Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
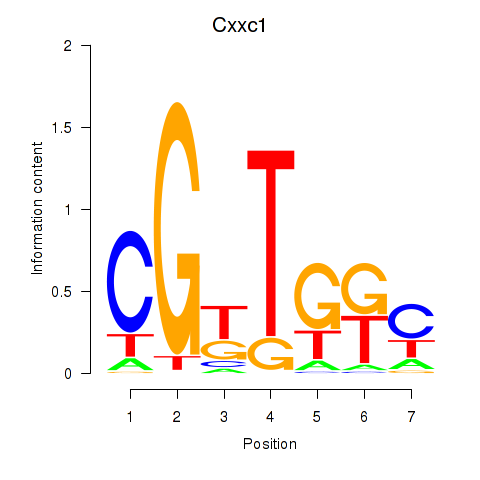
Results for Cxxc1
Z-value: 0.37
Transcription factors associated with Cxxc1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Cxxc1
|
ENSMUSG00000024560.8 | Cxxc1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Cxxc1 | mm39_v1_chr18_+_74349189_74349217 | 0.01 | 9.4e-01 | Click! |
Activity profile of Cxxc1 motif
Sorted Z-values of Cxxc1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Cxxc1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_135287372 | 2.80 |
ENSMUST00000050471.4
|
Pbp2
|
phosphatidylethanolamine binding protein 2 |
| chr7_-_45546081 | 1.85 |
ENSMUST00000120299.4
ENSMUST00000039049.16 |
Syngr4
|
synaptogyrin 4 |
| chr5_+_14075281 | 1.81 |
ENSMUST00000073957.8
|
Sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr3_-_146205429 | 1.67 |
ENSMUST00000029839.11
|
Spata1
|
spermatogenesis associated 1 |
| chr8_+_93084253 | 1.63 |
ENSMUST00000210246.2
ENSMUST00000034184.12 |
Irx5
|
Iroquois homeobox 5 |
| chr13_-_55979191 | 1.57 |
ENSMUST00000021968.7
|
Pitx1
|
paired-like homeodomain transcription factor 1 |
| chr5_+_25721059 | 1.51 |
ENSMUST00000045016.9
|
Cct8l1
|
chaperonin containing TCP1, subunit 8 (theta)-like 1 |
| chr1_+_86354045 | 1.35 |
ENSMUST00000046004.6
|
Tex44
|
testis expressed 44 |
| chr4_-_42853888 | 1.02 |
ENSMUST00000107979.2
|
Fam205a1
|
family with sequence similarity 205, member A1 |
| chr10_+_75790348 | 0.92 |
ENSMUST00000099577.4
|
Gm5134
|
predicted gene 5134 |
| chr5_-_25305621 | 0.87 |
ENSMUST00000030784.14
|
Prkag2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr9_+_78016468 | 0.86 |
ENSMUST00000118869.8
ENSMUST00000125615.2 |
Cilk1
|
ciliogenesis associated kinase 1 |
| chrX_+_5959507 | 0.79 |
ENSMUST00000103007.4
|
Nudt11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11 |
| chr7_-_126944754 | 0.77 |
ENSMUST00000205266.2
|
Zfp768
|
zinc finger protein 768 |
| chr3_-_41696906 | 0.72 |
ENSMUST00000026866.15
|
Sclt1
|
sodium channel and clathrin linker 1 |
| chr7_-_129867967 | 0.67 |
ENSMUST00000117872.8
ENSMUST00000120187.9 |
Fgfr2
|
fibroblast growth factor receptor 2 |
| chr9_+_64940004 | 0.66 |
ENSMUST00000167773.2
|
Dpp8
|
dipeptidylpeptidase 8 |
| chr18_-_43610829 | 0.62 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr7_-_129867921 | 0.61 |
ENSMUST00000122054.8
|
Fgfr2
|
fibroblast growth factor receptor 2 |
| chr4_+_108857803 | 0.60 |
ENSMUST00000065977.11
ENSMUST00000102736.9 |
Nrd1
|
nardilysin, N-arginine dibasic convertase, NRD convertase 1 |
| chr19_-_4489415 | 0.60 |
ENSMUST00000235680.2
ENSMUST00000117462.2 ENSMUST00000048197.10 |
Rhod
|
ras homolog family member D |
| chr16_-_64591509 | 0.58 |
ENSMUST00000076991.7
|
4930453N24Rik
|
RIKEN cDNA 4930453N24 gene |
| chr4_+_108857967 | 0.58 |
ENSMUST00000106644.9
|
Nrd1
|
nardilysin, N-arginine dibasic convertase, NRD convertase 1 |
| chr7_-_28135616 | 0.48 |
ENSMUST00000208199.2
|
Samd4b
|
sterile alpha motif domain containing 4B |
| chr9_-_107483855 | 0.43 |
ENSMUST00000073448.12
ENSMUST00000194606.2 ENSMUST00000195662.6 |
Sema3b
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B |
| chr7_+_128346655 | 0.39 |
ENSMUST00000042942.10
|
Sec23ip
|
Sec23 interacting protein |
| chr1_+_59296065 | 0.33 |
ENSMUST00000160662.8
ENSMUST00000114248.3 |
Cdk15
|
cyclin-dependent kinase 15 |
| chr10_-_51507527 | 0.32 |
ENSMUST00000219286.2
ENSMUST00000020062.4 ENSMUST00000218684.2 |
Gprc6a
|
G protein-coupled receptor, family C, group 6, member A |
| chr11_-_88608958 | 0.30 |
ENSMUST00000107908.2
|
Msi2
|
musashi RNA-binding protein 2 |
| chr11_+_113540154 | 0.29 |
ENSMUST00000063776.14
|
Cog1
|
component of oligomeric golgi complex 1 |
| chr17_+_28076209 | 0.28 |
ENSMUST00000130810.2
|
Uhrf1bp1
|
UHRF1 (ICBP90) binding protein 1 |
| chr14_-_62718581 | 0.27 |
ENSMUST00000186010.8
|
Gm4131
|
predicted gene 4131 |
| chr17_-_87754282 | 0.25 |
ENSMUST00000234406.2
ENSMUST00000040440.7 |
Calm2
|
calmodulin 2 |
| chr17_-_56343531 | 0.24 |
ENSMUST00000233803.2
|
Sh3gl1
|
SH3-domain GRB2-like 1 |
| chr11_+_113539995 | 0.19 |
ENSMUST00000018805.15
|
Cog1
|
component of oligomeric golgi complex 1 |
| chr19_+_38919477 | 0.18 |
ENSMUST00000145051.2
|
Hells
|
helicase, lymphoid specific |
| chr18_+_37138256 | 0.17 |
ENSMUST00000115658.6
|
Pcdha11
|
protocadherin alpha 11 |
| chr10_-_78248771 | 0.15 |
ENSMUST00000062678.11
|
Rrp1
|
ribosomal RNA processing 1 |
| chr11_-_85125889 | 0.13 |
ENSMUST00000018625.10
|
Appbp2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chr2_+_151753119 | 0.09 |
ENSMUST00000028955.6
|
Angpt4
|
angiopoietin 4 |
| chr15_-_38518406 | 0.05 |
ENSMUST00000151319.8
|
Azin1
|
antizyme inhibitor 1 |
| chr11_+_60428788 | 0.02 |
ENSMUST00000044250.4
|
Alkbh5
|
alkB homolog 5, RNA demethylase |
| chr9_-_78016302 | 0.01 |
ENSMUST00000001402.14
|
Fbxo9
|
f-box protein 9 |
| chr2_+_146854916 | 0.00 |
ENSMUST00000028921.6
|
Xrn2
|
5'-3' exoribonuclease 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0035603 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) coronal suture morphogenesis(GO:0060365) |
| 0.4 | 1.6 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.2 | 0.5 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.1 | 1.6 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 | 0.8 | GO:1901908 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.7 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.9 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.0 | 1.2 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 2.4 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 | 0.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 1.5 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.0 | 0.2 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.0 | 0.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.9 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.4 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.5 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.7 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.6 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.5 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.8 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.9 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 1.9 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 2.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 1.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.8 | GO:0034431 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 0.9 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.2 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.0 | 1.5 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 2.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.6 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.5 | GO:0030371 | translation repressor activity(GO:0030371) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.3 | PID FGF PATHWAY | FGF signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 0.9 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 1.3 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |