Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
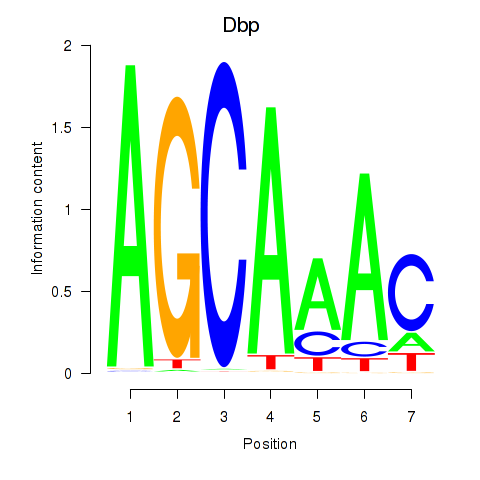
Results for Dbp
Z-value: 1.48
Transcription factors associated with Dbp
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Dbp
|
ENSMUSG00000059824.13 | Dbp |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Dbp | mm39_v1_chr7_+_45354512_45354735 | 0.23 | 5.2e-02 | Click! |
Activity profile of Dbp motif
Sorted Z-values of Dbp motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Dbp
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_26187883 | 14.26 |
ENSMUST00000108308.10
ENSMUST00000075054.10 |
Nlgn1
|
neuroligin 1 |
| chr18_-_15536747 | 13.17 |
ENSMUST00000079081.8
|
Aqp4
|
aquaporin 4 |
| chr16_-_34334314 | 12.37 |
ENSMUST00000151491.8
ENSMUST00000114960.9 |
Kalrn
|
kalirin, RhoGEF kinase |
| chr12_+_29584560 | 12.32 |
ENSMUST00000021009.10
|
Myt1l
|
myelin transcription factor 1-like |
| chr7_-_74958121 | 11.90 |
ENSMUST00000085164.7
|
Sv2b
|
synaptic vesicle glycoprotein 2 b |
| chr5_+_57879201 | 11.88 |
ENSMUST00000199310.2
|
Pcdh7
|
protocadherin 7 |
| chr8_+_55407872 | 11.54 |
ENSMUST00000033915.9
|
Gpm6a
|
glycoprotein m6a |
| chr1_+_66426127 | 11.53 |
ENSMUST00000145419.8
|
Map2
|
microtubule-associated protein 2 |
| chr3_+_14011445 | 10.33 |
ENSMUST00000192209.6
ENSMUST00000171075.8 ENSMUST00000108372.4 |
Ralyl
|
RALY RNA binding protein-like |
| chr16_+_39804711 | 9.93 |
ENSMUST00000187695.7
|
Lsamp
|
limbic system-associated membrane protein |
| chr16_-_34334454 | 9.43 |
ENSMUST00000089655.12
|
Kalrn
|
kalirin, RhoGEF kinase |
| chr12_+_49429574 | 9.20 |
ENSMUST00000179669.3
|
Foxg1
|
forkhead box G1 |
| chr7_-_84021655 | 8.99 |
ENSMUST00000208392.2
ENSMUST00000208232.2 ENSMUST00000208863.2 |
Arnt2
|
aryl hydrocarbon receptor nuclear translocator 2 |
| chr10_+_90412638 | 8.94 |
ENSMUST00000183136.8
ENSMUST00000182595.8 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr10_+_123099945 | 8.82 |
ENSMUST00000238972.2
ENSMUST00000050756.8 |
Tafa2
|
TAFA chemokine like family member 2 |
| chr10_+_90412570 | 8.74 |
ENSMUST00000182430.8
ENSMUST00000182960.8 ENSMUST00000182045.2 ENSMUST00000182083.2 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr2_+_164802729 | 8.57 |
ENSMUST00000202623.4
|
Slc12a5
|
solute carrier family 12, member 5 |
| chr4_+_28813152 | 8.52 |
ENSMUST00000108194.9
ENSMUST00000108191.2 |
Epha7
|
Eph receptor A7 |
| chr2_-_127363251 | 8.36 |
ENSMUST00000028850.15
ENSMUST00000103215.11 |
Kcnip3
|
Kv channel interacting protein 3, calsenilin |
| chr2_+_164802766 | 8.28 |
ENSMUST00000202223.4
|
Slc12a5
|
solute carrier family 12, member 5 |
| chr15_-_37459570 | 8.12 |
ENSMUST00000119730.8
ENSMUST00000120746.8 |
Ncald
|
neurocalcin delta |
| chr8_+_24159669 | 8.01 |
ENSMUST00000042352.11
ENSMUST00000207301.2 |
Zmat4
|
zinc finger, matrin type 4 |
| chr15_-_37458768 | 7.99 |
ENSMUST00000116445.9
|
Ncald
|
neurocalcin delta |
| chr4_+_28813125 | 7.86 |
ENSMUST00000080934.11
ENSMUST00000029964.12 |
Epha7
|
Eph receptor A7 |
| chr12_+_49429790 | 7.71 |
ENSMUST00000021333.5
|
Foxg1
|
forkhead box G1 |
| chr3_-_89230190 | 7.62 |
ENSMUST00000200436.2
ENSMUST00000029673.10 |
Efna3
|
ephrin A3 |
| chr11_+_97732108 | 7.40 |
ENSMUST00000155954.3
ENSMUST00000164364.2 ENSMUST00000170806.2 |
B230217C12Rik
|
RIKEN cDNA B230217C12 gene |
| chr4_+_123077515 | 7.36 |
ENSMUST00000152194.2
|
Hpcal4
|
hippocalcin-like 4 |
| chr18_-_25886908 | 7.35 |
ENSMUST00000115816.3
ENSMUST00000223704.2 |
Celf4
|
CUGBP, Elav-like family member 4 |
| chr16_+_6887689 | 7.35 |
ENSMUST00000229741.2
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr16_+_91022300 | 7.30 |
ENSMUST00000035608.10
|
Olig2
|
oligodendrocyte transcription factor 2 |
| chr10_+_90412432 | 7.30 |
ENSMUST00000182786.8
ENSMUST00000182600.8 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chrX_+_168468186 | 7.25 |
ENSMUST00000112107.8
ENSMUST00000112104.8 |
Mid1
|
midline 1 |
| chr2_+_49956441 | 7.24 |
ENSMUST00000112712.10
ENSMUST00000128451.8 ENSMUST00000053208.14 |
Lypd6
|
LY6/PLAUR domain containing 6 |
| chr5_+_115604321 | 7.10 |
ENSMUST00000145785.8
ENSMUST00000031495.11 ENSMUST00000112071.8 ENSMUST00000125568.2 |
Pla2g1b
|
phospholipase A2, group IB, pancreas |
| chr2_-_52448552 | 6.72 |
ENSMUST00000102760.10
ENSMUST00000102761.9 |
Cacnb4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chrX_-_42256694 | 6.65 |
ENSMUST00000115058.8
ENSMUST00000115059.8 |
Tenm1
|
teneurin transmembrane protein 1 |
| chr10_+_90412114 | 6.59 |
ENSMUST00000182427.8
ENSMUST00000182053.8 ENSMUST00000182113.8 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr5_-_103359117 | 6.51 |
ENSMUST00000112846.8
ENSMUST00000170792.9 ENSMUST00000112847.9 ENSMUST00000238446.3 ENSMUST00000133069.8 |
Mapk10
|
mitogen-activated protein kinase 10 |
| chr6_+_87350292 | 6.47 |
ENSMUST00000032128.6
|
Gkn2
|
gastrokine 2 |
| chr10_+_90412539 | 6.47 |
ENSMUST00000182284.8
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr9_-_112016966 | 6.42 |
ENSMUST00000178410.2
ENSMUST00000172380.10 |
Arpp21
|
cyclic AMP-regulated phosphoprotein, 21 |
| chr5_-_24806960 | 6.32 |
ENSMUST00000030791.12
|
Smarcd3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr17_+_44114894 | 6.20 |
ENSMUST00000044895.13
|
Rcan2
|
regulator of calcineurin 2 |
| chr10_-_63926044 | 6.15 |
ENSMUST00000105439.2
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr3_+_64856617 | 6.14 |
ENSMUST00000238901.2
|
Kcnab1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr7_-_126620378 | 6.09 |
ENSMUST00000159916.5
|
Prrt2
|
proline-rich transmembrane protein 2 |
| chr18_+_23937019 | 6.06 |
ENSMUST00000025127.5
|
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr5_+_146392371 | 6.06 |
ENSMUST00000238592.2
|
Wasf3
|
WASP family, member 3 |
| chr7_-_126620249 | 6.03 |
ENSMUST00000202045.2
|
Prrt2
|
proline-rich transmembrane protein 2 |
| chr9_+_32135540 | 5.88 |
ENSMUST00000168954.9
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr3_+_18108313 | 5.86 |
ENSMUST00000026120.8
|
Bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr11_-_41891662 | 5.83 |
ENSMUST00000070725.11
|
Gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 2 |
| chr17_-_31363245 | 5.73 |
ENSMUST00000024826.8
|
Tff2
|
trefoil factor 2 (spasmolytic protein 1) |
| chrX_-_58211440 | 5.71 |
ENSMUST00000119306.2
|
Fgf13
|
fibroblast growth factor 13 |
| chr9_-_49710190 | 5.70 |
ENSMUST00000114476.8
ENSMUST00000193547.6 |
Ncam1
|
neural cell adhesion molecule 1 |
| chr2_+_70393782 | 5.68 |
ENSMUST00000123330.3
|
Gad1
|
glutamate decarboxylase 1 |
| chr3_+_76500857 | 5.68 |
ENSMUST00000162471.2
|
Fstl5
|
follistatin-like 5 |
| chr2_-_57942844 | 5.65 |
ENSMUST00000090940.6
|
Ermn
|
ermin, ERM-like protein |
| chr19_-_46315543 | 5.53 |
ENSMUST00000223917.2
ENSMUST00000224447.2 ENSMUST00000041391.5 ENSMUST00000096029.12 |
Psd
|
pleckstrin and Sec7 domain containing |
| chr14_-_88708782 | 5.51 |
ENSMUST00000192557.2
ENSMUST00000061628.7 |
Pcdh20
|
protocadherin 20 |
| chr15_+_4404965 | 5.50 |
ENSMUST00000061925.5
|
Plcxd3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr6_-_53797748 | 5.47 |
ENSMUST00000127748.5
|
Tril
|
TLR4 interactor with leucine-rich repeats |
| chr18_+_37637317 | 5.46 |
ENSMUST00000052179.8
|
Pcdhb20
|
protocadherin beta 20 |
| chr7_-_16651107 | 5.38 |
ENSMUST00000173139.2
|
Calm3
|
calmodulin 3 |
| chr9_-_4796217 | 5.37 |
ENSMUST00000027020.13
ENSMUST00000163309.2 |
Gria4
|
glutamate receptor, ionotropic, AMPA4 (alpha 4) |
| chr12_+_61570669 | 5.30 |
ENSMUST00000055815.14
ENSMUST00000119481.2 |
Lrfn5
|
leucine rich repeat and fibronectin type III domain containing 5 |
| chr2_+_107120934 | 5.28 |
ENSMUST00000037012.3
|
Kcna4
|
potassium voltage-gated channel, shaker-related subfamily, member 4 |
| chr19_-_40502117 | 5.27 |
ENSMUST00000225148.2
ENSMUST00000224667.2 ENSMUST00000225153.2 ENSMUST00000238831.2 ENSMUST00000226047.2 ENSMUST00000224247.2 |
Sorbs1
|
sorbin and SH3 domain containing 1 |
| chrX_-_43879022 | 5.26 |
ENSMUST00000115056.2
|
Dcaf12l1
|
DDB1 and CUL4 associated factor 12-like 1 |
| chr9_-_49710058 | 5.10 |
ENSMUST00000192584.2
ENSMUST00000166811.9 |
Ncam1
|
neural cell adhesion molecule 1 |
| chr1_+_11484313 | 5.03 |
ENSMUST00000171690.9
ENSMUST00000048613.14 |
A830018L16Rik
|
RIKEN cDNA A830018L16 gene |
| chr5_+_63957805 | 5.02 |
ENSMUST00000162757.4
|
Nwd2
|
NACHT and WD repeat domain containing 2 |
| chr9_+_37450551 | 4.98 |
ENSMUST00000002008.7
ENSMUST00000215957.2 ENSMUST00000215271.2 |
Vsig2
|
V-set and immunoglobulin domain containing 2 |
| chr2_+_65676111 | 4.93 |
ENSMUST00000122912.8
|
Csrnp3
|
cysteine-serine-rich nuclear protein 3 |
| chrX_+_70093766 | 4.91 |
ENSMUST00000239162.2
ENSMUST00000114629.4 ENSMUST00000082088.10 |
Mamld1
|
mastermind-like domain containing 1 |
| chr14_+_120513106 | 4.90 |
ENSMUST00000227012.2
ENSMUST00000167459.3 |
Mbnl2
|
muscleblind like splicing factor 2 |
| chr14_-_24053994 | 4.90 |
ENSMUST00000225431.2
ENSMUST00000188210.8 ENSMUST00000224787.2 ENSMUST00000225315.2 ENSMUST00000225556.2 ENSMUST00000223727.2 ENSMUST00000223655.2 ENSMUST00000224077.2 ENSMUST00000224812.2 ENSMUST00000224285.2 ENSMUST00000225471.2 ENSMUST00000224232.2 ENSMUST00000223749.2 ENSMUST00000224025.2 |
Kcnma1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr2_+_20727274 | 4.86 |
ENSMUST00000114607.8
|
Etl4
|
enhancer trap locus 4 |
| chr1_-_132470672 | 4.83 |
ENSMUST00000086521.11
|
Cntn2
|
contactin 2 |
| chr12_+_69954506 | 4.82 |
ENSMUST00000223456.2
|
Atl1
|
atlastin GTPase 1 |
| chr7_+_57240894 | 4.81 |
ENSMUST00000039697.14
|
Gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 3 |
| chr15_+_18819033 | 4.80 |
ENSMUST00000166873.9
|
Cdh10
|
cadherin 10 |
| chr15_+_16728842 | 4.77 |
ENSMUST00000228307.2
|
Cdh9
|
cadherin 9 |
| chr7_+_45434755 | 4.71 |
ENSMUST00000233503.2
ENSMUST00000120005.10 ENSMUST00000211609.2 |
Lmtk3
|
lemur tyrosine kinase 3 |
| chr13_-_49301407 | 4.68 |
ENSMUST00000162581.8
ENSMUST00000110097.9 ENSMUST00000049265.15 ENSMUST00000035538.13 ENSMUST00000110096.8 ENSMUST00000091623.10 |
Wnk2
|
WNK lysine deficient protein kinase 2 |
| chr3_-_50398027 | 4.68 |
ENSMUST00000029297.6
ENSMUST00000194462.6 |
Slc7a11
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 11 |
| chr5_+_107645626 | 4.68 |
ENSMUST00000152474.8
ENSMUST00000060553.8 |
Btbd8
|
BTB (POZ) domain containing 8 |
| chrX_-_43879055 | 4.64 |
ENSMUST00000060481.9
|
Dcaf12l1
|
DDB1 and CUL4 associated factor 12-like 1 |
| chr11_+_77821626 | 4.64 |
ENSMUST00000093995.10
ENSMUST00000000646.14 |
Sez6
|
seizure related gene 6 |
| chr3_+_13536696 | 4.64 |
ENSMUST00000191806.3
ENSMUST00000193117.3 |
Ralyl
|
RALY RNA binding protein-like |
| chr9_-_112016834 | 4.55 |
ENSMUST00000111872.9
ENSMUST00000164754.9 |
Arpp21
|
cyclic AMP-regulated phosphoprotein, 21 |
| chr5_+_71857261 | 4.47 |
ENSMUST00000031122.9
|
Gabrb1
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 1 |
| chr3_-_146487102 | 4.45 |
ENSMUST00000005164.12
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr9_+_32135781 | 4.44 |
ENSMUST00000183121.2
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr9_-_4796142 | 4.39 |
ENSMUST00000063508.15
|
Gria4
|
glutamate receptor, ionotropic, AMPA4 (alpha 4) |
| chr6_-_91784405 | 4.38 |
ENSMUST00000162300.8
|
Grip2
|
glutamate receptor interacting protein 2 |
| chr2_-_140513320 | 4.37 |
ENSMUST00000056760.4
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3 |
| chr18_+_37847792 | 4.36 |
ENSMUST00000192511.2
ENSMUST00000193476.2 |
Pcdhga7
|
protocadherin gamma subfamily A, 7 |
| chr1_+_140173787 | 4.36 |
ENSMUST00000239229.2
ENSMUST00000120709.8 ENSMUST00000120796.8 ENSMUST00000119786.8 |
Kcnt2
|
potassium channel, subfamily T, member 2 |
| chr8_+_12999394 | 4.32 |
ENSMUST00000110867.9
|
Mcf2l
|
mcf.2 transforming sequence-like |
| chrX_-_164110372 | 4.19 |
ENSMUST00000058787.9
|
Glra2
|
glycine receptor, alpha 2 subunit |
| chr4_-_87724533 | 4.15 |
ENSMUST00000126353.8
ENSMUST00000149357.8 |
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chrX_-_71699740 | 4.12 |
ENSMUST00000055966.13
|
Gabra3
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 3 |
| chr14_+_70314727 | 4.10 |
ENSMUST00000225200.2
|
Egr3
|
early growth response 3 |
| chr14_-_121976273 | 3.99 |
ENSMUST00000212376.3
ENSMUST00000239077.2 |
Dock9
|
dedicator of cytokinesis 9 |
| chr2_-_77533596 | 3.93 |
ENSMUST00000171063.8
|
Zfp385b
|
zinc finger protein 385B |
| chr18_+_37617848 | 3.91 |
ENSMUST00000053856.6
|
Pcdhb17
|
protocadherin beta 17 |
| chr13_+_117738972 | 3.86 |
ENSMUST00000006991.9
|
Hcn1
|
hyperpolarization activated cyclic nucleotide gated potassium channel 1 |
| chr2_-_140513382 | 3.84 |
ENSMUST00000110057.3
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3 |
| chr6_+_51500881 | 3.81 |
ENSMUST00000049152.15
|
Snx10
|
sorting nexin 10 |
| chr6_-_113478779 | 3.80 |
ENSMUST00000101059.4
ENSMUST00000204268.3 ENSMUST00000205170.2 ENSMUST00000205075.2 ENSMUST00000204134.3 |
Prrt3
|
proline-rich transmembrane protein 3 |
| chr2_+_67948057 | 3.79 |
ENSMUST00000112346.3
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr2_+_65676176 | 3.77 |
ENSMUST00000053910.10
|
Csrnp3
|
cysteine-serine-rich nuclear protein 3 |
| chr12_+_108572015 | 3.68 |
ENSMUST00000109854.9
|
Evl
|
Ena-vasodilator stimulated phosphoprotein |
| chr1_-_181841969 | 3.61 |
ENSMUST00000193074.6
|
Enah
|
ENAH actin regulator |
| chr17_+_69144053 | 3.60 |
ENSMUST00000178545.3
|
Tmem200c
|
transmembrane protein 200C |
| chr18_+_37630044 | 3.57 |
ENSMUST00000059571.7
|
Pcdhb19
|
protocadherin beta 19 |
| chr10_+_85222677 | 3.56 |
ENSMUST00000105307.8
ENSMUST00000020231.10 |
Btbd11
|
BTB (POZ) domain containing 11 |
| chr6_-_138404076 | 3.54 |
ENSMUST00000203435.3
|
Lmo3
|
LIM domain only 3 |
| chr5_+_150119860 | 3.50 |
ENSMUST00000202600.4
|
Fry
|
FRY microtubule binding protein |
| chr13_-_94383040 | 3.47 |
ENSMUST00000153558.8
|
Scamp1
|
secretory carrier membrane protein 1 |
| chr19_+_22670134 | 3.47 |
ENSMUST00000237470.2
ENSMUST00000099564.10 ENSMUST00000099569.10 ENSMUST00000099566.5 ENSMUST00000235712.2 |
Trpm3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr13_+_110063364 | 3.47 |
ENSMUST00000117420.8
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr18_+_57605525 | 3.45 |
ENSMUST00000209786.2
|
Ctxn3
|
cortexin 3 |
| chr11_-_37126709 | 3.43 |
ENSMUST00000102801.8
|
Tenm2
|
teneurin transmembrane protein 2 |
| chr18_+_63841756 | 3.43 |
ENSMUST00000072726.7
ENSMUST00000235648.2 ENSMUST00000236879.2 |
Wdr7
|
WD repeat domain 7 |
| chr18_-_25886750 | 3.42 |
ENSMUST00000224553.2
ENSMUST00000025117.14 |
Celf4
|
CUGBP, Elav-like family member 4 |
| chrX_+_152281200 | 3.41 |
ENSMUST00000060714.10
|
Ubqln2
|
ubiquilin 2 |
| chr6_+_106095726 | 3.37 |
ENSMUST00000113258.8
ENSMUST00000079416.6 |
Cntn4
|
contactin 4 |
| chrX_+_47712614 | 3.34 |
ENSMUST00000114936.8
|
Slc25a14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr13_+_83723743 | 3.32 |
ENSMUST00000198217.5
ENSMUST00000199210.5 |
Mef2c
|
myocyte enhancer factor 2C |
| chr9_-_60557076 | 3.29 |
ENSMUST00000053171.14
|
Lrrc49
|
leucine rich repeat containing 49 |
| chr1_+_143516402 | 3.27 |
ENSMUST00000038252.4
|
B3galt2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chrX_+_47712676 | 3.26 |
ENSMUST00000177710.2
|
Slc25a14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr6_+_15185399 | 3.26 |
ENSMUST00000115474.8
ENSMUST00000115472.8 ENSMUST00000031545.14 |
Foxp2
|
forkhead box P2 |
| chr1_+_179788675 | 3.24 |
ENSMUST00000076687.12
ENSMUST00000097450.10 ENSMUST00000212756.2 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr16_+_17327076 | 3.17 |
ENSMUST00000232242.2
|
Lztr1
|
leucine-zipper-like transcriptional regulator, 1 |
| chr5_+_98328723 | 3.15 |
ENSMUST00000112959.4
|
Prdm8
|
PR domain containing 8 |
| chr13_+_118851214 | 3.14 |
ENSMUST00000022246.9
|
Fgf10
|
fibroblast growth factor 10 |
| chr8_-_85500998 | 3.12 |
ENSMUST00000109762.8
|
Nfix
|
nuclear factor I/X |
| chr18_+_37610858 | 3.11 |
ENSMUST00000051442.7
|
Pcdhb16
|
protocadherin beta 16 |
| chr15_+_41694317 | 3.11 |
ENSMUST00000166917.3
ENSMUST00000230127.2 ENSMUST00000230131.2 |
Oxr1
|
oxidation resistance 1 |
| chr5_-_92190489 | 3.08 |
ENSMUST00000113140.5
ENSMUST00000113143.8 |
Cdkl2
|
cyclin-dependent kinase-like 2 (CDC2-related kinase) |
| chr6_-_91784299 | 3.05 |
ENSMUST00000159684.8
|
Grip2
|
glutamate receptor interacting protein 2 |
| chr19_+_56450062 | 3.02 |
ENSMUST00000178590.9
ENSMUST00000039666.8 |
Plekhs1
|
pleckstrin homology domain containing, family S member 1 |
| chr1_+_127234441 | 3.01 |
ENSMUST00000171405.2
|
Mgat5
|
mannoside acetylglucosaminyltransferase 5 |
| chr10_-_30531832 | 2.99 |
ENSMUST00000217138.2
ENSMUST00000217644.2 ENSMUST00000216172.2 |
Ncoa7
|
nuclear receptor coactivator 7 |
| chr2_-_102231208 | 2.97 |
ENSMUST00000102573.8
|
Trim44
|
tripartite motif-containing 44 |
| chr13_-_43457626 | 2.95 |
ENSMUST00000055341.7
|
Gfod1
|
glucose-fructose oxidoreductase domain containing 1 |
| chr5_+_150183201 | 2.95 |
ENSMUST00000087204.9
|
Fry
|
FRY microtubule binding protein |
| chr6_+_36364990 | 2.94 |
ENSMUST00000172278.8
|
Chrm2
|
cholinergic receptor, muscarinic 2, cardiac |
| chr3_-_89671888 | 2.92 |
ENSMUST00000200558.5
ENSMUST00000029562.5 |
Chrnb2
|
cholinergic receptor, nicotinic, beta polypeptide 2 (neuronal) |
| chr10_+_94386714 | 2.91 |
ENSMUST00000148910.3
ENSMUST00000117460.8 |
Tmcc3
|
transmembrane and coiled coil domains 3 |
| chr10_-_30531768 | 2.89 |
ENSMUST00000092610.12
|
Ncoa7
|
nuclear receptor coactivator 7 |
| chr7_+_30411634 | 2.89 |
ENSMUST00000005692.14
ENSMUST00000170371.2 |
Atp4a
|
ATPase, H+/K+ exchanging, gastric, alpha polypeptide |
| chr7_-_78228116 | 2.84 |
ENSMUST00000206268.2
ENSMUST00000039431.14 |
Ntrk3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr13_+_41013230 | 2.81 |
ENSMUST00000110191.10
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr10_+_12936248 | 2.80 |
ENSMUST00000193426.6
|
Plagl1
|
pleiomorphic adenoma gene-like 1 |
| chr7_-_144761806 | 2.74 |
ENSMUST00000208788.2
|
Smim38
|
small integral membrane protein 38 |
| chr5_-_44956932 | 2.73 |
ENSMUST00000199261.2
ENSMUST00000199534.5 |
Ldb2
|
LIM domain binding 2 |
| chr3_+_133942244 | 2.69 |
ENSMUST00000181904.3
|
Cxxc4
|
CXXC finger 4 |
| chr5_+_99002293 | 2.68 |
ENSMUST00000031278.6
ENSMUST00000200388.2 |
Bmp3
|
bone morphogenetic protein 3 |
| chrX_-_151820545 | 2.63 |
ENSMUST00000051484.5
|
Mageh1
|
MAGE family member H1 |
| chr14_-_55163311 | 2.62 |
ENSMUST00000022813.8
|
Efs
|
embryonal Fyn-associated substrate |
| chr7_+_108549545 | 2.58 |
ENSMUST00000207583.2
|
Tub
|
tubby bipartite transcription factor |
| chr18_-_12429048 | 2.57 |
ENSMUST00000234212.2
|
Ankrd29
|
ankyrin repeat domain 29 |
| chr2_+_105505823 | 2.57 |
ENSMUST00000167211.9
ENSMUST00000111083.10 |
Pax6
|
paired box 6 |
| chr3_+_109481223 | 2.56 |
ENSMUST00000106576.3
|
Vav3
|
vav 3 oncogene |
| chr4_-_87724512 | 2.50 |
ENSMUST00000148059.2
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr18_+_37488174 | 2.50 |
ENSMUST00000192867.2
ENSMUST00000051163.3 |
Pcdhb8
|
protocadherin beta 8 |
| chr11_+_58197947 | 2.50 |
ENSMUST00000049353.9
|
Zfp692
|
zinc finger protein 692 |
| chr11_+_24028022 | 2.49 |
ENSMUST00000000881.13
|
Bcl11a
|
B cell CLL/lymphoma 11A (zinc finger protein) |
| chr1_-_134162231 | 2.48 |
ENSMUST00000169927.2
|
Adora1
|
adenosine A1 receptor |
| chrX_-_87159237 | 2.48 |
ENSMUST00000113966.8
ENSMUST00000113964.2 |
Il1rapl1
|
interleukin 1 receptor accessory protein-like 1 |
| chr5_-_77262968 | 2.46 |
ENSMUST00000081964.7
|
Hopx
|
HOP homeobox |
| chr19_-_55229668 | 2.45 |
ENSMUST00000069183.8
|
Gucy2g
|
guanylate cyclase 2g |
| chr2_-_6889685 | 2.43 |
ENSMUST00000183091.8
ENSMUST00000182851.8 |
Celf2
|
CUGBP, Elav-like family member 2 |
| chr17_+_47451868 | 2.40 |
ENSMUST00000190080.9
|
Trerf1
|
transcriptional regulating factor 1 |
| chr9_+_22011488 | 2.39 |
ENSMUST00000213607.2
|
Cnn1
|
calponin 1 |
| chr13_-_47168286 | 2.37 |
ENSMUST00000052747.4
|
Nhlrc1
|
NHL repeat containing 1 |
| chr18_+_37622518 | 2.37 |
ENSMUST00000055949.4
|
Pcdhb18
|
protocadherin beta 18 |
| chr6_-_40562700 | 2.36 |
ENSMUST00000177178.2
ENSMUST00000129948.9 ENSMUST00000101491.11 |
Clec5a
|
C-type lectin domain family 5, member a |
| chr1_+_131566223 | 2.36 |
ENSMUST00000112411.2
|
Ctse
|
cathepsin E |
| chr2_-_6889783 | 2.35 |
ENSMUST00000170438.8
ENSMUST00000114924.10 ENSMUST00000114934.11 |
Celf2
|
CUGBP, Elav-like family member 2 |
| chr14_+_70314652 | 2.35 |
ENSMUST00000035908.3
|
Egr3
|
early growth response 3 |
| chr6_-_94677118 | 2.34 |
ENSMUST00000101126.3
ENSMUST00000032105.11 |
Lrig1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr1_-_37535202 | 2.30 |
ENSMUST00000143636.8
|
Mgat4a
|
mannoside acetylglucosaminyltransferase 4, isoenzyme A |
| chr4_+_43669266 | 2.29 |
ENSMUST00000107864.8
|
Tmem8b
|
transmembrane protein 8B |
| chr6_-_141801918 | 2.29 |
ENSMUST00000163678.2
|
Slco1a4
|
solute carrier organic anion transporter family, member 1a4 |
| chr11_+_24028173 | 2.26 |
ENSMUST00000109514.8
|
Bcl11a
|
B cell CLL/lymphoma 11A (zinc finger protein) |
| chr4_+_80828883 | 2.25 |
ENSMUST00000055922.4
|
Lurap1l
|
leucine rich adaptor protein 1-like |
| chr11_-_113956996 | 2.20 |
ENSMUST00000041627.14
|
Sdk2
|
sidekick cell adhesion molecule 2 |
| chr5_-_44957016 | 2.20 |
ENSMUST00000199256.5
|
Ldb2
|
LIM domain binding 2 |
| chr6_-_141801897 | 2.19 |
ENSMUST00000165990.8
|
Slco1a4
|
solute carrier organic anion transporter family, member 1a4 |
| chr10_+_80692948 | 2.17 |
ENSMUST00000220091.2
|
Sppl2b
|
signal peptide peptidase like 2B |
| chr7_-_118304930 | 2.16 |
ENSMUST00000207323.2
ENSMUST00000038791.15 |
Gde1
|
glycerophosphodiester phosphodiesterase 1 |
| chr5_-_124327776 | 2.15 |
ENSMUST00000159677.8
|
Pitpnm2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chr2_+_35581905 | 2.14 |
ENSMUST00000065001.12
|
Dab2ip
|
disabled 2 interacting protein |
| chr11_+_24028111 | 2.14 |
ENSMUST00000109516.8
|
Bcl11a
|
B cell CLL/lymphoma 11A (zinc finger protein) |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.6 | 16.9 | GO:0021852 | pyramidal neuron migration(GO:0021852) |
| 5.6 | 16.8 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 4.8 | 14.3 | GO:0098942 | cytoskeletal matrix organization at active zone(GO:0048789) neurexin clustering involved in presynaptic membrane assembly(GO:0097115) retrograde trans-synaptic signaling by trans-synaptic protein complex(GO:0098942) |
| 3.5 | 38.0 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 2.7 | 21.8 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 2.7 | 10.8 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 2.6 | 13.2 | GO:0070295 | renal water absorption(GO:0070295) |
| 2.4 | 7.3 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 2.4 | 7.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 1.7 | 6.9 | GO:1904799 | regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) |
| 1.7 | 3.4 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 1.6 | 16.4 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 1.6 | 6.4 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 1.3 | 3.9 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 1.2 | 3.7 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleoside transport(GO:0032242) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 1.2 | 4.8 | GO:0060168 | regulation of adenosine receptor signaling pathway(GO:0060167) positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 1.0 | 3.1 | GO:0061033 | bronchiole development(GO:0060435) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 1.0 | 8.2 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 1.0 | 6.1 | GO:0034465 | response to carbon monoxide(GO:0034465) eye blink reflex(GO:0060082) |
| 1.0 | 2.0 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) |
| 1.0 | 7.6 | GO:1905247 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.9 | 2.8 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.9 | 6.5 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.9 | 3.7 | GO:0003322 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 0.9 | 6.3 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.8 | 10.8 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.8 | 5.7 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.8 | 5.7 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.8 | 5.7 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.8 | 7.1 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.8 | 2.3 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.7 | 2.9 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.7 | 4.7 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.7 | 15.1 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.6 | 7.4 | GO:0014824 | artery smooth muscle contraction(GO:0014824) |
| 0.6 | 7.3 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.6 | 1.8 | GO:0021629 | olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 0.6 | 2.9 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.6 | 5.9 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.6 | 9.0 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.5 | 6.4 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.5 | 4.2 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.5 | 4.7 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.5 | 12.9 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.5 | 3.1 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.5 | 6.7 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.5 | 6.2 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.5 | 1.4 | GO:2000299 | negative regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000299) |
| 0.4 | 1.8 | GO:0060912 | central nervous system myelin formation(GO:0032289) cardiac cell fate specification(GO:0060912) |
| 0.4 | 2.1 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.4 | 1.2 | GO:0021660 | rhombomere formation(GO:0021594) rhombomere 3 formation(GO:0021660) rhombomere 5 morphogenesis(GO:0021664) rhombomere 5 formation(GO:0021666) |
| 0.4 | 2.5 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.4 | 1.6 | GO:0036343 | psychomotor behavior(GO:0036343) motor behavior(GO:0061744) |
| 0.4 | 1.6 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.4 | 4.7 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.4 | 9.9 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.4 | 1.5 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.4 | 1.1 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.4 | 1.1 | GO:1903406 | regulation of sodium:potassium-exchanging ATPase activity(GO:1903406) |
| 0.4 | 1.4 | GO:0045186 | zonula adherens assembly(GO:0045186) |
| 0.4 | 2.5 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.4 | 1.8 | GO:1900222 | negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.3 | 4.5 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.3 | 6.4 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.3 | 6.6 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.3 | 4.4 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.3 | 0.9 | GO:2001178 | mediator complex assembly(GO:0036034) regulation of mediator complex assembly(GO:2001176) positive regulation of mediator complex assembly(GO:2001178) |
| 0.3 | 2.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.3 | 1.5 | GO:0048687 | regulation of sprouting of injured axon(GO:0048686) positive regulation of sprouting of injured axon(GO:0048687) regulation of axon extension involved in regeneration(GO:0048690) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.3 | 8.3 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.3 | 13.0 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.3 | 11.0 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.3 | 1.7 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.3 | 1.4 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.3 | 7.3 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.3 | 3.5 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.3 | 2.4 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.3 | 3.8 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.3 | 9.8 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.2 | 6.2 | GO:0007614 | short-term memory(GO:0007614) |
| 0.2 | 8.4 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.2 | 2.5 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.2 | 4.1 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.2 | 2.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.2 | 13.0 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.2 | 1.5 | GO:0048069 | eye pigmentation(GO:0048069) melanosome assembly(GO:1903232) |
| 0.2 | 4.5 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.2 | 7.5 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 5.9 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.2 | 4.7 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.2 | 0.6 | GO:1904635 | glomerular visceral epithelial cell apoptotic process(GO:1903210) regulation of glomerular visceral epithelial cell apoptotic process(GO:1904633) positive regulation of glomerular visceral epithelial cell apoptotic process(GO:1904635) positive regulation of progesterone biosynthetic process(GO:2000184) |
| 0.2 | 1.0 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.2 | 1.8 | GO:0021814 | cell motility involved in cerebral cortex radial glia guided migration(GO:0021814) modulation of microtubule cytoskeleton involved in cerebral cortex radial glia guided migration(GO:0021815) |
| 0.2 | 2.6 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.2 | 1.3 | GO:0038095 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Fc-epsilon receptor signaling pathway(GO:0038095) Kit signaling pathway(GO:0038109) |
| 0.2 | 1.0 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.2 | 1.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.2 | 3.2 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.2 | 0.8 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.2 | 11.0 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.2 | 0.5 | GO:1903367 | positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.2 | 2.9 | GO:0010155 | regulation of proton transport(GO:0010155) |
| 0.1 | 5.8 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.1 | 24.4 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.1 | 4.1 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 3.2 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.1 | 1.4 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.1 | 1.4 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.1 | 1.1 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.1 | 3.1 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 13.4 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 3.8 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 0.3 | GO:0072277 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.1 | 1.3 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.1 | 2.1 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 14.3 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.1 | 6.9 | GO:1903078 | positive regulation of protein localization to plasma membrane(GO:1903078) |
| 0.1 | 5.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 2.6 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.1 | 0.3 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.1 | 1.3 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 4.5 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 1.7 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 2.3 | GO:0033033 | negative regulation of myeloid cell apoptotic process(GO:0033033) |
| 0.1 | 6.2 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 1.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 0.5 | GO:0090306 | spindle assembly involved in meiosis(GO:0090306) |
| 0.1 | 5.9 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.1 | 9.2 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.1 | 1.9 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 4.6 | GO:0002718 | regulation of cytokine production involved in immune response(GO:0002718) |
| 0.1 | 0.6 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.1 | 4.8 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.1 | 8.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.6 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 1.0 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.1 | 0.7 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.1 | 0.3 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.1 | 0.5 | GO:0071313 | cellular response to caffeine(GO:0071313) cellular response to ATP(GO:0071318) cellular response to purine-containing compound(GO:0071415) |
| 0.1 | 0.2 | GO:1903923 | protein processing in phagocytic vesicle(GO:1900756) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 0.1 | 0.5 | GO:0021554 | optic nerve development(GO:0021554) |
| 0.1 | 0.3 | GO:0060023 | soft palate development(GO:0060023) |
| 0.1 | 0.2 | GO:0038016 | insulin receptor internalization(GO:0038016) |
| 0.0 | 2.2 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.8 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.2 | GO:0000105 | histidine biosynthetic process(GO:0000105) 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.0 | 4.0 | GO:1903955 | positive regulation of establishment of protein localization to mitochondrion(GO:1903749) positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.7 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 7.8 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 7.8 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 1.8 | GO:0021766 | hippocampus development(GO:0021766) |
| 0.0 | 1.1 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.0 | 2.8 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.6 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 4.6 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 | 0.7 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.4 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.6 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0021747 | cochlear nucleus development(GO:0021747) |
| 0.0 | 2.5 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 1.6 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.5 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) |
| 0.0 | 0.7 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 3.7 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 0.9 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 3.5 | GO:0006839 | mitochondrial transport(GO:0006839) |
| 0.0 | 3.4 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.4 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.0 | 1.6 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.5 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 2.2 | GO:0045665 | negative regulation of neuron differentiation(GO:0045665) |
| 0.0 | 3.1 | GO:0007264 | small GTPase mediated signal transduction(GO:0007264) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 11.9 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 1.3 | 3.9 | GO:0098855 | HCN channel complex(GO:0098855) |
| 1.3 | 11.5 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 1.1 | 6.9 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.7 | 19.2 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.7 | 5.5 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.6 | 14.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.6 | 2.8 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.5 | 4.3 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.5 | 2.1 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.5 | 3.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.5 | 3.8 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.4 | 1.3 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.4 | 8.9 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.4 | 16.8 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.4 | 6.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.4 | 5.4 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.4 | 1.8 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.3 | 4.8 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.3 | 1.6 | GO:0098890 | extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.3 | 22.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.3 | 7.8 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.3 | 2.2 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.3 | 4.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.3 | 102.5 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.2 | 4.8 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.2 | 5.7 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 1.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.2 | 0.6 | GO:1990843 | subsarcolemmal mitochondrion(GO:1990843) interfibrillar mitochondrion(GO:1990844) |
| 0.2 | 18.1 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.2 | 11.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.2 | 2.0 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.2 | 17.9 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.2 | 44.7 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.1 | 2.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 4.6 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 6.1 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 15.4 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 8.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 0.5 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.1 | 21.2 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.1 | 0.2 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 6.6 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 2.0 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 2.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 16.4 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 0.9 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.1 | 1.5 | GO:0031082 | BLOC complex(GO:0031082) |
| 0.1 | 4.7 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 11.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.6 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 9.4 | GO:0031252 | cell leading edge(GO:0031252) |
| 0.0 | 0.5 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 3.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.8 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 7.3 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 15.5 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.8 | GO:0090568 | nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 2.4 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 39.0 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 2.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 3.8 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.1 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.6 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.4 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.9 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.3 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.2 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 22.0 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 1.8 | 7.1 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 1.5 | 16.9 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 1.3 | 5.4 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 1.3 | 13.2 | GO:0015288 | porin activity(GO:0015288) |
| 1.3 | 6.5 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 1.3 | 11.5 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 1.2 | 9.8 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 1.2 | 4.7 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 1.0 | 2.9 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.9 | 9.3 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.8 | 5.7 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.8 | 6.1 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.8 | 3.0 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.8 | 14.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.7 | 2.9 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.7 | 12.3 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.7 | 7.2 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.7 | 24.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.7 | 6.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.7 | 45.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.7 | 4.7 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.7 | 2.0 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.6 | 3.9 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.6 | 9.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.6 | 5.7 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.6 | 4.2 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.5 | 1.6 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.5 | 1.4 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.5 | 10.0 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.4 | 1.3 | GO:0052692 | raffinose alpha-galactosidase activity(GO:0052692) |
| 0.4 | 6.7 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.4 | 7.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.4 | 2.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.4 | 10.8 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.4 | 5.7 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.4 | 3.7 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.4 | 4.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.4 | 18.6 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.3 | 1.4 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.3 | 1.7 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.3 | 14.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.3 | 3.5 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.3 | 1.9 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.3 | 6.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.3 | 1.8 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.2 | 8.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.2 | 4.9 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.2 | 6.1 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.2 | 1.4 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.2 | 1.8 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.2 | 5.3 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.2 | 1.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.2 | 14.8 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.2 | 2.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.2 | 2.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.2 | 8.0 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.2 | 2.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.2 | 2.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.2 | 4.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.2 | 1.5 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.2 | 1.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.2 | 7.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 4.1 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.2 | 5.5 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.2 | 9.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.2 | 0.8 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 2.9 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.1 | 1.6 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 0.7 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.1 | 2.8 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 6.9 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 0.5 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.1 | 2.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.3 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 13.2 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 10.7 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 2.5 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 1.8 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 1.1 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 4.5 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 8.4 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 5.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 7.4 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.1 | 1.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 2.4 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 3.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 4.1 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 1.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.5 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.1 | 0.1 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 1.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.5 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.1 | 1.0 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 5.1 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 11.3 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 3.1 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 5.7 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 1.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.6 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.3 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.2 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 1.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 3.3 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 0.3 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 11.8 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 14.0 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 8.3 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 1.0 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 2.6 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 1.4 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 2.6 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.4 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 3.5 | GO:0005261 | cation channel activity(GO:0005261) |
| 0.0 | 0.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 6.2 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.8 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 7.5 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 3.3 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.4 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 2.1 | GO:0051015 | actin filament binding(GO:0051015) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 21.8 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.6 | 29.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.2 | 17.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.2 | 11.2 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.2 | 18.1 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.2 | 6.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 1.5 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 4.5 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 9.6 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 6.5 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 8.0 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 11.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 3.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 3.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 6.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 2.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 4.2 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.1 | 1.4 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 7.4 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 2.4 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 2.1 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.1 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.6 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.7 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.1 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 23.6 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.8 | 19.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.7 | 17.2 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.5 | 7.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.3 | 4.2 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.3 | 8.6 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.3 | 20.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.3 | 5.7 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.3 | 9.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.3 | 4.9 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.2 | 23.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.2 | 5.7 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.2 | 5.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.2 | 2.9 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.2 | 9.0 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.2 | 6.9 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.2 | 7.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 11.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 3.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.2 | 7.0 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 21.2 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 5.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.5 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.1 | 1.5 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 1.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 1.9 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.5 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 2.8 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 1.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 2.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 2.1 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 1.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.9 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 3.9 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 1.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.6 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 1.3 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.5 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 1.1 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 0.3 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 2.4 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 1.3 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |