Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Crx_Gsc
Z-value: 0.79
Transcription factors associated with Crx_Gsc
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
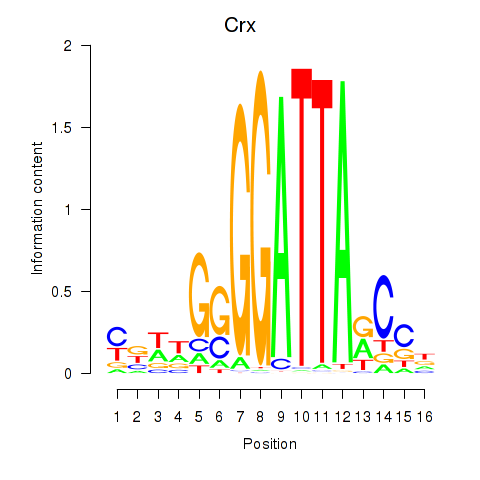
Crx
|
ENSMUSG00000041578.16 | Crx |
|
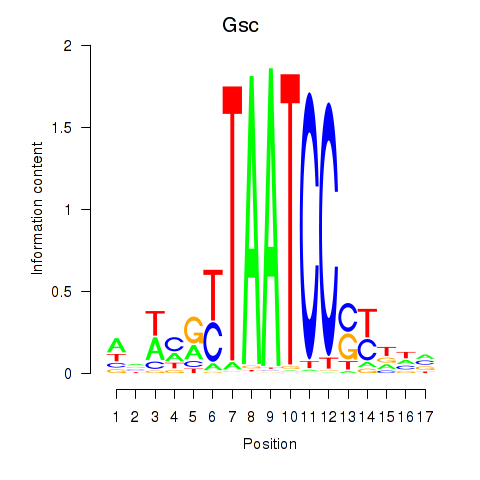
Gsc
|
ENSMUSG00000021095.6 | Gsc |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gsc | mm39_v1_chr12_-_104439589_104439633 | -0.32 | 6.8e-03 | Click! |
| Crx | mm39_v1_chr7_-_15613769_15613893 | -0.08 | 5.3e-01 | Click! |
Activity profile of Crx_Gsc motif
Sorted Z-values of Crx_Gsc motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Crx_Gsc
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_33614378 | 7.18 |
ENSMUST00000115044.8
|
Muc13
|
mucin 13, epithelial transmembrane |
| chr6_-_69553484 | 6.88 |
ENSMUST00000103357.4
|
Igkv4-57
|
immunoglobulin kappa variable 4-57 |
| chr12_-_114621406 | 6.83 |
ENSMUST00000192077.2
|
Ighv1-15
|
immunoglobulin heavy variable 1-15 |
| chr6_-_69204417 | 5.72 |
ENSMUST00000103346.3
|
Igkv4-72
|
immunoglobulin kappa chain variable 4-72 |
| chr14_-_68819544 | 5.47 |
ENSMUST00000022641.9
|
Adamdec1
|
ADAM-like, decysin 1 |
| chr6_-_69245427 | 5.10 |
ENSMUST00000103348.3
|
Igkv4-70
|
immunoglobulin kappa chain variable 4-70 |
| chr6_-_68713748 | 4.65 |
ENSMUST00000183936.2
ENSMUST00000196863.2 |
Igkv19-93
|
immunoglobulin kappa chain variable 19-93 |
| chr6_-_59403264 | 4.47 |
ENSMUST00000051065.6
|
Gprin3
|
GPRIN family member 3 |
| chr7_-_79497940 | 4.32 |
ENSMUST00000107392.8
|
Anpep
|
alanyl (membrane) aminopeptidase |
| chr7_-_79497922 | 4.31 |
ENSMUST00000205502.2
|
Anpep
|
alanyl (membrane) aminopeptidase |
| chr16_+_33614715 | 4.25 |
ENSMUST00000023520.7
|
Muc13
|
mucin 13, epithelial transmembrane |
| chr6_+_68247469 | 4.07 |
ENSMUST00000103321.3
|
Igkv1-110
|
immunoglobulin kappa variable 1-110 |
| chr10_+_127478844 | 3.99 |
ENSMUST00000092074.12
ENSMUST00000120279.2 |
Stat6
|
signal transducer and activator of transcription 6 |
| chr12_-_113379925 | 3.95 |
ENSMUST00000194162.6
ENSMUST00000192250.2 |
Ighd
|
immunoglobulin heavy constant delta |
| chr6_-_68746087 | 3.82 |
ENSMUST00000103333.4
|
Igkv4-91
|
immunoglobulin kappa chain variable 4-91 |
| chr12_-_114646685 | 3.67 |
ENSMUST00000194350.6
ENSMUST00000103504.3 |
Ighv1-18
|
immunoglobulin heavy variable V1-18 |
| chr12_-_114752425 | 3.57 |
ENSMUST00000103510.2
|
Ighv1-26
|
immunoglobulin heavy variable 1-26 |
| chr12_-_114263874 | 3.54 |
ENSMUST00000103482.2
ENSMUST00000194159.2 |
Ighv9-4
|
immunoglobulin heavy variable 9-4 |
| chr3_-_16060545 | 3.39 |
ENSMUST00000194367.6
|
Gm5150
|
predicted gene 5150 |
| chr12_+_79177523 | 3.35 |
ENSMUST00000021550.7
|
Arg2
|
arginase type II |
| chr12_-_113823290 | 3.29 |
ENSMUST00000103459.5
|
Ighv5-17
|
immunoglobulin heavy variable 5-17 |
| chr8_+_22224506 | 3.28 |
ENSMUST00000080533.6
|
Defa24
|
defensin, alpha, 24 |
| chr12_-_115587215 | 3.16 |
ENSMUST00000199933.5
ENSMUST00000103539.3 |
Ighv1-69
|
immunoglobulin heavy variable 1-69 |
| chr3_-_16060491 | 3.10 |
ENSMUST00000108347.3
|
Gm5150
|
predicted gene 5150 |
| chr11_+_70396070 | 2.92 |
ENSMUST00000019063.3
|
Tm4sf5
|
transmembrane 4 superfamily member 5 |
| chr9_-_46146558 | 2.79 |
ENSMUST00000121916.8
ENSMUST00000034586.9 |
Apoc3
|
apolipoprotein C-III |
| chr3_-_144555062 | 2.76 |
ENSMUST00000159989.2
|
Clca3b
|
chloride channel accessory 3B |
| chr14_+_51193449 | 2.74 |
ENSMUST00000095925.5
|
Pnp2
|
purine-nucleoside phosphorylase 2 |
| chr12_-_115157739 | 2.72 |
ENSMUST00000103525.3
|
Ighv1-54
|
immunoglobulin heavy variable V1-54 |
| chr15_-_76501041 | 2.52 |
ENSMUST00000073428.7
|
Slc39a4
|
solute carrier family 39 (zinc transporter), member 4 |
| chr1_+_93789068 | 2.40 |
ENSMUST00000094663.4
|
Gal3st2
|
galactose-3-O-sulfotransferase 2 |
| chr9_-_46146928 | 2.37 |
ENSMUST00000118649.8
|
Apoc3
|
apolipoprotein C-III |
| chr1_-_87029312 | 2.35 |
ENSMUST00000113270.3
|
Alpi
|
alkaline phosphatase, intestinal |
| chr17_+_34416707 | 2.25 |
ENSMUST00000025196.9
|
Psmb8
|
proteasome (prosome, macropain) subunit, beta type 8 (large multifunctional peptidase 7) |
| chr12_-_114901026 | 2.21 |
ENSMUST00000103516.2
ENSMUST00000191868.2 |
Ighv1-42
|
immunoglobulin heavy variable V1-42 |
| chr5_+_53195423 | 2.18 |
ENSMUST00000170523.8
|
Slc34a2
|
solute carrier family 34 (sodium phosphate), member 2 |
| chr15_-_75766321 | 2.12 |
ENSMUST00000023237.8
|
Naprt
|
nicotinate phosphoribosyltransferase |
| chr19_+_43678109 | 2.11 |
ENSMUST00000081079.6
|
Entpd7
|
ectonucleoside triphosphate diphosphohydrolase 7 |
| chr5_-_143963413 | 2.07 |
ENSMUST00000031622.13
|
Ocm
|
oncomodulin |
| chr10_-_75617245 | 2.07 |
ENSMUST00000001715.10
|
Gstt3
|
glutathione S-transferase, theta 3 |
| chr17_+_34416689 | 2.06 |
ENSMUST00000173441.9
|
Psmb8
|
proteasome (prosome, macropain) subunit, beta type 8 (large multifunctional peptidase 7) |
| chr3_+_28752050 | 2.05 |
ENSMUST00000029240.14
|
Slc2a2
|
solute carrier family 2 (facilitated glucose transporter), member 2 |
| chr1_+_93855759 | 2.03 |
ENSMUST00000177958.2
|
Gal3st2b
|
galactose-3-O-sulfotransferase 2B |
| chr1_+_172328768 | 1.98 |
ENSMUST00000111228.2
|
Tagln2
|
transgelin 2 |
| chr11_+_3981769 | 1.98 |
ENSMUST00000019512.8
|
Sec14l4
|
SEC14-like lipid binding 4 |
| chr17_+_87943401 | 1.98 |
ENSMUST00000235125.2
ENSMUST00000053577.9 ENSMUST00000234009.2 |
Epcam
|
epithelial cell adhesion molecule |
| chr1_+_93789548 | 1.97 |
ENSMUST00000187896.2
|
Gal3st2
|
galactose-3-O-sulfotransferase 2 |
| chr3_-_73615535 | 1.96 |
ENSMUST00000138216.8
|
Bche
|
butyrylcholinesterase |
| chr16_+_23109213 | 1.93 |
ENSMUST00000115335.2
|
St6gal1
|
beta galactoside alpha 2,6 sialyltransferase 1 |
| chr13_+_19362068 | 1.92 |
ENSMUST00000103553.3
|
Trgv7
|
T cell receptor gamma, variable 7 |
| chr2_+_3771709 | 1.85 |
ENSMUST00000177037.2
|
Fam107b
|
family with sequence similarity 107, member B |
| chr17_-_35077089 | 1.81 |
ENSMUST00000153400.8
|
Cfb
|
complement factor B |
| chr6_+_123206802 | 1.80 |
ENSMUST00000112554.9
ENSMUST00000024118.11 ENSMUST00000117130.8 |
Clec4n
|
C-type lectin domain family 4, member n |
| chr6_+_123206880 | 1.76 |
ENSMUST00000205129.2
|
Clec4n
|
C-type lectin domain family 4, member n |
| chr15_-_76501525 | 1.74 |
ENSMUST00000230977.2
|
Slc39a4
|
solute carrier family 39 (zinc transporter), member 4 |
| chr14_-_87378641 | 1.72 |
ENSMUST00000168889.3
ENSMUST00000022599.14 |
Diaph3
|
diaphanous related formin 3 |
| chr6_-_68887957 | 1.68 |
ENSMUST00000200454.2
|
Igkv4-86
|
immunoglobulin kappa variable 4-86 |
| chr3_-_73615732 | 1.67 |
ENSMUST00000029367.6
|
Bche
|
butyrylcholinesterase |
| chr6_+_119213468 | 1.67 |
ENSMUST00000238905.2
ENSMUST00000037434.13 |
Cacna2d4
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
| chr18_-_73948501 | 1.66 |
ENSMUST00000025439.5
|
Me2
|
malic enzyme 2, NAD(+)-dependent, mitochondrial |
| chr5_+_117258185 | 1.65 |
ENSMUST00000111978.8
|
Taok3
|
TAO kinase 3 |
| chr2_+_24275321 | 1.62 |
ENSMUST00000056641.15
ENSMUST00000142522.8 ENSMUST00000131930.2 |
Psd4
|
pleckstrin and Sec7 domain containing 4 |
| chr3_+_103767581 | 1.61 |
ENSMUST00000029433.9
|
Ptpn22
|
protein tyrosine phosphatase, non-receptor type 22 (lymphoid) |
| chr6_-_68887922 | 1.58 |
ENSMUST00000103337.3
|
Igkv4-86
|
immunoglobulin kappa variable 4-86 |
| chr5_-_104125270 | 1.58 |
ENSMUST00000112803.3
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr8_+_79235946 | 1.58 |
ENSMUST00000209490.2
ENSMUST00000034111.10 |
Slc10a7
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 7 |
| chr11_-_116080361 | 1.57 |
ENSMUST00000148601.2
|
Acox1
|
acyl-Coenzyme A oxidase 1, palmitoyl |
| chr11_-_16958647 | 1.56 |
ENSMUST00000102881.10
|
Plek
|
pleckstrin |
| chr11_-_101998648 | 1.53 |
ENSMUST00000177304.8
ENSMUST00000017455.15 |
Pyy
|
peptide YY |
| chr5_+_104447037 | 1.52 |
ENSMUST00000031246.9
|
Ibsp
|
integrin binding sialoprotein |
| chr3_+_128993609 | 1.52 |
ENSMUST00000173645.4
|
Pitx2
|
paired-like homeodomain transcription factor 2 |
| chr11_-_40624200 | 1.51 |
ENSMUST00000020579.9
|
Hmmr
|
hyaluronan mediated motility receptor (RHAMM) |
| chr5_-_104125192 | 1.45 |
ENSMUST00000120320.8
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr17_+_34124078 | 1.45 |
ENSMUST00000172817.2
|
Smim40
|
small integral membrane protein 40 |
| chr17_-_87573294 | 1.44 |
ENSMUST00000145895.8
ENSMUST00000129616.8 ENSMUST00000155904.2 ENSMUST00000151155.8 ENSMUST00000144236.9 ENSMUST00000024963.11 |
Mcfd2
|
multiple coagulation factor deficiency 2 |
| chr5_-_28442871 | 1.44 |
ENSMUST00000120068.2
|
Cnpy1
|
canopy FGF signaling regulator 1 |
| chr10_+_75778885 | 1.43 |
ENSMUST00000121151.2
|
Vpreb3
|
pre-B lymphocyte gene 3 |
| chr18_+_70058613 | 1.39 |
ENSMUST00000080050.6
|
Ccdc68
|
coiled-coil domain containing 68 |
| chr11_-_70350783 | 1.39 |
ENSMUST00000019064.9
|
Cxcl16
|
chemokine (C-X-C motif) ligand 16 |
| chr11_-_99176086 | 1.39 |
ENSMUST00000017255.4
|
Krt24
|
keratin 24 |
| chr14_+_54431947 | 1.37 |
ENSMUST00000103715.2
|
Traj26
|
T cell receptor alpha joining 26 |
| chr7_+_99827886 | 1.37 |
ENSMUST00000207358.2
ENSMUST00000207995.2 ENSMUST00000049333.13 ENSMUST00000170954.10 ENSMUST00000179842.3 ENSMUST00000208260.2 |
Kcne3
|
potassium voltage-gated channel, Isk-related subfamily, gene 3 |
| chr4_-_140805613 | 1.37 |
ENSMUST00000030760.15
|
Necap2
|
NECAP endocytosis associated 2 |
| chr1_+_171265103 | 1.36 |
ENSMUST00000043839.5
|
F11r
|
F11 receptor |
| chr13_-_74918745 | 1.35 |
ENSMUST00000223033.2
ENSMUST00000222588.2 |
Cast
|
calpastatin |
| chr1_-_126665904 | 1.35 |
ENSMUST00000160693.2
|
Nckap5
|
NCK-associated protein 5 |
| chr12_-_114528632 | 1.34 |
ENSMUST00000195337.2
ENSMUST00000103497.2 |
Ighv15-2
|
immunoglobulin heavy variable V15-2 |
| chr6_+_122898764 | 1.33 |
ENSMUST00000060484.9
|
Clec4a1
|
C-type lectin domain family 4, member a1 |
| chr9_+_123596276 | 1.32 |
ENSMUST00000166236.9
ENSMUST00000111454.4 ENSMUST00000168910.2 |
Ccr9
|
chemokine (C-C motif) receptor 9 |
| chr7_-_101765970 | 1.31 |
ENSMUST00000211408.2
|
Chrna10
|
cholinergic receptor, nicotinic, alpha polypeptide 10 |
| chr9_+_88209250 | 1.31 |
ENSMUST00000034992.8
|
Nt5e
|
5' nucleotidase, ecto |
| chr11_+_83553400 | 1.30 |
ENSMUST00000019074.4
|
Ccl4
|
chemokine (C-C motif) ligand 4 |
| chr13_+_21995906 | 1.30 |
ENSMUST00000104941.4
|
H4c17
|
H4 clustered histone 17 |
| chr19_+_3373285 | 1.29 |
ENSMUST00000025835.6
|
Cpt1a
|
carnitine palmitoyltransferase 1a, liver |
| chr9_-_50528727 | 1.28 |
ENSMUST00000131351.8
ENSMUST00000171462.8 |
Nkapd1
|
NKAP domain containing 1 |
| chr19_+_11469386 | 1.26 |
ENSMUST00000079855.11
|
Gm8369
|
predicted gene 8369 |
| chrX_-_101232978 | 1.26 |
ENSMUST00000033683.8
|
Rps4x
|
ribosomal protein S4, X-linked |
| chr19_-_5416626 | 1.25 |
ENSMUST00000237167.2
|
Banf1
|
BAF nuclear assembly factor 1 |
| chr5_-_143963461 | 1.25 |
ENSMUST00000110702.2
|
Ocm
|
oncomodulin |
| chr12_-_115206715 | 1.23 |
ENSMUST00000103527.2
ENSMUST00000194071.2 |
Ighv1-56
|
immunoglobulin heavy variable 1-56 |
| chr16_+_36004452 | 1.22 |
ENSMUST00000114858.2
|
Cstdc4
|
cystatin domain containing 4 |
| chr18_+_70058533 | 1.22 |
ENSMUST00000043929.11
|
Ccdc68
|
coiled-coil domain containing 68 |
| chr1_+_178014983 | 1.21 |
ENSMUST00000161075.8
ENSMUST00000027783.14 |
Desi2
|
desumoylating isopeptidase 2 |
| chr6_+_41165156 | 1.21 |
ENSMUST00000103277.2
|
Trbv20
|
T cell receptor beta, variable 20 |
| chr6_+_119213541 | 1.20 |
ENSMUST00000186622.3
|
Cacna2d4
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
| chr3_-_113198765 | 1.20 |
ENSMUST00000179568.3
|
Amy2a4
|
amylase 2a4 |
| chr9_+_108880158 | 1.18 |
ENSMUST00000198708.5
|
Shisa5
|
shisa family member 5 |
| chr4_+_130640436 | 1.17 |
ENSMUST00000151698.8
|
Laptm5
|
lysosomal-associated protein transmembrane 5 |
| chr5_-_104125226 | 1.16 |
ENSMUST00000048118.15
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr5_+_77413282 | 1.15 |
ENSMUST00000080359.12
|
Rest
|
RE1-silencing transcription factor |
| chr10_-_59452489 | 1.13 |
ENSMUST00000020312.13
|
Mcu
|
mitochondrial calcium uniporter |
| chr19_-_5416339 | 1.12 |
ENSMUST00000170010.3
|
Banf1
|
BAF nuclear assembly factor 1 |
| chr19_+_5416769 | 1.11 |
ENSMUST00000025759.9
|
Eif1ad
|
eukaryotic translation initiation factor 1A domain containing |
| chr9_-_50528530 | 1.09 |
ENSMUST00000147671.2
ENSMUST00000145139.8 ENSMUST00000155435.8 |
Nkapd1
|
NKAP domain containing 1 |
| chr5_-_66309244 | 1.09 |
ENSMUST00000167950.8
|
Rbm47
|
RNA binding motif protein 47 |
| chr7_-_110581652 | 1.08 |
ENSMUST00000005751.13
|
Irag1
|
inositol 1,4,5-triphosphate receptor associated 1 |
| chr13_-_74918701 | 1.07 |
ENSMUST00000223126.2
|
Cast
|
calpastatin |
| chr5_+_122348140 | 1.05 |
ENSMUST00000196187.5
ENSMUST00000100747.3 |
Hvcn1
|
hydrogen voltage-gated channel 1 |
| chr6_-_113717689 | 1.03 |
ENSMUST00000032440.6
|
Sec13
|
SEC13 homolog, nuclear pore and COPII coat complex component |
| chr13_-_64645606 | 1.02 |
ENSMUST00000180282.2
|
Fam240b
|
family with sequence similarity 240 member B |
| chr7_+_19753097 | 1.01 |
ENSMUST00000117909.2
|
Nlrp9b
|
NLR family, pyrin domain containing 9B |
| chr1_+_178015287 | 1.01 |
ENSMUST00000159284.2
|
Desi2
|
desumoylating isopeptidase 2 |
| chr4_-_133360749 | 1.00 |
ENSMUST00000084238.5
|
Zdhhc18
|
zinc finger, DHHC domain containing 18 |
| chr10_-_86843878 | 0.98 |
ENSMUST00000035288.17
|
Stab2
|
stabilin 2 |
| chr11_-_109364046 | 0.98 |
ENSMUST00000070872.13
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
| chr4_+_73931679 | 0.97 |
ENSMUST00000098006.9
ENSMUST00000084474.6 |
Frmd3
|
FERM domain containing 3 |
| chr6_+_34843896 | 0.96 |
ENSMUST00000202999.2
|
Tmem140
|
transmembrane protein 140 |
| chr19_-_4189603 | 0.95 |
ENSMUST00000025761.8
|
Cabp4
|
calcium binding protein 4 |
| chr6_+_117877272 | 0.94 |
ENSMUST00000177743.8
ENSMUST00000167182.8 |
Hnrnpf
|
heterogeneous nuclear ribonucleoprotein F |
| chr5_-_65248927 | 0.94 |
ENSMUST00000043352.8
|
Tmem156
|
transmembrane protein 156 |
| chr5_+_64316771 | 0.92 |
ENSMUST00000043893.13
|
Tbc1d1
|
TBC1 domain family, member 1 |
| chr1_-_165021879 | 0.92 |
ENSMUST00000043338.10
|
Sft2d2
|
SFT2 domain containing 2 |
| chr17_-_13211374 | 0.90 |
ENSMUST00000159551.8
ENSMUST00000160781.8 |
Wtap
|
Wilms tumour 1-associating protein |
| chr2_-_144112444 | 0.88 |
ENSMUST00000028909.5
|
Snx5
|
sorting nexin 5 |
| chr11_+_5470652 | 0.84 |
ENSMUST00000063084.16
|
Xbp1
|
X-box binding protein 1 |
| chr10_-_51466567 | 0.84 |
ENSMUST00000020064.5
|
Fam162b
|
family with sequence similarity 162, member B |
| chr1_-_189420270 | 0.84 |
ENSMUST00000171929.8
ENSMUST00000165962.2 |
Cenpf
|
centromere protein F |
| chr1_+_24717968 | 0.83 |
ENSMUST00000095062.10
|
Lmbrd1
|
LMBR1 domain containing 1 |
| chr5_-_137529465 | 0.83 |
ENSMUST00000150063.9
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr1_-_74233790 | 0.83 |
ENSMUST00000053389.5
|
Cxcr1
|
chemokine (C-X-C motif) receptor 1 |
| chr7_+_92524460 | 0.82 |
ENSMUST00000076052.8
|
Prcp
|
prolylcarboxypeptidase (angiotensinase C) |
| chr6_-_5193757 | 0.82 |
ENSMUST00000177159.9
ENSMUST00000176945.2 |
Pon1
|
paraoxonase 1 |
| chr7_+_24584197 | 0.81 |
ENSMUST00000156372.8
ENSMUST00000124035.2 |
Rps19
|
ribosomal protein S19 |
| chr11_-_66059270 | 0.81 |
ENSMUST00000108691.2
|
Dnah9
|
dynein, axonemal, heavy chain 9 |
| chr7_+_28510350 | 0.78 |
ENSMUST00000174477.8
|
Hnrnpl
|
heterogeneous nuclear ribonucleoprotein L |
| chr15_-_76524097 | 0.78 |
ENSMUST00000168185.8
|
Tonsl
|
tonsoku-like, DNA repair protein |
| chr18_-_70274639 | 0.77 |
ENSMUST00000121693.8
|
Rab27b
|
RAB27B, member RAS oncogene family |
| chr1_-_149798090 | 0.77 |
ENSMUST00000111926.9
|
Pla2g4a
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr16_-_16687119 | 0.75 |
ENSMUST00000075017.5
|
Vpreb1
|
pre-B lymphocyte gene 1 |
| chr6_-_69220672 | 0.75 |
ENSMUST00000196201.2
|
Igkv4-71
|
immunoglobulin kappa chain variable 4-71 |
| chr9_-_106768601 | 0.73 |
ENSMUST00000069036.14
|
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr7_+_92524495 | 0.71 |
ENSMUST00000207594.2
|
Prcp
|
prolylcarboxypeptidase (angiotensinase C) |
| chr2_+_121188195 | 0.71 |
ENSMUST00000125812.8
ENSMUST00000078222.9 ENSMUST00000125221.3 ENSMUST00000150271.8 |
Ckmt1
|
creatine kinase, mitochondrial 1, ubiquitous |
| chr13_+_108182941 | 0.71 |
ENSMUST00000224361.2
ENSMUST00000224390.2 |
Smim15
|
small integral membrane protein 15 |
| chr18_-_74340885 | 0.71 |
ENSMUST00000177604.2
|
Ska1
|
spindle and kinetochore associated complex subunit 1 |
| chr1_+_24717793 | 0.70 |
ENSMUST00000186190.2
|
Lmbrd1
|
LMBR1 domain containing 1 |
| chr1_-_153363354 | 0.69 |
ENSMUST00000186380.7
ENSMUST00000188345.2 ENSMUST00000042141.12 |
Dhx9
|
DEAH (Asp-Glu-Ala-His) box polypeptide 9 |
| chr5_-_92426014 | 0.69 |
ENSMUST00000159345.8
ENSMUST00000113102.10 |
Naaa
|
N-acylethanolamine acid amidase |
| chr6_+_113508636 | 0.69 |
ENSMUST00000036340.12
ENSMUST00000204827.3 |
Fancd2
|
Fanconi anemia, complementation group D2 |
| chr10_+_76284907 | 0.69 |
ENSMUST00000092406.12
|
2610028H24Rik
|
RIKEN cDNA 2610028H24 gene |
| chr7_-_63588610 | 0.69 |
ENSMUST00000063694.10
|
Klf13
|
Kruppel-like factor 13 |
| chr6_-_5193816 | 0.68 |
ENSMUST00000002663.12
|
Pon1
|
paraoxonase 1 |
| chr18_+_84869883 | 0.68 |
ENSMUST00000163083.2
|
Cyb5a
|
cytochrome b5 type A (microsomal) |
| chr18_-_74340842 | 0.66 |
ENSMUST00000040188.16
|
Ska1
|
spindle and kinetochore associated complex subunit 1 |
| chr11_+_96355413 | 0.66 |
ENSMUST00000103154.11
ENSMUST00000100521.10 ENSMUST00000100519.11 |
Skap1
|
src family associated phosphoprotein 1 |
| chr7_+_28510310 | 0.66 |
ENSMUST00000038572.15
|
Hnrnpl
|
heterogeneous nuclear ribonucleoprotein L |
| chr7_-_110581376 | 0.65 |
ENSMUST00000154466.2
|
Irag1
|
inositol 1,4,5-triphosphate receptor associated 1 |
| chr6_-_134543876 | 0.65 |
ENSMUST00000032322.15
ENSMUST00000126836.4 |
Lrp6
|
low density lipoprotein receptor-related protein 6 |
| chr11_+_117090465 | 0.64 |
ENSMUST00000093907.11
|
Septin9
|
septin 9 |
| chr4_+_33062999 | 0.64 |
ENSMUST00000108162.8
ENSMUST00000024035.9 |
Gabrr2
|
gamma-aminobutyric acid (GABA) C receptor, subunit rho 2 |
| chr14_+_54183465 | 0.64 |
ENSMUST00000197130.5
ENSMUST00000103677.3 |
Trdv2-1
|
T cell receptor delta variable 2-1 |
| chr12_-_84447625 | 0.64 |
ENSMUST00000117286.2
|
Entpd5
|
ectonucleoside triphosphate diphosphohydrolase 5 |
| chr1_+_24717722 | 0.63 |
ENSMUST00000186096.7
|
Lmbrd1
|
LMBR1 domain containing 1 |
| chr19_+_4189786 | 0.62 |
ENSMUST00000096338.5
|
Gpr152
|
G protein-coupled receptor 152 |
| chr8_-_73246435 | 0.61 |
ENSMUST00000152080.8
|
Slc35e1
|
solute carrier family 35, member E1 |
| chr7_-_45375205 | 0.61 |
ENSMUST00000094424.7
|
Spaca4
|
sperm acrosome associated 4 |
| chr11_-_120489172 | 0.59 |
ENSMUST00000026125.3
|
Alyref
|
Aly/REF export factor |
| chr7_+_99243812 | 0.59 |
ENSMUST00000162290.2
|
Arrb1
|
arrestin, beta 1 |
| chr14_-_64654397 | 0.59 |
ENSMUST00000210428.2
|
Msra
|
methionine sulfoxide reductase A |
| chr4_+_80752535 | 0.58 |
ENSMUST00000102831.8
|
Tyrp1
|
tyrosinase-related protein 1 |
| chr1_+_24717711 | 0.57 |
ENSMUST00000191471.7
|
Lmbrd1
|
LMBR1 domain containing 1 |
| chr11_-_66059330 | 0.57 |
ENSMUST00000080665.10
|
Dnah9
|
dynein, axonemal, heavy chain 9 |
| chr14_+_26722319 | 0.57 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr12_-_118162652 | 0.57 |
ENSMUST00000084806.7
|
Dnah11
|
dynein, axonemal, heavy chain 11 |
| chrX_+_72527208 | 0.57 |
ENSMUST00000033741.15
ENSMUST00000169489.2 |
Bgn
|
biglycan |
| chr1_-_119576347 | 0.56 |
ENSMUST00000027632.14
ENSMUST00000187194.2 |
Epb41l5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr11_-_109363406 | 0.56 |
ENSMUST00000168740.3
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
| chr10_-_75673175 | 0.56 |
ENSMUST00000220440.2
|
Gstt2
|
glutathione S-transferase, theta 2 |
| chr12_+_85645801 | 0.56 |
ENSMUST00000177587.9
|
Jdp2
|
Jun dimerization protein 2 |
| chr8_-_108129829 | 0.55 |
ENSMUST00000003947.9
|
Nqo1
|
NAD(P)H dehydrogenase, quinone 1 |
| chr7_+_26534730 | 0.54 |
ENSMUST00000005685.15
|
Cyp2a5
|
cytochrome P450, family 2, subfamily a, polypeptide 5 |
| chr5_-_123859153 | 0.54 |
ENSMUST00000196282.5
|
Zcchc8
|
zinc finger, CCHC domain containing 8 |
| chrX_-_99670174 | 0.53 |
ENSMUST00000015812.12
|
Pdzd11
|
PDZ domain containing 11 |
| chr16_+_91184661 | 0.53 |
ENSMUST00000139503.2
|
Ifnar2
|
interferon (alpha and beta) receptor 2 |
| chr7_-_103492361 | 0.53 |
ENSMUST00000063957.6
|
Hbb-bh1
|
hemoglobin Z, beta-like embryonic chain |
| chrX_+_73468140 | 0.53 |
ENSMUST00000135165.8
ENSMUST00000114128.8 ENSMUST00000004330.10 ENSMUST00000114133.9 ENSMUST00000114129.9 ENSMUST00000132749.2 |
Ikbkg
|
inhibitor of kappaB kinase gamma |
| chr6_-_93889483 | 0.52 |
ENSMUST00000205116.3
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr4_-_82777746 | 0.50 |
ENSMUST00000156055.2
ENSMUST00000030110.15 |
Zdhhc21
|
zinc finger, DHHC domain containing 21 |
| chr11_+_96355441 | 0.50 |
ENSMUST00000071510.14
ENSMUST00000107662.9 |
Skap1
|
src family associated phosphoprotein 1 |
| chr14_+_8555284 | 0.48 |
ENSMUST00000144914.3
|
Gm281
|
predicted gene 281 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.2 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 1.0 | 4.0 | GO:0035771 | T-helper 1 cell lineage commitment(GO:0002296) interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.9 | 4.3 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.8 | 1.6 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.8 | 2.4 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 0.7 | 3.7 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.7 | 2.7 | GO:0038016 | insulin receptor internalization(GO:0038016) |
| 0.7 | 11.4 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.7 | 3.4 | GO:0010958 | regulation of amino acid import(GO:0010958) L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.6 | 3.6 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.5 | 1.6 | GO:0010925 | regulation of inositol-polyphosphate 5-phosphatase activity(GO:0010924) positive regulation of inositol-polyphosphate 5-phosphatase activity(GO:0010925) phospholipase C-inhibiting G-protein coupled receptor signaling pathway(GO:0030845) regulation of cell diameter(GO:0060305) |
| 0.5 | 1.5 | GO:1902617 | response to fluoride(GO:1902617) |
| 0.4 | 1.3 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.4 | 2.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.4 | 2.0 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.4 | 1.9 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.4 | 1.5 | GO:0002353 | plasma kallikrein-kinin cascade(GO:0002353) |
| 0.4 | 8.0 | GO:0035814 | negative regulation of renal sodium excretion(GO:0035814) |
| 0.4 | 2.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.3 | 1.4 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.3 | 1.0 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 0.3 | 1.3 | GO:0046086 | adenosine biosynthetic process(GO:0046086) |
| 0.3 | 1.3 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.3 | 0.3 | GO:0060701 | negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) |
| 0.3 | 1.5 | GO:0060578 | subthalamic nucleus development(GO:0021763) superior vena cava morphogenesis(GO:0060578) |
| 0.3 | 4.4 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.3 | 1.2 | GO:2000705 | regulation of dense core granule biogenesis(GO:2000705) |
| 0.3 | 0.8 | GO:0009087 | methionine catabolic process(GO:0009087) |
| 0.3 | 1.6 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.2 | 0.7 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.2 | 0.6 | GO:0061354 | planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) |
| 0.2 | 31.9 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.2 | 0.8 | GO:1903487 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) regulation of lactation(GO:1903487) |
| 0.2 | 1.7 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.2 | 0.8 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.2 | 4.4 | GO:0051923 | sulfation(GO:0051923) |
| 0.2 | 1.6 | GO:0051025 | negative regulation of immunoglobulin secretion(GO:0051025) |
| 0.2 | 2.1 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.2 | 2.4 | GO:0007343 | egg activation(GO:0007343) |
| 0.2 | 2.1 | GO:0009191 | ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.1 | 0.4 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.1 | 0.4 | GO:1904092 | regulation of autophagic cell death(GO:1904092) negative regulation of autophagic cell death(GO:1904093) |
| 0.1 | 33.1 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 0.7 | GO:0035546 | interferon-beta secretion(GO:0035546) regulation of interferon-beta secretion(GO:0035547) positive regulation of interferon-beta secretion(GO:0035549) |
| 0.1 | 0.4 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.1 | 2.9 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 1.4 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.1 | 0.5 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.1 | 0.8 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 0.8 | GO:0031394 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 3.6 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.1 | 1.0 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.1 | 0.4 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.1 | 1.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 3.3 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.1 | 0.7 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.1 | 0.3 | GO:1903413 | response to bile acid(GO:1903412) cellular response to bile acid(GO:1903413) |
| 0.1 | 0.5 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 0.4 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.1 | 0.4 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.1 | 1.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 1.3 | GO:0009437 | carnitine metabolic process(GO:0009437) positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.1 | 0.3 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.1 | 0.2 | GO:0060854 | patterning of lymph vessels(GO:0060854) |
| 0.1 | 1.0 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 | 0.5 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 1.4 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 1.9 | GO:0097066 | response to thyroid hormone(GO:0097066) |
| 0.1 | 0.4 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 0.4 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.1 | 0.3 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 0.1 | 0.3 | GO:0000101 | sulfur amino acid transport(GO:0000101) |
| 0.1 | 0.9 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 0.3 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 0.2 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.1 | 0.3 | GO:1901297 | positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.1 | 0.5 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 1.4 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.1 | 0.6 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.1 | 0.2 | GO:0046356 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) acetyl-CoA catabolic process(GO:0046356) |
| 0.1 | 0.2 | GO:0060995 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.1 | 0.9 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.7 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.6 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.4 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 1.2 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 1.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.4 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.2 | GO:0046502 | uroporphyrinogen III metabolic process(GO:0046502) |
| 0.0 | 0.5 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.7 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.4 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.4 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 4.3 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 2.7 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.2 | GO:0045583 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.0 | 0.3 | GO:0032202 | telomere assembly(GO:0032202) scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.9 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.0 | 0.4 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 2.9 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 4.2 | GO:0046889 | positive regulation of lipid biosynthetic process(GO:0046889) |
| 0.0 | 0.7 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.6 | GO:0045821 | positive regulation of glycolytic process(GO:0045821) |
| 0.0 | 0.5 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.9 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.0 | 0.5 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.3 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.4 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.3 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.0 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 0.2 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 1.5 | GO:0042755 | eating behavior(GO:0042755) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.9 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.5 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.8 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.1 | GO:0090209 | negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.3 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.0 | 0.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.1 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.8 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 0.2 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 1.0 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.6 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.0 | 0.2 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.3 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.0 | 0.2 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.1 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 | 0.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.2 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 1.9 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 1.7 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.4 | GO:0035411 | catenin import into nucleus(GO:0035411) |
| 0.0 | 0.1 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.6 | 5.2 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.6 | 8.6 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.4 | 4.0 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.3 | 31.6 | GO:0042571 | immunoglobulin complex(GO:0019814) immunoglobulin complex, circulating(GO:0042571) |
| 0.2 | 3.6 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.2 | 0.8 | GO:0035101 | FACT complex(GO:0035101) |
| 0.2 | 0.6 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.2 | 1.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.2 | 0.5 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.1 | 2.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 1.9 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 0.9 | GO:0036396 | MIS complex(GO:0036396) |
| 0.1 | 1.0 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 1.4 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.1 | 0.7 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 0.9 | GO:0097422 | extrinsic component of endosome membrane(GO:0031313) tubular endosome(GO:0097422) |
| 0.1 | 1.0 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 0.8 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.1 | 0.4 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 2.2 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 1.4 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 0.4 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 2.7 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 0.5 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 0.4 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.1 | 1.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.2 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.1 | 1.6 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 1.0 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.2 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 4.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.3 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.4 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.3 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.4 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 1.4 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 1.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.5 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 2.0 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 14.7 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.1 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.0 | 0.5 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.4 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.3 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.6 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.4 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 1.7 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 2.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 41.1 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.5 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.2 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.2 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 2.2 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.3 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 1.4 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.0 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.0 | 0.3 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 1.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 3.6 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 0.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 2.7 | GO:0009897 | external side of plasma membrane(GO:0009897) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.2 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 1.2 | 3.6 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.9 | 2.7 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.7 | 2.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.6 | 4.4 | GO:0050694 | galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.6 | 1.7 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.4 | 2.2 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.4 | 1.3 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.4 | 1.3 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.4 | 1.3 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.4 | 2.1 | GO:0055056 | dehydroascorbic acid transporter activity(GO:0033300) D-glucose transmembrane transporter activity(GO:0055056) |
| 0.4 | 1.1 | GO:0015292 | uniporter activity(GO:0015292) |
| 0.4 | 1.5 | GO:0004063 | aryldialkylphosphatase activity(GO:0004063) |
| 0.3 | 2.4 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.3 | 3.7 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.3 | 2.3 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.3 | 8.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.3 | 1.0 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.3 | 35.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.2 | 2.7 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.2 | 0.7 | GO:0045142 | ATP-dependent DNA/RNA helicase activity(GO:0033680) triplex DNA binding(GO:0045142) |
| 0.2 | 1.6 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.2 | 0.6 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.2 | 0.8 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.2 | 4.7 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.2 | 1.1 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.2 | 1.2 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.2 | 1.0 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.1 | 0.6 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.1 | 0.4 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.1 | 2.4 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 0.5 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 0.1 | 0.6 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.1 | 1.4 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.1 | 0.5 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.1 | 0.5 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.1 | 2.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.1 | 0.5 | GO:0004904 | interferon receptor activity(GO:0004904) type I interferon receptor activity(GO:0004905) type I interferon binding(GO:0019962) |
| 0.1 | 4.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.7 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.9 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.8 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.1 | 0.6 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.2 | GO:0001607 | neuromedin U receptor activity(GO:0001607) |
| 0.1 | 1.7 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 0.5 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 2.7 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 0.8 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 2.0 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.7 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.1 | 0.4 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.1 | 1.3 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 2.0 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 1.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 1.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.2 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 1.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.2 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 1.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 0.4 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.1 | 2.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 1.6 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.3 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.4 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.3 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.0 | 0.2 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 1.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.4 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.2 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.7 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.5 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 4.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.8 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 1.9 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 1.6 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0070736 | protein-glycine ligase activity, initiating(GO:0070736) |
| 0.0 | 0.4 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 2.0 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 1.8 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.8 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.3 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.3 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.3 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.1 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.0 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 1.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 1.7 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.9 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.7 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.8 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.5 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 1.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.6 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.1 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.0 | 0.4 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 1.5 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.7 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.3 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.4 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 1.4 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.2 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 1.2 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.3 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 1.8 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 1.4 | GO:1990782 | protein tyrosine kinase binding(GO:1990782) |
| 0.0 | 1.7 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.5 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.4 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.1 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.2 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 1.9 | GO:0015293 | symporter activity(GO:0015293) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.6 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.1 | 4.0 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 11.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 8.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.5 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 1.5 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.9 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 1.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.3 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.8 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.7 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.5 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 3.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.7 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 13.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.3 | 2.4 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.3 | 5.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 2.5 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 3.6 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.2 | 4.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 2.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 5.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 0.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 1.0 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 1.5 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 1.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 4.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 1.6 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 1.0 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 1.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 1.3 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 0.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.8 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.5 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 1.4 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 2.1 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 1.0 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.8 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 3.0 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.6 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.6 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 2.3 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.3 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 1.5 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.7 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 1.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |