Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
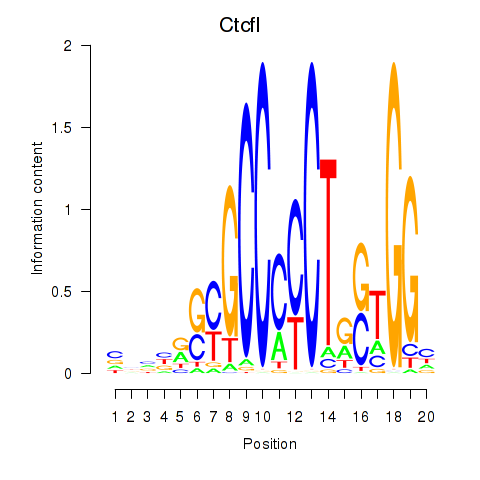
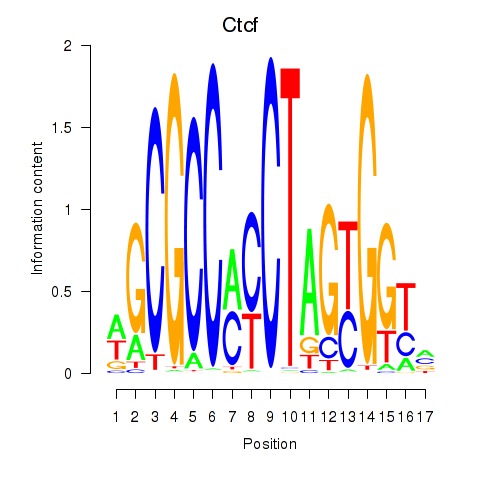
Results for Ctcfl_Ctcf
Z-value: 1.48
Transcription factors associated with Ctcfl_Ctcf
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ctcfl
|
ENSMUSG00000070495.12 | Ctcfl |
|
Ctcf
|
ENSMUSG00000005698.16 | Ctcf |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ctcfl | mm39_v1_chr2_-_172961168_172961318 | -0.30 | 9.5e-03 | Click! |
| Ctcf | mm39_v1_chr8_+_106363141_106363221 | 0.21 | 7.0e-02 | Click! |
Activity profile of Ctcfl_Ctcf motif
Sorted Z-values of Ctcfl_Ctcf motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Ctcfl_Ctcf
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_32243295 | 20.78 |
ENSMUST00000091089.12
ENSMUST00000078352.12 ENSMUST00000113350.8 ENSMUST00000202578.4 ENSMUST00000113365.8 |
Dnm1
|
dynamin 1 |
| chr2_-_32243246 | 18.52 |
ENSMUST00000201433.4
ENSMUST00000113352.9 ENSMUST00000201494.4 |
Dnm1
|
dynamin 1 |
| chr13_-_58056089 | 17.93 |
ENSMUST00000185502.7
ENSMUST00000186271.7 ENSMUST00000185905.2 ENSMUST00000187852.7 |
Spock1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 1 |
| chr8_-_110894836 | 15.83 |
ENSMUST00000003754.8
ENSMUST00000212297.2 |
Calb2
|
calbindin 2 |
| chr16_-_18445172 | 15.66 |
ENSMUST00000231335.2
ENSMUST00000232653.2 |
Gm49601
Septin5
|
predicted gene, 49601 septin 5 |
| chr19_+_6468761 | 15.11 |
ENSMUST00000113462.8
ENSMUST00000077182.13 ENSMUST00000236635.2 ENSMUST00000113461.8 |
Nrxn2
|
neurexin II |
| chr16_+_96268806 | 13.03 |
ENSMUST00000061739.9
|
Pcp4
|
Purkinje cell protein 4 |
| chr2_-_117173190 | 12.95 |
ENSMUST00000173541.8
ENSMUST00000172901.8 ENSMUST00000173252.2 |
Rasgrp1
|
RAS guanyl releasing protein 1 |
| chr2_-_53975501 | 12.94 |
ENSMUST00000100089.3
|
Rprm
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr11_+_3464861 | 12.26 |
ENSMUST00000094469.6
|
Selenom
|
selenoprotein M |
| chr2_+_65451100 | 11.42 |
ENSMUST00000144254.6
ENSMUST00000028377.14 |
Scn2a
|
sodium channel, voltage-gated, type II, alpha |
| chr7_+_30758767 | 11.14 |
ENSMUST00000039775.9
|
Lgi4
|
leucine-rich repeat LGI family, member 4 |
| chr11_-_102338473 | 10.68 |
ENSMUST00000049057.5
|
Fam171a2
|
family with sequence similarity 171, member A2 |
| chr11_-_76468527 | 10.65 |
ENSMUST00000176179.8
|
Abr
|
active BCR-related gene |
| chr19_-_45804446 | 10.61 |
ENSMUST00000079431.10
ENSMUST00000026247.13 ENSMUST00000162528.9 |
Kcnip2
|
Kv channel-interacting protein 2 |
| chr2_+_157756535 | 9.91 |
ENSMUST00000109523.2
|
Vstm2l
|
V-set and transmembrane domain containing 2-like |
| chr11_+_119158713 | 9.22 |
ENSMUST00000106259.9
|
Gaa
|
glucosidase, alpha, acid |
| chr5_-_5430172 | 9.02 |
ENSMUST00000030763.13
|
Cdk14
|
cyclin-dependent kinase 14 |
| chr11_+_119158829 | 8.99 |
ENSMUST00000026666.13
ENSMUST00000106258.2 |
Gaa
|
glucosidase, alpha, acid |
| chr13_-_54209669 | 8.89 |
ENSMUST00000021932.6
ENSMUST00000221470.2 |
Drd1
|
dopamine receptor D1 |
| chr16_-_30369378 | 8.88 |
ENSMUST00000140402.8
|
Tmem44
|
transmembrane protein 44 |
| chr18_+_73996743 | 8.76 |
ENSMUST00000134847.2
|
Mro
|
maestro |
| chr11_-_3864664 | 8.74 |
ENSMUST00000109995.2
ENSMUST00000051207.2 |
Slc35e4
|
solute carrier family 35, member E4 |
| chr1_-_135095344 | 8.56 |
ENSMUST00000027682.9
|
Gpr37l1
|
G protein-coupled receptor 37-like 1 |
| chr7_+_25016492 | 8.53 |
ENSMUST00000128119.2
|
Megf8
|
multiple EGF-like-domains 8 |
| chr19_-_10217968 | 8.50 |
ENSMUST00000189897.2
ENSMUST00000186056.7 ENSMUST00000088013.12 |
Myrf
|
myelin regulatory factor |
| chr5_-_135107078 | 8.38 |
ENSMUST00000067935.11
ENSMUST00000076203.3 |
Vps37d
|
vacuolar protein sorting 37D |
| chr13_-_54914366 | 8.34 |
ENSMUST00000036825.14
|
Sncb
|
synuclein, beta |
| chr11_+_116809669 | 8.30 |
ENSMUST00000103027.10
|
Mgat5b
|
mannoside acetylglucosaminyltransferase 5, isoenzyme B |
| chr5_-_137739863 | 8.22 |
ENSMUST00000061789.14
|
Nyap1
|
neuronal tyrosine-phosphorylated phosphoinositide 3-kinase adaptor 1 |
| chr6_+_103674695 | 8.18 |
ENSMUST00000205098.2
|
Chl1
|
cell adhesion molecule L1-like |
| chr5_+_135835713 | 7.97 |
ENSMUST00000126232.8
|
Srrm3
|
serine/arginine repetitive matrix 3 |
| chr19_+_5100815 | 7.93 |
ENSMUST00000224178.2
ENSMUST00000225799.3 ENSMUST00000025818.8 |
Rin1
|
Ras and Rab interactor 1 |
| chr1_+_191873078 | 7.89 |
ENSMUST00000078470.12
ENSMUST00000110844.3 |
Kcnh1
|
potassium voltage-gated channel, subfamily H (eag-related), member 1 |
| chr13_-_58056138 | 7.87 |
ENSMUST00000189373.8
|
Spock1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 1 |
| chr18_-_36648850 | 7.74 |
ENSMUST00000025363.7
|
Hbegf
|
heparin-binding EGF-like growth factor |
| chr5_-_36555434 | 7.64 |
ENSMUST00000037370.14
ENSMUST00000070720.8 |
Sorcs2
|
sortilin-related VPS10 domain containing receptor 2 |
| chr18_+_37085673 | 7.49 |
ENSMUST00000192512.6
ENSMUST00000192295.2 ENSMUST00000115661.5 |
Pcdha4
Gm42416
|
protocadherin alpha 4 predicted gene, 42416 |
| chr5_-_142536720 | 7.31 |
ENSMUST00000129212.2
ENSMUST00000110785.2 ENSMUST00000063635.15 |
Radil
|
Ras association and DIL domains |
| chr19_-_10847121 | 7.30 |
ENSMUST00000120524.2
ENSMUST00000025645.14 |
Tmem132a
|
transmembrane protein 132A |
| chr16_-_20244631 | 7.30 |
ENSMUST00000077867.10
|
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr5_-_123270702 | 7.21 |
ENSMUST00000031401.6
|
Rhof
|
ras homolog family member F (in filopodia) |
| chr2_-_65397850 | 7.14 |
ENSMUST00000238483.2
ENSMUST00000100069.9 |
Scn3a
|
sodium channel, voltage-gated, type III, alpha |
| chr15_-_44651411 | 7.12 |
ENSMUST00000090057.6
ENSMUST00000110269.8 ENSMUST00000228639.2 |
Sybu
|
syntabulin (syntaxin-interacting) |
| chr3_-_96615838 | 7.12 |
ENSMUST00000029742.9
ENSMUST00000200387.2 ENSMUST00000171249.6 |
Nudt17
|
nudix (nucleoside diphosphate linked moiety X)-type motif 17 |
| chr19_+_5100475 | 7.00 |
ENSMUST00000225427.2
|
Rin1
|
Ras and Rab interactor 1 |
| chr14_-_70855980 | 7.00 |
ENSMUST00000228001.2
|
Dmtn
|
dematin actin binding protein |
| chr18_+_37606591 | 6.95 |
ENSMUST00000050034.3
|
Pcdhb15
|
protocadherin beta 15 |
| chr9_-_102231884 | 6.74 |
ENSMUST00000035129.14
ENSMUST00000085169.12 ENSMUST00000149800.3 |
Ephb1
|
Eph receptor B1 |
| chr2_-_91793847 | 6.67 |
ENSMUST00000028667.10
|
Dgkz
|
diacylglycerol kinase zeta |
| chr2_+_181356797 | 6.64 |
ENSMUST00000071585.10
ENSMUST00000238942.2 ENSMUST00000148334.8 ENSMUST00000108763.8 |
Oprl1
|
opioid receptor-like 1 |
| chr16_-_20245071 | 6.55 |
ENSMUST00000115547.9
ENSMUST00000096199.5 |
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr19_-_10282218 | 6.50 |
ENSMUST00000039327.11
|
Dagla
|
diacylglycerol lipase, alpha |
| chr6_+_87864796 | 6.32 |
ENSMUST00000113607.10
ENSMUST00000049966.6 |
Copg1
|
coatomer protein complex, subunit gamma 1 |
| chr2_-_155356716 | 6.23 |
ENSMUST00000029131.11
|
Ggt7
|
gamma-glutamyltransferase 7 |
| chr13_+_58954374 | 6.23 |
ENSMUST00000225488.2
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr11_-_114686712 | 6.18 |
ENSMUST00000000206.4
|
Btbd17
|
BTB (POZ) domain containing 17 |
| chr7_+_64151435 | 6.16 |
ENSMUST00000032732.15
|
Apba2
|
amyloid beta (A4) precursor protein-binding, family A, member 2 |
| chr13_+_58954447 | 6.10 |
ENSMUST00000224259.2
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr18_+_37544717 | 6.08 |
ENSMUST00000051126.4
|
Pcdhb10
|
protocadherin beta 10 |
| chr8_+_83589979 | 6.05 |
ENSMUST00000078525.7
|
Rnf150
|
ring finger protein 150 |
| chr2_+_25293056 | 5.99 |
ENSMUST00000071442.12
|
Npdc1
|
neural proliferation, differentiation and control 1 |
| chr19_+_46292759 | 5.98 |
ENSMUST00000237330.2
|
Nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B cells 2, p49/p100 |
| chr8_+_67150055 | 5.97 |
ENSMUST00000039303.7
|
Npy1r
|
neuropeptide Y receptor Y1 |
| chr15_-_98705791 | 5.96 |
ENSMUST00000075444.8
|
Ddn
|
dendrin |
| chr15_+_99122742 | 5.91 |
ENSMUST00000041415.5
|
Kcnh3
|
potassium voltage-gated channel, subfamily H (eag-related), member 3 |
| chr12_+_16703383 | 5.84 |
ENSMUST00000221596.2
ENSMUST00000111064.3 ENSMUST00000220892.2 |
Ntsr2
|
neurotensin receptor 2 |
| chr11_-_102469896 | 5.84 |
ENSMUST00000107080.2
|
Gm11627
|
predicted gene 11627 |
| chr18_+_37898633 | 5.81 |
ENSMUST00000044851.8
|
Pcdhga12
|
protocadherin gamma subfamily A, 12 |
| chr16_+_17093941 | 5.81 |
ENSMUST00000164950.11
|
Tmem191c
|
transmembrane protein 191C |
| chr7_+_4122523 | 5.81 |
ENSMUST00000119661.8
ENSMUST00000129423.8 |
Ttyh1
|
tweety family member 1 |
| chr13_+_117738972 | 5.80 |
ENSMUST00000006991.9
|
Hcn1
|
hyperpolarization activated cyclic nucleotide gated potassium channel 1 |
| chr9_-_108329576 | 5.78 |
ENSMUST00000035232.13
ENSMUST00000195435.6 |
Klhdc8b
|
kelch domain containing 8B |
| chr5_-_5315968 | 5.72 |
ENSMUST00000115451.8
ENSMUST00000115452.8 ENSMUST00000131392.8 |
Cdk14
|
cyclin-dependent kinase 14 |
| chr11_-_100650768 | 5.72 |
ENSMUST00000107363.3
|
Kcnh4
|
potassium voltage-gated channel, subfamily H (eag-related), member 4 |
| chr18_+_37533899 | 5.69 |
ENSMUST00000057228.2
|
Pcdhb9
|
protocadherin beta 9 |
| chr1_-_5140504 | 5.65 |
ENSMUST00000147158.2
ENSMUST00000118000.8 |
Rgs20
|
regulator of G-protein signaling 20 |
| chr12_-_80807454 | 5.64 |
ENSMUST00000073251.8
|
Ccdc177
|
coiled-coil domain containing 177 |
| chr13_-_49301407 | 5.63 |
ENSMUST00000162581.8
ENSMUST00000110097.9 ENSMUST00000049265.15 ENSMUST00000035538.13 ENSMUST00000110096.8 ENSMUST00000091623.10 |
Wnk2
|
WNK lysine deficient protein kinase 2 |
| chr17_+_44112679 | 5.60 |
ENSMUST00000229744.2
|
Rcan2
|
regulator of calcineurin 2 |
| chr17_-_8046125 | 5.59 |
ENSMUST00000239425.2
ENSMUST00000167580.8 |
Fndc1
|
fibronectin type III domain containing 1 |
| chr1_+_163607143 | 5.58 |
ENSMUST00000077642.12
ENSMUST00000027877.7 |
Kifap3
|
kinesin-associated protein 3 |
| chr11_-_95405368 | 5.55 |
ENSMUST00000058866.8
|
Nxph3
|
neurexophilin 3 |
| chr13_-_113182891 | 5.54 |
ENSMUST00000231962.2
ENSMUST00000022282.6 |
Gpx8
|
glutathione peroxidase 8 (putative) |
| chr16_-_20245138 | 5.51 |
ENSMUST00000079158.13
|
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr2_+_3119442 | 5.50 |
ENSMUST00000091505.11
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr2_+_109522781 | 5.49 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr8_+_106412905 | 5.49 |
ENSMUST00000213019.2
|
Carmil2
|
capping protein regulator and myosin 1 linker 2 |
| chr13_-_12121831 | 5.45 |
ENSMUST00000021750.15
ENSMUST00000170156.3 ENSMUST00000220597.2 |
Ryr2
|
ryanodine receptor 2, cardiac |
| chr3_-_89153135 | 5.35 |
ENSMUST00000041022.15
|
Trim46
|
tripartite motif-containing 46 |
| chrX_-_74621828 | 5.34 |
ENSMUST00000033545.6
|
Rab39b
|
RAB39B, member RAS oncogene family |
| chr7_+_4122555 | 5.31 |
ENSMUST00000079415.12
|
Ttyh1
|
tweety family member 1 |
| chr9_-_66951234 | 5.21 |
ENSMUST00000113690.8
|
Tpm1
|
tropomyosin 1, alpha |
| chr15_-_36496880 | 5.19 |
ENSMUST00000228601.2
ENSMUST00000057486.9 |
Ankrd46
|
ankyrin repeat domain 46 |
| chr18_+_37093321 | 5.15 |
ENSMUST00000192168.2
|
Pcdha5
|
protocadherin alpha 5 |
| chr5_-_137739364 | 5.14 |
ENSMUST00000149512.3
|
Nyap1
|
neuronal tyrosine-phosphorylated phosphoinositide 3-kinase adaptor 1 |
| chr8_+_67149815 | 5.12 |
ENSMUST00000212588.2
|
Npy1r
|
neuropeptide Y receptor Y1 |
| chr7_-_30750856 | 5.05 |
ENSMUST00000073892.6
|
Fxyd7
|
FXYD domain-containing ion transport regulator 7 |
| chr10_-_18421796 | 5.03 |
ENSMUST00000214548.2
ENSMUST00000020000.7 |
Hebp2
|
heme binding protein 2 |
| chr4_-_110209106 | 5.01 |
ENSMUST00000106603.9
|
Elavl4
|
ELAV like RNA binding protein 4 |
| chr4_+_126156118 | 5.00 |
ENSMUST00000030660.9
|
Trappc3
|
trafficking protein particle complex 3 |
| chr2_-_65397809 | 4.98 |
ENSMUST00000066432.12
|
Scn3a
|
sodium channel, voltage-gated, type III, alpha |
| chr11_-_109188892 | 4.97 |
ENSMUST00000106706.8
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr2_+_25318642 | 4.97 |
ENSMUST00000102919.4
|
Abca2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr2_-_66240408 | 4.93 |
ENSMUST00000112366.8
|
Scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr11_+_49792627 | 4.89 |
ENSMUST00000093141.12
ENSMUST00000093142.12 |
Rasgef1c
|
RasGEF domain family, member 1C |
| chr6_+_86005663 | 4.88 |
ENSMUST00000204059.3
|
Add2
|
adducin 2 (beta) |
| chr9_+_111140741 | 4.84 |
ENSMUST00000078626.8
|
Trank1
|
tetratricopeptide repeat and ankyrin repeat containing 1 |
| chr14_-_70405288 | 4.82 |
ENSMUST00000129174.8
ENSMUST00000125300.3 |
Pdlim2
|
PDZ and LIM domain 2 |
| chr6_-_124441731 | 4.81 |
ENSMUST00000008297.5
|
Clstn3
|
calsyntenin 3 |
| chr7_-_108769719 | 4.80 |
ENSMUST00000208136.2
ENSMUST00000036992.9 |
Lmo1
|
LIM domain only 1 |
| chr5_-_115791032 | 4.80 |
ENSMUST00000121746.8
ENSMUST00000118576.8 |
Bicdl1
|
BICD family like cargo adaptor 1 |
| chr8_-_85663976 | 4.77 |
ENSMUST00000109741.9
ENSMUST00000119820.2 |
Mast1
|
microtubule associated serine/threonine kinase 1 |
| chr1_-_37758863 | 4.77 |
ENSMUST00000160589.2
|
Cracdl
|
capping protein inhibiting regulator of actin like |
| chr11_-_109188947 | 4.64 |
ENSMUST00000020920.10
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr12_+_108389075 | 4.61 |
ENSMUST00000109860.8
|
Eml1
|
echinoderm microtubule associated protein like 1 |
| chr11_+_104122216 | 4.55 |
ENSMUST00000106992.10
|
Mapt
|
microtubule-associated protein tau |
| chr5_+_120787253 | 4.52 |
ENSMUST00000156722.2
|
Rasal1
|
RAS protein activator like 1 (GAP1 like) |
| chr9_+_106088798 | 4.48 |
ENSMUST00000216850.2
|
Twf2
|
twinfilin actin binding protein 2 |
| chr6_-_87312743 | 4.45 |
ENSMUST00000042025.12
ENSMUST00000205033.2 |
Antxr1
|
anthrax toxin receptor 1 |
| chr5_-_114961521 | 4.43 |
ENSMUST00000140374.2
|
1500011B03Rik
|
RIKEN cDNA 1500011B03 gene |
| chr2_+_3119371 | 4.37 |
ENSMUST00000115099.9
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr6_+_83142902 | 4.35 |
ENSMUST00000077407.12
ENSMUST00000113913.8 ENSMUST00000130212.8 |
Dctn1
|
dynactin 1 |
| chr2_-_12306722 | 4.34 |
ENSMUST00000028106.11
|
Itga8
|
integrin alpha 8 |
| chr10_+_74802996 | 4.28 |
ENSMUST00000037813.5
|
Gnaz
|
guanine nucleotide binding protein, alpha z subunit |
| chr7_-_30750828 | 4.26 |
ENSMUST00000206341.2
|
Fxyd7
|
FXYD domain-containing ion transport regulator 7 |
| chr4_-_149391963 | 4.25 |
ENSMUST00000055647.15
ENSMUST00000030806.6 ENSMUST00000238956.2 ENSMUST00000060537.13 |
Kif1b
|
kinesin family member 1B |
| chr1_+_191307748 | 4.22 |
ENSMUST00000045450.7
|
Ints7
|
integrator complex subunit 7 |
| chr7_+_27967222 | 4.17 |
ENSMUST00000059596.8
|
Eid2
|
EP300 interacting inhibitor of differentiation 2 |
| chr2_+_3119558 | 4.17 |
ENSMUST00000062934.7
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr19_-_10857734 | 4.15 |
ENSMUST00000133303.8
|
Tmem109
|
transmembrane protein 109 |
| chr9_-_75506406 | 4.12 |
ENSMUST00000215036.2
|
Tmod2
|
tropomodulin 2 |
| chr7_-_67409432 | 4.11 |
ENSMUST00000208815.2
ENSMUST00000074233.12 ENSMUST00000208231.2 ENSMUST00000051389.10 |
Synm
|
synemin, intermediate filament protein |
| chr10_-_18421628 | 4.08 |
ENSMUST00000213153.2
|
Hebp2
|
heme binding protein 2 |
| chr10_+_88215079 | 4.07 |
ENSMUST00000130301.8
ENSMUST00000020251.10 |
Gnptab
|
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chr5_+_147894121 | 4.07 |
ENSMUST00000085558.11
ENSMUST00000129092.2 |
Mtus2
|
microtubule associated tumor suppressor candidate 2 |
| chr9_-_58111589 | 4.07 |
ENSMUST00000217578.2
ENSMUST00000114144.9 ENSMUST00000214649.2 |
Islr
Islr2
|
immunoglobulin superfamily containing leucine-rich repeat immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr5_+_91665474 | 4.04 |
ENSMUST00000040576.10
|
Parm1
|
prostate androgen-regulated mucin-like protein 1 |
| chr18_+_37554471 | 4.03 |
ENSMUST00000053073.6
|
Pcdhb11
|
protocadherin beta 11 |
| chr12_-_87037204 | 4.03 |
ENSMUST00000222543.2
|
Zdhhc22
|
zinc finger, DHHC-type containing 22 |
| chr12_+_16703709 | 4.03 |
ENSMUST00000221049.2
|
Ntsr2
|
neurotensin receptor 2 |
| chr13_-_53440087 | 4.02 |
ENSMUST00000021918.10
|
Ror2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr4_+_43381979 | 4.00 |
ENSMUST00000035645.12
ENSMUST00000144911.8 |
Rusc2
|
RUN and SH3 domain containing 2 |
| chr6_-_136150491 | 3.99 |
ENSMUST00000111905.8
ENSMUST00000152012.8 ENSMUST00000143943.8 ENSMUST00000125905.2 |
Grin2b
|
glutamate receptor, ionotropic, NMDA2B (epsilon 2) |
| chr11_-_59855762 | 3.97 |
ENSMUST00000062405.8
|
Rasd1
|
RAS, dexamethasone-induced 1 |
| chr15_+_100659622 | 3.93 |
ENSMUST00000023776.13
|
Slc4a8
|
solute carrier family 4 (anion exchanger), member 8 |
| chr15_-_44978223 | 3.90 |
ENSMUST00000022967.7
|
Kcnv1
|
potassium channel, subfamily V, member 1 |
| chr1_-_16590244 | 3.87 |
ENSMUST00000144138.4
ENSMUST00000145092.8 ENSMUST00000131257.9 ENSMUST00000153966.9 ENSMUST00000162435.8 |
Stau2
|
staufen double-stranded RNA binding protein 2 |
| chr10_-_31321938 | 3.85 |
ENSMUST00000000305.7
|
Tpd52l1
|
tumor protein D52-like 1 |
| chr12_-_110669076 | 3.82 |
ENSMUST00000155242.8
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr4_-_132939790 | 3.81 |
ENSMUST00000052090.9
|
Gpr3
|
G-protein coupled receptor 3 |
| chr11_-_97464866 | 3.81 |
ENSMUST00000207653.2
ENSMUST00000107593.8 |
Srcin1
|
SRC kinase signaling inhibitor 1 |
| chr7_-_83735702 | 3.80 |
ENSMUST00000064174.12
|
Cemip
|
cell migration inducing protein, hyaluronan binding |
| chr6_+_29853745 | 3.78 |
ENSMUST00000064872.13
ENSMUST00000152581.8 ENSMUST00000176265.8 ENSMUST00000154079.8 |
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr2_+_29759495 | 3.77 |
ENSMUST00000047521.7
ENSMUST00000134152.2 |
Cercam
|
cerebral endothelial cell adhesion molecule |
| chr11_+_63023893 | 3.73 |
ENSMUST00000108700.2
|
Pmp22
|
peripheral myelin protein 22 |
| chr9_-_66951114 | 3.72 |
ENSMUST00000113686.8
|
Tpm1
|
tropomyosin 1, alpha |
| chr17_-_27784640 | 3.72 |
ENSMUST00000232276.2
ENSMUST00000145183.3 ENSMUST00000232203.2 ENSMUST00000231753.2 ENSMUST00000154473.9 |
AI413582
|
expressed sequence AI413582 |
| chr18_+_38429792 | 3.71 |
ENSMUST00000237211.2
|
Rnf14
|
ring finger protein 14 |
| chr11_-_45835737 | 3.71 |
ENSMUST00000129820.8
|
Lsm11
|
U7 snRNP-specific Sm-like protein LSM11 |
| chr11_+_104122399 | 3.68 |
ENSMUST00000132977.8
ENSMUST00000132245.8 ENSMUST00000100347.11 |
Mapt
|
microtubule-associated protein tau |
| chr9_+_120132962 | 3.67 |
ENSMUST00000048121.13
|
Myrip
|
myosin VIIA and Rab interacting protein |
| chr1_-_150268470 | 3.67 |
ENSMUST00000006167.13
ENSMUST00000097547.10 |
Odr4
|
odr4 GPCR localization factor homolog |
| chr1_+_90131702 | 3.66 |
ENSMUST00000065587.5
ENSMUST00000159654.2 |
Ackr3
|
atypical chemokine receptor 3 |
| chr17_+_27904155 | 3.65 |
ENSMUST00000231669.2
ENSMUST00000097360.3 ENSMUST00000231236.2 |
Pacsin1
|
protein kinase C and casein kinase substrate in neurons 1 |
| chr5_-_103174794 | 3.64 |
ENSMUST00000128869.8
|
Mapk10
|
mitogen-activated protein kinase 10 |
| chr7_-_45750153 | 3.64 |
ENSMUST00000180081.3
|
Kcnj11
|
potassium inwardly rectifying channel, subfamily J, member 11 |
| chr10_+_88214989 | 3.59 |
ENSMUST00000127615.8
|
Gnptab
|
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chr2_-_114005752 | 3.58 |
ENSMUST00000102543.5
ENSMUST00000043160.13 |
Aqr
|
aquarius |
| chr11_+_102993865 | 3.52 |
ENSMUST00000152971.2
|
Acbd4
|
acyl-Coenzyme A binding domain containing 4 |
| chr14_-_24537067 | 3.51 |
ENSMUST00000026322.9
|
Polr3a
|
polymerase (RNA) III (DNA directed) polypeptide A |
| chr2_+_126850613 | 3.50 |
ENSMUST00000110394.8
ENSMUST00000002063.15 |
Ap4e1
|
adaptor-related protein complex AP-4, epsilon 1 |
| chr19_-_6191440 | 3.49 |
ENSMUST00000025893.7
|
Arl2
|
ADP-ribosylation factor-like 2 |
| chr8_-_70346625 | 3.48 |
ENSMUST00000152938.9
|
Yjefn3
|
YjeF N-terminal domain containing 3 |
| chr5_-_129907878 | 3.47 |
ENSMUST00000026617.13
|
Phkg1
|
phosphorylase kinase gamma 1 |
| chr15_-_76702170 | 3.44 |
ENSMUST00000175843.3
ENSMUST00000177026.3 ENSMUST00000176736.3 ENSMUST00000036176.16 ENSMUST00000176219.9 ENSMUST00000239134.2 ENSMUST00000239003.2 ENSMUST00000077821.10 |
Arhgap39
|
Rho GTPase activating protein 39 |
| chr9_+_30338329 | 3.42 |
ENSMUST00000164099.3
|
Snx19
|
sorting nexin 19 |
| chr11_+_63023395 | 3.36 |
ENSMUST00000108701.8
|
Pmp22
|
peripheral myelin protein 22 |
| chr10_+_52566616 | 3.35 |
ENSMUST00000105473.3
|
Slc35f1
|
solute carrier family 35, member F1 |
| chr6_-_87312681 | 3.34 |
ENSMUST00000204805.3
|
Antxr1
|
anthrax toxin receptor 1 |
| chr18_-_43526411 | 3.34 |
ENSMUST00000025379.14
|
Dpysl3
|
dihydropyrimidinase-like 3 |
| chr6_-_41613322 | 3.34 |
ENSMUST00000031902.7
|
Trpv6
|
transient receptor potential cation channel, subfamily V, member 6 |
| chr6_-_137146708 | 3.33 |
ENSMUST00000117919.8
|
Rerg
|
RAS-like, estrogen-regulated, growth-inhibitor |
| chr14_+_62793175 | 3.33 |
ENSMUST00000039064.8
|
Fam124a
|
family with sequence similarity 124, member A |
| chr16_+_21023505 | 3.33 |
ENSMUST00000006112.7
|
Ephb3
|
Eph receptor B3 |
| chr6_-_137146677 | 3.31 |
ENSMUST00000119610.4
ENSMUST00000149100.3 |
Rerg
|
RAS-like, estrogen-regulated, growth-inhibitor |
| chr18_+_37568647 | 3.29 |
ENSMUST00000055495.6
|
Pcdhb12
|
protocadherin beta 12 |
| chrX_-_72830487 | 3.29 |
ENSMUST00000052761.9
|
Idh3g
|
isocitrate dehydrogenase 3 (NAD+), gamma |
| chr4_-_144921567 | 3.28 |
ENSMUST00000036579.14
|
Vps13d
|
vacuolar protein sorting 13D |
| chr15_-_89080643 | 3.28 |
ENSMUST00000078953.9
|
Dennd6b
|
DENN/MADD domain containing 6B |
| chr19_+_7394951 | 3.26 |
ENSMUST00000159348.3
|
2700081O15Rik
|
RIKEN cDNA 2700081O15 gene |
| chr4_+_124978927 | 3.25 |
ENSMUST00000154689.8
ENSMUST00000055213.11 ENSMUST00000106171.9 |
Meaf6
|
MYST/Esa1-associated factor 6 |
| chr19_-_36034740 | 3.22 |
ENSMUST00000164639.8
ENSMUST00000166074.2 ENSMUST00000099505.4 |
Htr7
|
5-hydroxytryptamine (serotonin) receptor 7 |
| chr5_-_5799315 | 3.20 |
ENSMUST00000015796.9
|
Steap1
|
six transmembrane epithelial antigen of the prostate 1 |
| chr2_-_121185765 | 3.17 |
ENSMUST00000134796.3
ENSMUST00000110628.8 ENSMUST00000110627.8 ENSMUST00000110625.8 |
Ppip5k1
|
diphosphoinositol pentakisphosphate kinase 1 |
| chr9_+_65008735 | 3.16 |
ENSMUST00000213533.2
ENSMUST00000035499.5 ENSMUST00000077696.13 ENSMUST00000166273.2 |
Igdcc4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr11_+_104122341 | 3.15 |
ENSMUST00000106993.10
|
Mapt
|
microtubule-associated protein tau |
| chrX_-_132799144 | 3.14 |
ENSMUST00000087557.12
|
Tspan6
|
tetraspanin 6 |
| chr10_-_31321793 | 3.13 |
ENSMUST00000213639.2
ENSMUST00000215515.2 ENSMUST00000214644.2 ENSMUST00000213528.2 |
Tpd52l1
|
tumor protein D52-like 1 |
| chr11_-_97464755 | 3.12 |
ENSMUST00000126287.2
ENSMUST00000107590.9 |
Srcin1
|
SRC kinase signaling inhibitor 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.5 | 39.3 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 6.1 | 18.2 | GO:0043181 | vacuolar sequestering(GO:0043181) |
| 3.3 | 9.8 | GO:0051542 | elastin biosynthetic process(GO:0051542) |
| 3.1 | 12.3 | GO:0010269 | response to selenium ion(GO:0010269) |
| 2.6 | 15.8 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 2.6 | 7.7 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 2.4 | 12.1 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 2.3 | 13.7 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 2.1 | 8.5 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 2.1 | 12.3 | GO:0099540 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 1.9 | 5.8 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 1.8 | 5.5 | GO:0061193 | taste bud development(GO:0061193) |
| 1.8 | 5.5 | GO:2000813 | negative regulation of barbed-end actin filament capping(GO:2000813) |
| 1.8 | 5.4 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) |
| 1.6 | 6.5 | GO:0099541 | diacylglycerol catabolic process(GO:0046340) trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 1.6 | 6.4 | GO:0098963 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 1.5 | 10.6 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 1.5 | 22.7 | GO:0010917 | negative regulation of mitochondrial membrane potential(GO:0010917) |
| 1.5 | 8.9 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 1.4 | 4.2 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 1.3 | 10.7 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 1.3 | 6.6 | GO:1904059 | positive regulation of sensory perception of pain(GO:1904058) regulation of locomotor rhythm(GO:1904059) |
| 1.3 | 7.8 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 1.3 | 7.8 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 1.2 | 7.0 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 1.1 | 4.4 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 1.1 | 3.2 | GO:0051385 | response to mineralocorticoid(GO:0051385) |
| 1.1 | 14.9 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 1.0 | 11.4 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 1.0 | 9.1 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 1.0 | 3.0 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 1.0 | 3.0 | GO:0060168 | positive regulation of adenosine receptor signaling pathway(GO:0060168) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 1.0 | 2.9 | GO:0030827 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 1.0 | 6.7 | GO:2001015 | negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.9 | 2.8 | GO:0070256 | positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleoside transport(GO:0032242) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 0.9 | 13.0 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.9 | 15.4 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.9 | 5.4 | GO:0032439 | endosome localization(GO:0032439) |
| 0.9 | 4.3 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.8 | 4.1 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.8 | 2.4 | GO:0002501 | MHC protein complex assembly(GO:0002396) peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.8 | 4.0 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.8 | 11.1 | GO:0030432 | peristalsis(GO:0030432) |
| 0.8 | 2.3 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.7 | 5.0 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.7 | 4.8 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.7 | 4.7 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.7 | 2.7 | GO:0072276 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.6 | 1.8 | GO:2001055 | positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 0.6 | 2.4 | GO:2000983 | regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.6 | 5.3 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.5 | 3.8 | GO:0045583 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.5 | 15.1 | GO:0098596 | vocal learning(GO:0042297) imitative learning(GO:0098596) |
| 0.5 | 4.8 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.5 | 6.3 | GO:0051683 | establishment of Golgi localization(GO:0051683) |
| 0.5 | 1.6 | GO:0072425 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.5 | 3.6 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.5 | 1.0 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) |
| 0.5 | 1.4 | GO:0031990 | RNA import into nucleus(GO:0006404) mRNA export from nucleus in response to heat stress(GO:0031990) negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.5 | 0.9 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.5 | 1.8 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.5 | 8.6 | GO:0021940 | positive regulation of cerebellar granule cell precursor proliferation(GO:0021940) |
| 0.5 | 4.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.4 | 4.5 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.4 | 3.0 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.4 | 4.6 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.4 | 4.9 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.4 | 11.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.4 | 1.6 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.4 | 3.2 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.4 | 4.8 | GO:1901374 | acetylcholine transport(GO:0015870) acetate ester transport(GO:1901374) |
| 0.4 | 2.7 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) |
| 0.4 | 2.7 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.4 | 2.6 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.4 | 5.6 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.4 | 3.9 | GO:0033605 | positive regulation of catecholamine secretion(GO:0033605) |
| 0.3 | 3.7 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.3 | 2.0 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.3 | 5.3 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.3 | 6.7 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.3 | 5.5 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.3 | 3.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.3 | 4.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.3 | 2.5 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.3 | 2.8 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.3 | 3.7 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.3 | 2.1 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.3 | 1.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.3 | 0.8 | GO:1990164 | histone H3-S28 phosphorylation(GO:0043988) histone H2A phosphorylation(GO:1990164) |
| 0.3 | 6.7 | GO:0007614 | short-term memory(GO:0007614) |
| 0.3 | 3.3 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.3 | 11.4 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.3 | 2.4 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.3 | 3.7 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.3 | 4.0 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.3 | 3.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.3 | 7.1 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.3 | 0.8 | GO:0061738 | mitotic cytokinesis checkpoint(GO:0044878) late endosomal microautophagy(GO:0061738) |
| 0.3 | 2.3 | GO:0034651 | cortisol biosynthetic process(GO:0034651) |
| 0.3 | 3.3 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.2 | 18.9 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.2 | 1.0 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.2 | 1.0 | GO:0097017 | renal protein absorption(GO:0097017) protein processing in phagocytic vesicle(GO:1900756) positive regulation of keratinocyte apoptotic process(GO:1902174) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 0.2 | 0.7 | GO:0003274 | endocardial cushion fusion(GO:0003274) |
| 0.2 | 3.3 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.2 | 3.3 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.2 | 9.2 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.2 | 0.9 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.2 | 3.0 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.2 | 0.7 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.2 | 1.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.2 | 1.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.2 | 7.2 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.2 | 1.9 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.2 | 1.2 | GO:2001023 | carbon catabolite regulation of transcription from RNA polymerase II promoter(GO:0000429) carbon catabolite activation of transcription from RNA polymerase II promoter(GO:0000436) regulation of response to drug(GO:2001023) |
| 0.2 | 5.6 | GO:0010663 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.2 | 0.6 | GO:0060825 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.2 | 0.6 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.2 | 2.9 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.2 | 3.1 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.2 | 4.2 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.2 | 6.2 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.2 | 1.4 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.2 | 1.6 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.2 | 1.6 | GO:0015675 | nickel cation transport(GO:0015675) |
| 0.2 | 7.8 | GO:0042417 | dopamine metabolic process(GO:0042417) |
| 0.2 | 2.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.2 | 8.2 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.2 | 3.0 | GO:0007625 | grooming behavior(GO:0007625) |
| 0.2 | 1.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.2 | 1.7 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.2 | 2.1 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.2 | 11.8 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.2 | 0.9 | GO:0014028 | notochord formation(GO:0014028) |
| 0.2 | 6.0 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.2 | 9.4 | GO:0061098 | positive regulation of protein tyrosine kinase activity(GO:0061098) |
| 0.2 | 8.0 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.2 | 0.9 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.2 | 5.5 | GO:0033198 | response to ATP(GO:0033198) |
| 0.2 | 2.0 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.2 | 1.5 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.2 | 1.2 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.2 | 5.0 | GO:0060285 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.2 | 9.3 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.2 | 1.0 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.2 | 28.0 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.2 | 0.8 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.2 | 4.5 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.2 | 14.9 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.2 | 0.9 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.2 | 1.1 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.1 | 0.6 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.1 | 0.7 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) |
| 0.1 | 1.3 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 2.5 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.1 | 1.7 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.1 | 2.8 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 1.5 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.1 | 1.4 | GO:0032026 | response to magnesium ion(GO:0032026) |
| 0.1 | 1.1 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 0.5 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.1 | 0.5 | GO:0009305 | protein biotinylation(GO:0009305) response to biotin(GO:0070781) histone biotinylation(GO:0071110) |
| 0.1 | 1.9 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.1 | 1.2 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.1 | 0.6 | GO:1903599 | positive regulation of mitophagy(GO:1903599) |
| 0.1 | 3.8 | GO:0040020 | regulation of meiotic nuclear division(GO:0040020) |
| 0.1 | 1.0 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 3.0 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.1 | 4.8 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.1 | 0.9 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.1 | 3.4 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) |
| 0.1 | 0.8 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.1 | 0.6 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 | 2.9 | GO:0070200 | establishment of protein localization to telomere(GO:0070200) |
| 0.1 | 0.9 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.1 | 4.3 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.1 | 4.0 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.1 | 0.4 | GO:0048702 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) embryonic neurocranium morphogenesis(GO:0048702) |
| 0.1 | 1.5 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 2.2 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.1 | 2.7 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 5.8 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 0.9 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 3.1 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 13.1 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.1 | 0.9 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 0.3 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 0.7 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.1 | 1.7 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.1 | 2.2 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 0.3 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.1 | 3.3 | GO:0006734 | NADH metabolic process(GO:0006734) |
| 0.1 | 2.8 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.1 | 2.5 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 1.4 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 1.1 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 0.2 | GO:1903028 | asymmetric Golgi ribbon formation(GO:0090164) regulation of opsonization(GO:1903027) positive regulation of opsonization(GO:1903028) |
| 0.1 | 1.7 | GO:0030818 | negative regulation of cAMP biosynthetic process(GO:0030818) |
| 0.1 | 2.1 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.1 | 1.8 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.1 | 1.4 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.1 | 2.6 | GO:0003299 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.1 | 1.0 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.1 | 0.6 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.1 | 1.2 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.1 | 2.7 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.1 | 0.6 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 0.2 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.1 | 1.7 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.1 | 1.0 | GO:0048820 | hair follicle maturation(GO:0048820) |
| 0.1 | 0.8 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.1 | 1.7 | GO:0051703 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.1 | 1.9 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.1 | 1.6 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.1 | 3.7 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.1 | 0.2 | GO:0060982 | coronary artery morphogenesis(GO:0060982) regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 3.6 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 | 2.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 3.6 | GO:0031623 | receptor internalization(GO:0031623) |
| 0.0 | 6.2 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 5.2 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.5 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.0 | 0.8 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 5.2 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 5.3 | GO:0014065 | phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.0 | 0.8 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 2.2 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 3.6 | GO:0050808 | synapse organization(GO:0050808) |
| 0.0 | 2.7 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 | 0.4 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.3 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 1.3 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.4 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 | 0.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 1.4 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.3 | GO:0000423 | macromitophagy(GO:0000423) response to mitochondrial depolarisation(GO:0098780) |
| 0.0 | 0.1 | GO:0021594 | rhombomere formation(GO:0021594) rhombomere 3 formation(GO:0021660) rhombomere 5 morphogenesis(GO:0021664) rhombomere 5 formation(GO:0021666) |
| 0.0 | 0.1 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 2.1 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 1.2 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.2 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 2.9 | GO:0007612 | learning(GO:0007612) |
| 0.0 | 1.4 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.0 | 10.4 | GO:0015711 | organic anion transport(GO:0015711) |
| 0.0 | 1.1 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 0.5 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.2 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.5 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.7 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 1.2 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 1.2 | GO:0000725 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.0 | 0.3 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.4 | GO:0045109 | intermediate filament organization(GO:0045109) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 13.0 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 2.2 | 11.1 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 1.9 | 5.8 | GO:0098855 | HCN channel complex(GO:0098855) |
| 1.9 | 5.6 | GO:0016939 | kinesin II complex(GO:0016939) |
| 1.6 | 45.8 | GO:0043196 | varicosity(GO:0043196) |
| 1.6 | 1.6 | GO:0097635 | extrinsic component of autophagosome membrane(GO:0097635) |
| 1.5 | 13.6 | GO:0045298 | tubulin complex(GO:0045298) |
| 1.4 | 36.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 1.3 | 5.3 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 1.1 | 3.3 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 1.1 | 15.8 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 1.0 | 7.0 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.9 | 13.0 | GO:0005883 | neurofilament(GO:0005883) |
| 0.8 | 8.9 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.8 | 12.7 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.8 | 25.8 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.8 | 4.7 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.6 | 7.1 | GO:0097433 | dense body(GO:0097433) |
| 0.6 | 3.8 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.6 | 3.7 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.6 | 5.5 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.6 | 8.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.6 | 5.9 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.5 | 8.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.5 | 1.6 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.5 | 3.5 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.5 | 7.8 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.5 | 4.6 | GO:0030681 | ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.5 | 6.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.4 | 3.4 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.4 | 4.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.4 | 16.9 | GO:0005921 | gap junction(GO:0005921) |
| 0.4 | 1.9 | GO:1990131 | Iml1 complex(GO:1990130) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.4 | 2.9 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.4 | 4.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.4 | 2.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.4 | 3.3 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.4 | 2.8 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.3 | 2.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.3 | 6.7 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.3 | 1.3 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.3 | 0.3 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.3 | 5.0 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.3 | 1.9 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.3 | 0.9 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.3 | 3.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.3 | 2.7 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.3 | 26.7 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.3 | 4.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.3 | 5.5 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.3 | 3.4 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.3 | 6.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.3 | 16.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.3 | 18.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.3 | 3.6 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.3 | 1.5 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.2 | 4.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.2 | 1.7 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.2 | 2.6 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 0.7 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.2 | 5.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 0.7 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.2 | 5.0 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.2 | 1.2 | GO:0031523 | Myb complex(GO:0031523) |
| 0.2 | 2.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 1.0 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.2 | 2.7 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.2 | 8.0 | GO:0097546 | ciliary base(GO:0097546) |
| 0.2 | 3.0 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 1.4 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.2 | 5.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 5.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.2 | 0.5 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.2 | 0.6 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.1 | 3.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 2.1 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 0.4 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 3.8 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.1 | 4.6 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 1.8 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 11.6 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 27.8 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 2.3 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 15.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 8.6 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 2.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 1.4 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 11.4 | GO:0016234 | inclusion body(GO:0016234) |
| 0.1 | 3.8 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 1.6 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 7.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 2.1 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.8 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 0.8 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.1 | 3.3 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.1 | 0.9 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 1.0 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.9 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 2.4 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 5.6 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 3.8 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.1 | 1.0 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 1.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 7.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 2.1 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 1.2 | GO:0033176 | proton-transporting V-type ATPase complex(GO:0033176) |
| 0.1 | 10.0 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 2.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 7.6 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 3.8 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 11.8 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.1 | 0.8 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 5.9 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 1.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 8.9 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.1 | 1.0 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 101.5 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 0.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 2.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 0.6 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 1.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 2.7 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.7 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.5 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.9 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.9 | GO:0031082 | BLOC complex(GO:0031082) |
| 0.0 | 2.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 2.9 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.1 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.0 | 0.8 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 1.8 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.8 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.7 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.5 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 2.4 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 1.9 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.7 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.5 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 3.1 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 7.3 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.3 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.4 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.9 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 8.3 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 3.0 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.0 | 19.1 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 0.8 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.3 | GO:0005684 | U2-type spliceosomal complex(GO:0005684) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 18.2 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 3.9 | 43.3 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 3.3 | 9.9 | GO:0016492 | G-protein coupled neurotensin receptor activity(GO:0016492) |
| 2.8 | 11.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 2.7 | 13.6 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 2.3 | 13.7 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 2.2 | 8.9 | GO:0031750 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) D3 dopamine receptor binding(GO:0031750) |
| 2.1 | 10.6 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 2.1 | 8.3 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 2.1 | 12.3 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 1.9 | 7.7 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 1.8 | 5.5 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 1.6 | 11.1 | GO:0001601 | peptide YY receptor activity(GO:0001601) pancreatic polypeptide receptor activity(GO:0001602) |
| 1.4 | 10.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 1.4 | 5.4 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 1.2 | 3.7 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 1.2 | 36.4 | GO:0031402 | sodium ion binding(GO:0031402) |
| 1.2 | 25.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 1.1 | 13.0 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 1.0 | 2.9 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.9 | 6.6 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.9 | 10.4 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.9 | 3.7 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.9 | 2.7 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.8 | 2.4 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.8 | 2.4 | GO:0019002 | GMP binding(GO:0019002) |
| 0.8 | 3.8 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.8 | 3.0 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 0.7 | 3.6 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.7 | 5.5 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.7 | 3.3 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.6 | 5.6 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.6 | 1.8 | GO:0031753 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.6 | 4.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.6 | 2.3 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.5 | 3.8 | GO:0002135 | CTP binding(GO:0002135) sulfonylurea receptor binding(GO:0017098) |
| 0.5 | 4.3 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.5 | 4.8 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.5 | 1.6 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 0.5 | 3.2 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.5 | 3.7 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.5 | 3.5 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.5 | 4.6 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.5 | 1.4 | GO:0016213 | linoleoyl-CoA desaturase activity(GO:0016213) |
| 0.4 | 1.3 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.4 | 2.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.4 | 3.2 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.4 | 6.7 | GO:0001727 | lipid kinase activity(GO:0001727) diacylglycerol kinase activity(GO:0004143) |
| 0.4 | 6.2 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.4 | 1.2 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.4 | 2.3 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.4 | 3.0 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.4 | 4.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.4 | 1.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.3 | 1.4 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.3 | 2.4 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.3 | 4.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.3 | 1.4 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.3 | 3.3 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.3 | 4.8 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.3 | 20.4 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.3 | 0.9 | GO:0016501 | prostacyclin receptor activity(GO:0016501) |
| 0.3 | 1.9 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.3 | 1.5 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.3 | 2.0 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.3 | 1.6 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.3 | 14.8 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.2 | 1.0 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.2 | 1.0 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.2 | 13.4 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.2 | 3.2 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.2 | 7.9 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.2 | 10.2 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.2 | 1.3 | GO:0034618 | arginine binding(GO:0034618) |
| 0.2 | 1.2 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.2 | 16.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.2 | 1.6 | GO:0015099 | nickel cation transmembrane transporter activity(GO:0015099) |
| 0.2 | 1.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 5.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.2 | 6.0 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.2 | 3.7 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.2 | 2.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.2 | 2.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.2 | 2.4 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.2 | 1.7 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) potassium ion antiporter activity(GO:0022821) |
| 0.2 | 11.9 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.2 | 4.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 5.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.2 | 3.4 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.2 | 3.0 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 1.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 8.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.2 | 2.5 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.2 | 25.9 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.2 | 2.2 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.2 | 2.8 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 0.9 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.1 | 12.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 1.2 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.7 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.8 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 0.5 | GO:0004077 | biotin-[acetyl-CoA-carboxylase] ligase activity(GO:0004077) biotin-[methylcrotonoyl-CoA-carboxylase] ligase activity(GO:0004078) biotin-[methylmalonyl-CoA-carboxytransferase] ligase activity(GO:0004079) biotin-[propionyl-CoA-carboxylase (ATP-hydrolyzing)] ligase activity(GO:0004080) biotin-protein ligase activity(GO:0018271) |
| 0.1 | 54.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 4.4 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 1.7 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 2.8 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.1 | 5.5 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 5.0 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 10.9 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 0.7 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.1 | 8.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.4 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 2.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 0.7 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 0.6 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 1.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 1.4 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.1 | 0.9 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.1 | 1.0 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 1.5 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 4.9 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.3 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 4.8 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 7.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.9 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.1 | 5.2 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.3 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.1 | 0.5 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 1.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.5 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 1.6 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 0.7 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 0.6 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.1 | 2.2 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 0.3 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.1 | 1.0 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 6.5 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
| 0.1 | 0.4 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 5.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 13.5 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 0.5 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 2.6 | GO:1990939 | ATP-dependent microtubule motor activity(GO:1990939) |
| 0.0 | 1.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.5 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 1.7 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 1.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.7 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 1.0 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.4 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 3.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 3.8 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 2.6 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 2.4 | GO:0005244 | voltage-gated ion channel activity(GO:0005244) voltage-gated channel activity(GO:0022832) |
| 0.0 | 1.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 1.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 9.9 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.8 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 4.2 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.3 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 1.8 | GO:0019209 | kinase activator activity(GO:0019209) |
| 0.0 | 3.3 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 3.3 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.5 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.2 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.5 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.9 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 1.6 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.8 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 1.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 1.7 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.0 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 2.7 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 2.6 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 2.5 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 4.5 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 1.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 6.3 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 1.0 | GO:0051082 | unfolded protein binding(GO:0051082) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 46.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.8 | 13.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.5 | 25.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.5 | 26.0 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.4 | 10.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.3 | 11.8 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.3 | 4.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.3 | 9.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.3 | 23.6 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.3 | 11.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.2 | 19.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.2 | 19.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 5.4 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 7.1 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 4.4 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 2.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 1.4 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 1.0 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 3.0 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 2.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 1.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 2.7 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 3.2 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.1 | 1.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.6 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.8 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.8 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.9 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 9.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.4 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 5.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.3 | PID P73PATHWAY | p73 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 40.5 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 1.1 | 19.4 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.4 | 26.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.4 | 19.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.4 | 14.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.4 | 18.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.3 | 4.8 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.3 | 13.0 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.3 | 12.1 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.3 | 3.6 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.2 | 3.8 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.2 | 5.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 3.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.2 | 5.1 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.2 | 8.1 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.2 | 3.0 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.2 | 2.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 8.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.2 | 2.8 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.2 | 12.6 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.2 | 12.4 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.2 | 2.6 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.2 | 5.5 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.2 | 28.6 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.2 | 3.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 4.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 4.0 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 3.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 11.3 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 1.3 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.1 | 0.6 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 2.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 0.4 | REACTOME SIGNALING BY ERBB4 | Genes involved in Signaling by ERBB4 |
| 0.1 | 2.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.4 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 16.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.0 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 2.7 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.1 | 1.0 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 2.9 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 0.7 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.1 | 0.8 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 1.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 1.2 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 6.7 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 0.9 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 1.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 2.5 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.9 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 1.2 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.7 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 2.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.8 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 1.2 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.8 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 1.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.9 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.9 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.4 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 0.4 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 2.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.0 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.6 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.7 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |