.12 ENSMUST00000173347.8 ENSMUST00000173727.8 ENSMUST00000173009.8 ENSMUST00000131537.9 ENSMUST00000173722.8 |
Lig3
|
ligase III, DNA, ATP-dependent |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:0006592 | ornithine biosynthetic process(GO:0006592) |
| 1.3 | 4.0 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 1.1 | 3.2 | GO:0001966 | thigmotaxis(GO:0001966) |
| 1.1 | 6.4 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.9 | 5.6 | GO:1903438 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.9 | 3.5 | GO:0009609 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) response to selenium ion(GO:0010269) |
| 0.9 | 10.4 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.8 | 2.4 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.8 | 4.6 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.7 | 4.2 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.7 | 2.6 | GO:0035666 | TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.6 | 11.5 | GO:0015816 | glycine transport(GO:0015816) |
| 0.5 | 1.6 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.5 | 1.5 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.5 | 1.5 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.5 | 5.3 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.4 | 1.3 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) |
| 0.4 | 2.1 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.4 | 1.7 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.4 | 2.5 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.4 | 2.1 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.4 | 1.2 | GO:1902287 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.4 | 2.5 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.4 | 1.8 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.3 | 1.7 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.3 | 1.7 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.3 | 13.9 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) |
| 0.3 | 4.0 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.3 | 2.8 | GO:2000320 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.3 | 1.3 | GO:0033387 | putrescine biosynthetic process(GO:0009446) putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.2 | 0.7 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.2 | 4.8 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.2 | 0.6 | GO:0042262 | DNA protection(GO:0042262) |
| 0.2 | 0.9 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.2 | 1.0 | GO:0060526 | prostate glandular acinus morphogenesis(GO:0060526) prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis(GO:0060527) |
| 0.2 | 8.4 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.2 | 5.5 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.2 | 6.4 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.2 | 3.0 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.2 | 1.1 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.1 | 1.0 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.1 | 1.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.1 | 4.8 | GO:0043496 | regulation of protein homodimerization activity(GO:0043496) |
| 0.1 | 2.7 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.1 | 1.9 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 2.6 | GO:0046653 | tetrahydrofolate metabolic process(GO:0046653) |
| 0.1 | 0.6 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.1 | 1.0 | GO:0051683 | establishment of Golgi localization(GO:0051683) |
| 0.1 | 2.2 | GO:0032402 | melanosome transport(GO:0032402) |
| 0.1 | 0.2 | GO:0051695 | actin filament uncapping(GO:0051695) negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.1 | 1.3 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 4.4 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 0.8 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.1 | 2.9 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.1 | 1.3 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.1 | 3.7 | GO:0009409 | response to cold(GO:0009409) |
| 0.1 | 4.2 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.1 | 0.7 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 1.1 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.1 | 1.5 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 3.0 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.1 | 0.3 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.1 | 4.8 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.1 | 0.6 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.1 | 0.5 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 4.2 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 0.9 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 1.1 | GO:0036474 | cell death in response to hydrogen peroxide(GO:0036474) regulation of hydrogen peroxide-induced cell death(GO:1903205) |
| 0.0 | 11.2 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 1.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.9 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.7 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.7 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 | 3.0 | GO:0055072 | iron ion homeostasis(GO:0055072) |
| 0.0 | 0.2 | GO:0009099 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 | 1.4 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 2.1 | GO:0007045 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) |
| 0.0 | 0.3 | GO:0071578 | iron ion transport(GO:0006826) zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.4 | GO:0009251 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.5 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.3 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 0.3 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.6 | 4.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.4 | 3.5 | GO:0097413 | Lewy body(GO:0097413) |
| 0.4 | 2.1 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.3 | 3.0 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.3 | 1.5 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.3 | 2.6 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.3 | 1.0 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.2 | 5.6 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.2 | 1.7 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 2.8 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 1.8 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.1 | 4.0 | GO:0045120 | pronucleus(GO:0045120) |
| 0.1 | 6.4 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 0.7 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.2 | GO:0097144 | BAX complex(GO:0097144) |
| 0.1 | 1.0 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 4.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 5.4 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 0.3 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 0.7 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 2.2 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.9 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 1.8 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 2.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.8 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 4.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.2 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 3.1 | GO:0015934 | large ribosomal subunit(GO:0015934) |
| 0.0 | 4.7 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.9 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 2.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.5 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.9 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 21.2 | GO:0031410 | cytoplasmic vesicle(GO:0031410) |
| 0.0 | 25.0 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 10.6 | GO:0005794 | Golgi apparatus(GO:0005794) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:0017084 | glutamate 5-kinase activity(GO:0004349) glutamate-5-semialdehyde dehydrogenase activity(GO:0004350) delta1-pyrroline-5-carboxylate synthetase activity(GO:0017084) amino acid kinase activity(GO:0019202) |
| 1.2 | 13.9 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 1.2 | 10.4 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.8 | 4.2 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.8 | 4.2 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.8 | 11.5 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.8 | 2.4 | GO:0004326 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) |
| 0.6 | 5.8 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.5 | 1.6 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) |
| 0.5 | 2.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.5 | 2.5 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.5 | 2.8 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.4 | 2.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.4 | 4.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.4 | 1.1 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.3 | 1.3 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.3 | 2.8 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.3 | 1.2 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.3 | 2.5 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.2 | 1.7 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.2 | 0.8 | GO:0004096 | catalase activity(GO:0004096) |
| 0.2 | 5.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 1.5 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.2 | 4.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 0.5 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.2 | 1.7 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.2 | 4.8 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 3.0 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 1.7 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 2.7 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 1.0 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.1 | 2.1 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.1 | 0.3 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.1 | 3.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.4 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 1.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 1.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 5.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 1.9 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.1 | 4.0 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.1 | 0.6 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 1.3 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 1.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 2.0 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 5.8 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 1.3 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 1.2 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.5 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 2.5 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.9 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 2.1 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.2 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.4 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 8.4 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.0 | 1.3 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.2 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.3 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.7 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 6.4 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 1.0 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 10.9 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.6 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.5 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.8 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 13.9 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 1.3 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 1.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 2.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 6.4 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 2.8 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 2.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 4.0 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 5.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 2.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 6.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.9 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.7 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 4.2 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 3.5 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.5 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.6 | PID FANCONI PATHWAY | Fanconi anemia pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 3.8 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.2 | 6.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 2.6 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.2 | 3.5 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.2 | 3.0 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 9.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 3.0 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 4.0 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 2.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 1.9 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 4.2 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 2.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 4.2 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 1.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 4.2 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 0.8 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 3.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 1.3 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.1 | 1.9 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 1.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 2.3 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 1.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 3.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 4.6 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.1 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 4.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 6.1 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 1.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.2 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 0.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.5 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.7 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |