Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
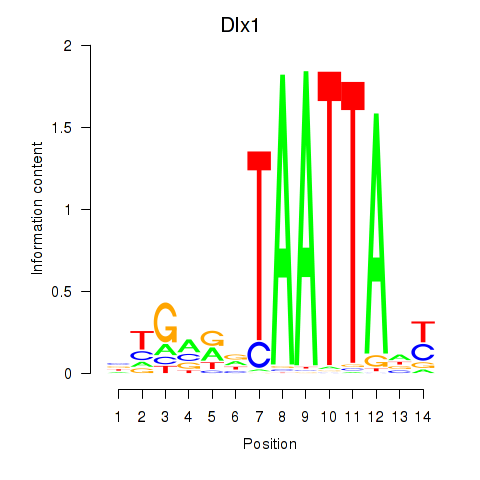
Results for Dlx1
Z-value: 1.95
Transcription factors associated with Dlx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Dlx1
|
ENSMUSG00000041911.4 | Dlx1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Dlx1 | mm39_v1_chr2_+_71359430_71359454 | -0.33 | 4.7e-03 | Click! |
Activity profile of Dlx1 motif
Sorted Z-values of Dlx1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Dlx1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_18904240 | 18.88 |
ENSMUST00000103746.3
|
Iglv1
|
immunoglobulin lambda variable 1 |
| chr10_+_115653152 | 18.29 |
ENSMUST00000080630.11
ENSMUST00000179196.3 ENSMUST00000035563.15 |
Tspan8
|
tetraspanin 8 |
| chr3_-_144680801 | 18.14 |
ENSMUST00000029923.10
ENSMUST00000238960.2 |
Clca4a
|
chloride channel accessory 4A |
| chr3_-_72875187 | 16.01 |
ENSMUST00000167334.8
|
Sis
|
sucrase isomaltase (alpha-glucosidase) |
| chr6_-_69204417 | 15.72 |
ENSMUST00000103346.3
|
Igkv4-72
|
immunoglobulin kappa chain variable 4-72 |
| chr6_+_70700207 | 15.05 |
ENSMUST00000103407.3
ENSMUST00000199487.2 |
Igkj3
|
immunoglobulin kappa joining 3 |
| chr6_+_78347636 | 14.56 |
ENSMUST00000204873.3
|
Reg3b
|
regenerating islet-derived 3 beta |
| chr6_+_78347844 | 14.22 |
ENSMUST00000096904.6
ENSMUST00000203266.2 |
Reg3b
|
regenerating islet-derived 3 beta |
| chr17_+_41121979 | 13.96 |
ENSMUST00000024721.8
ENSMUST00000233740.2 |
Rhag
|
Rhesus blood group-associated A glycoprotein |
| chr6_-_69415741 | 13.93 |
ENSMUST00000103354.3
|
Igkv4-59
|
immunoglobulin kappa variable 4-59 |
| chr6_-_69282389 | 13.81 |
ENSMUST00000103350.3
|
Igkv4-68
|
immunoglobulin kappa variable 4-68 |
| chr6_-_69678271 | 12.76 |
ENSMUST00000103363.2
|
Igkv4-50
|
immunoglobulin kappa variable 4-50 |
| chr12_-_114487525 | 12.51 |
ENSMUST00000103495.3
|
Ighv10-3
|
immunoglobulin heavy variable V10-3 |
| chr12_-_115425105 | 12.37 |
ENSMUST00000103532.3
|
Ighv1-62-3
|
immunoglobulin heavy variable 1-62-3 |
| chr12_-_114878652 | 12.23 |
ENSMUST00000103515.2
|
Ighv1-39
|
immunoglobulin heavy variable 1-39 |
| chr6_+_68247469 | 11.78 |
ENSMUST00000103321.3
|
Igkv1-110
|
immunoglobulin kappa variable 1-110 |
| chr6_-_69521891 | 11.68 |
ENSMUST00000103356.4
|
Igkv4-57-1
|
immunoglobulin kappa variable 4-57-1 |
| chr6_-_69245427 | 10.94 |
ENSMUST00000103348.3
|
Igkv4-70
|
immunoglobulin kappa chain variable 4-70 |
| chr6_+_70699822 | 10.53 |
ENSMUST00000198234.2
ENSMUST00000103406.2 |
Igkj2
|
immunoglobulin kappa joining 2 |
| chr3_-_106126794 | 9.95 |
ENSMUST00000082219.6
|
Chil4
|
chitinase-like 4 |
| chr11_-_106205320 | 9.85 |
ENSMUST00000167143.2
|
Cd79b
|
CD79B antigen |
| chr3_-_75177378 | 9.49 |
ENSMUST00000039047.5
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr6_+_41928559 | 9.32 |
ENSMUST00000031898.5
|
Sval1
|
seminal vesicle antigen-like 1 |
| chr6_-_69584812 | 9.24 |
ENSMUST00000103359.3
|
Igkv4-55
|
immunoglobulin kappa variable 4-55 |
| chr8_+_21555054 | 9.18 |
ENSMUST00000078121.4
|
Defa35
|
defensin, alpha, 35 |
| chr6_-_129077867 | 9.14 |
ENSMUST00000032258.8
|
Clec2e
|
C-type lectin domain family 2, member e |
| chr6_-_69394425 | 8.84 |
ENSMUST00000199160.2
|
Igkv4-61
|
immunoglobulin kappa chain variable 4-61 |
| chr17_+_24072493 | 8.84 |
ENSMUST00000061725.8
|
Prss32
|
protease, serine 32 |
| chr17_-_31348576 | 8.80 |
ENSMUST00000024827.5
|
Tff3
|
trefoil factor 3, intestinal |
| chr6_-_69626340 | 8.65 |
ENSMUST00000198328.2
|
Igkv4-53
|
immunoglobulin kappa variable 4-53 |
| chr16_+_45044678 | 8.47 |
ENSMUST00000102802.10
ENSMUST00000063654.6 |
Btla
|
B and T lymphocyte associated |
| chr6_-_69800923 | 8.46 |
ENSMUST00000103368.3
|
Igkv5-43
|
immunoglobulin kappa chain variable 5-43 |
| chr6_+_70675416 | 8.25 |
ENSMUST00000103403.3
|
Igkv3-2
|
immunoglobulin kappa variable 3-2 |
| chr6_-_69037208 | 8.14 |
ENSMUST00000103343.4
|
Igkv4-78
|
immunoglobulin kappa variable 4-78 |
| chr6_+_70648743 | 8.12 |
ENSMUST00000103401.3
|
Igkv3-4
|
immunoglobulin kappa variable 3-4 |
| chr3_+_92195873 | 8.09 |
ENSMUST00000090872.7
|
Sprr2a3
|
small proline-rich protein 2A3 |
| chr6_-_68713748 | 8.06 |
ENSMUST00000183936.2
ENSMUST00000196863.2 |
Igkv19-93
|
immunoglobulin kappa chain variable 19-93 |
| chr6_-_69792108 | 7.99 |
ENSMUST00000103367.3
|
Igkv12-44
|
immunoglobulin kappa variable 12-44 |
| chr14_-_122153185 | 7.89 |
ENSMUST00000055475.9
|
Gpr18
|
G protein-coupled receptor 18 |
| chr14_+_80237691 | 7.82 |
ENSMUST00000228749.2
ENSMUST00000088735.4 |
Olfm4
|
olfactomedin 4 |
| chr16_-_19079594 | 7.75 |
ENSMUST00000103752.3
ENSMUST00000197518.2 |
Iglv2
|
immunoglobulin lambda variable 2 |
| chr8_-_22193658 | 7.72 |
ENSMUST00000071886.7
|
Defa39
|
defensin, alpha, 39 |
| chr7_-_30523191 | 7.52 |
ENSMUST00000053156.10
|
Ffar2
|
free fatty acid receptor 2 |
| chr3_-_144638284 | 7.49 |
ENSMUST00000098549.4
|
Clca4b
|
chloride channel accessory 4B |
| chr3_-_14873406 | 7.26 |
ENSMUST00000181860.8
ENSMUST00000144327.3 |
Car1
|
carbonic anhydrase 1 |
| chr11_+_115802828 | 7.14 |
ENSMUST00000132961.2
|
Smim6
|
small integral membrane protein 6 |
| chr3_-_113325938 | 7.10 |
ENSMUST00000132353.2
|
Amy2a1
|
amylase 2a1 |
| chr10_-_75946790 | 7.09 |
ENSMUST00000120757.2
|
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr2_+_174292471 | 6.98 |
ENSMUST00000016399.6
|
Tubb1
|
tubulin, beta 1 class VI |
| chr1_+_40478926 | 6.97 |
ENSMUST00000173514.8
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chr11_+_58311921 | 6.90 |
ENSMUST00000013797.3
|
1810065E05Rik
|
RIKEN cDNA 1810065E05 gene |
| chr2_+_84818538 | 6.87 |
ENSMUST00000028466.12
|
Prg3
|
proteoglycan 3 |
| chr5_-_137856280 | 6.84 |
ENSMUST00000110978.7
ENSMUST00000199387.2 ENSMUST00000196195.2 |
Pilrb1
|
paired immunoglobin-like type 2 receptor beta 1 |
| chr13_+_52750883 | 6.84 |
ENSMUST00000055087.7
|
Syk
|
spleen tyrosine kinase |
| chr6_-_68746087 | 6.79 |
ENSMUST00000103333.4
|
Igkv4-91
|
immunoglobulin kappa chain variable 4-91 |
| chr8_+_21691577 | 6.78 |
ENSMUST00000110754.2
|
Defa41
|
defensin, alpha, 41 |
| chr10_-_43934774 | 6.78 |
ENSMUST00000239010.2
|
Crybg1
|
crystallin beta-gamma domain containing 1 |
| chr7_-_119058489 | 6.75 |
ENSMUST00000207887.3
ENSMUST00000239424.2 ENSMUST00000033255.8 |
Gp2
|
glycoprotein 2 (zymogen granule membrane) |
| chrX_+_149330371 | 6.75 |
ENSMUST00000066337.13
ENSMUST00000112715.2 |
Alas2
|
aminolevulinic acid synthase 2, erythroid |
| chr6_-_68994064 | 6.74 |
ENSMUST00000103341.4
|
Igkv4-80
|
immunoglobulin kappa variable 4-80 |
| chr6_-_69162381 | 6.72 |
ENSMUST00000103344.3
|
Igkv4-74
|
immunoglobulin kappa variable 4-74 |
| chr9_-_21223551 | 6.54 |
ENSMUST00000003397.9
ENSMUST00000213250.2 |
Ap1m2
|
adaptor protein complex AP-1, mu 2 subunit |
| chr12_-_75678092 | 6.51 |
ENSMUST00000238938.2
|
Rplp2-ps1
|
ribosomal protein, large P2, pseudogene 1 |
| chr6_-_70313491 | 6.34 |
ENSMUST00000103388.4
|
Igkv6-20
|
immunoglobulin kappa variable 6-20 |
| chr17_+_34482183 | 6.32 |
ENSMUST00000040828.7
ENSMUST00000237342.2 ENSMUST00000237866.2 |
H2-Ab1
|
histocompatibility 2, class II antigen A, beta 1 |
| chr6_-_69553484 | 6.26 |
ENSMUST00000103357.4
|
Igkv4-57
|
immunoglobulin kappa variable 4-57 |
| chr1_+_40478787 | 6.23 |
ENSMUST00000097772.10
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chr11_-_69786324 | 6.23 |
ENSMUST00000001631.7
|
Acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr13_-_95661726 | 6.08 |
ENSMUST00000022185.10
|
F2rl1
|
coagulation factor II (thrombin) receptor-like 1 |
| chr6_+_68495964 | 5.90 |
ENSMUST00000199510.5
ENSMUST00000103325.3 |
Igkv14-100
|
immunoglobulin kappa chain variable 14-100 |
| chr12_-_113733922 | 5.87 |
ENSMUST00000180013.3
|
Ighv2-9-1
|
immunoglobulin heavy variable 2-9-1 |
| chr3_+_138019040 | 5.76 |
ENSMUST00000013455.13
ENSMUST00000106247.2 |
Adh6a
|
alcohol dehydrogenase 6A (class V) |
| chr8_-_4829519 | 5.59 |
ENSMUST00000022945.9
|
Shcbp1
|
Shc SH2-domain binding protein 1 |
| chr15_-_58261093 | 5.54 |
ENSMUST00000227274.3
|
Anxa13
|
annexin A13 |
| chrX_+_159551171 | 5.46 |
ENSMUST00000112368.3
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr3_-_15902583 | 5.45 |
ENSMUST00000108354.8
ENSMUST00000108349.2 ENSMUST00000108352.9 ENSMUST00000108350.8 ENSMUST00000050623.11 |
Sirpb1c
|
signal-regulatory protein beta 1C |
| chr7_-_140597465 | 5.37 |
ENSMUST00000211330.2
|
Ifitm6
|
interferon induced transmembrane protein 6 |
| chr4_+_133795740 | 5.37 |
ENSMUST00000121391.8
|
Crybg2
|
crystallin beta-gamma domain containing 2 |
| chr6_+_70640233 | 5.30 |
ENSMUST00000103400.3
|
Igkv3-5
|
immunoglobulin kappa chain variable 3-5 |
| chr2_-_164198427 | 5.27 |
ENSMUST00000109367.10
|
Slpi
|
secretory leukocyte peptidase inhibitor |
| chr12_-_113928438 | 5.24 |
ENSMUST00000103478.4
|
Ighv3-1
|
immunoglobulin heavy variable 3-1 |
| chr3_+_106393348 | 5.24 |
ENSMUST00000183271.2
|
Dennd2d
|
DENN/MADD domain containing 2D |
| chr6_-_70318437 | 5.12 |
ENSMUST00000196599.2
|
Igkv8-19
|
immunoglobulin kappa variable 8-19 |
| chr12_-_115157739 | 5.05 |
ENSMUST00000103525.3
|
Ighv1-54
|
immunoglobulin heavy variable V1-54 |
| chr5_-_44139099 | 5.05 |
ENSMUST00000061299.9
|
Fgfbp1
|
fibroblast growth factor binding protein 1 |
| chr3_-_15491482 | 5.05 |
ENSMUST00000099201.9
ENSMUST00000194144.3 ENSMUST00000192700.3 |
Sirpb1a
|
signal-regulatory protein beta 1A |
| chr11_+_43046476 | 5.03 |
ENSMUST00000238415.2
|
Atp10b
|
ATPase, class V, type 10B |
| chrX_-_133012457 | 5.02 |
ENSMUST00000159259.3
ENSMUST00000113275.10 |
Nox1
|
NADPH oxidase 1 |
| chr6_+_135339929 | 5.02 |
ENSMUST00000032330.16
|
Emp1
|
epithelial membrane protein 1 |
| chr19_-_4240984 | 4.91 |
ENSMUST00000045864.4
|
Tbc1d10c
|
TBC1 domain family, member 10c |
| chr5_-_44139121 | 4.88 |
ENSMUST00000199894.2
|
Fgfbp1
|
fibroblast growth factor binding protein 1 |
| chr6_-_70116066 | 4.84 |
ENSMUST00000103379.3
ENSMUST00000197371.2 |
Igkv6-29
|
immunoglobulin kappa chain variable 6-29 |
| chr12_-_114451189 | 4.81 |
ENSMUST00000103493.3
|
Ighv1-4
|
immunoglobulin heavy variable 1-4 |
| chr5_-_137834470 | 4.80 |
ENSMUST00000110980.2
ENSMUST00000058897.11 ENSMUST00000199028.2 |
Pilra
|
paired immunoglobin-like type 2 receptor alpha |
| chr17_+_35133435 | 4.76 |
ENSMUST00000007249.15
|
Slc44a4
|
solute carrier family 44, member 4 |
| chr9_+_50466127 | 4.72 |
ENSMUST00000213916.2
|
Il18
|
interleukin 18 |
| chr3_-_15640045 | 4.71 |
ENSMUST00000192382.6
ENSMUST00000195778.3 ENSMUST00000091319.7 |
Sirpb1b
|
signal-regulatory protein beta 1B |
| chr8_-_62576140 | 4.61 |
ENSMUST00000034052.14
ENSMUST00000034054.9 |
Anxa10
|
annexin A10 |
| chr1_-_37535202 | 4.58 |
ENSMUST00000143636.8
|
Mgat4a
|
mannoside acetylglucosaminyltransferase 4, isoenzyme A |
| chr1_-_37535170 | 4.55 |
ENSMUST00000148047.2
|
Mgat4a
|
mannoside acetylglucosaminyltransferase 4, isoenzyme A |
| chr12_-_114012399 | 4.50 |
ENSMUST00000103468.3
|
Ighv11-2
|
immunoglobulin heavy variable V11-2 |
| chr9_-_21671571 | 4.46 |
ENSMUST00000217382.2
ENSMUST00000214149.2 ENSMUST00000098942.6 ENSMUST00000216057.2 |
Spc24
|
SPC24, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
| chr2_+_36120438 | 4.45 |
ENSMUST00000062069.6
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr6_-_34965339 | 4.45 |
ENSMUST00000201355.4
|
Slc23a4
|
solute carrier family 23 member 4 |
| chr10_-_44024843 | 4.44 |
ENSMUST00000200401.2
|
Crybg1
|
crystallin beta-gamma domain containing 1 |
| chr19_-_34143437 | 4.44 |
ENSMUST00000025686.9
|
Ankrd22
|
ankyrin repeat domain 22 |
| chr6_+_48849804 | 4.43 |
ENSMUST00000204856.2
|
Aoc1
|
amine oxidase, copper-containing 1 |
| chr10_+_101994841 | 4.38 |
ENSMUST00000020039.13
|
Mgat4c
|
MGAT4 family, member C |
| chr8_+_68729219 | 4.37 |
ENSMUST00000066594.4
|
Sh2d4a
|
SH2 domain containing 4A |
| chr16_-_75706161 | 4.36 |
ENSMUST00000114239.9
|
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chrX_+_159551009 | 4.32 |
ENSMUST00000033650.14
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr9_-_123507937 | 4.32 |
ENSMUST00000040960.13
|
Slc6a20a
|
solute carrier family 6 (neurotransmitter transporter), member 20A |
| chr17_-_36343573 | 4.30 |
ENSMUST00000102678.5
|
H2-T23
|
histocompatibility 2, T region locus 23 |
| chr10_-_62363217 | 4.15 |
ENSMUST00000160987.8
|
Srgn
|
serglycin |
| chr6_-_129449739 | 4.14 |
ENSMUST00000112076.9
ENSMUST00000184581.3 |
Clec7a
|
C-type lectin domain family 7, member a |
| chr3_-_88317601 | 4.11 |
ENSMUST00000193338.6
ENSMUST00000056370.13 |
Pmf1
|
polyamine-modulated factor 1 |
| chr1_+_34511793 | 4.11 |
ENSMUST00000188972.3
|
Ptpn18
|
protein tyrosine phosphatase, non-receptor type 18 |
| chr6_+_30541581 | 4.11 |
ENSMUST00000096066.5
|
Cpa2
|
carboxypeptidase A2, pancreatic |
| chr14_-_86986541 | 4.04 |
ENSMUST00000226254.2
|
Diaph3
|
diaphanous related formin 3 |
| chr12_-_115031622 | 4.04 |
ENSMUST00000194257.2
|
Ighv8-5
|
immunoglobulin heavy variable V8-5 |
| chr19_-_24178000 | 4.02 |
ENSMUST00000233658.3
|
Tjp2
|
tight junction protein 2 |
| chr11_+_58808830 | 4.00 |
ENSMUST00000020792.12
ENSMUST00000108818.4 |
Btnl10
|
butyrophilin-like 10 |
| chrX_+_106299484 | 4.00 |
ENSMUST00000101294.9
ENSMUST00000118820.8 ENSMUST00000120971.8 |
Gpr174
|
G protein-coupled receptor 174 |
| chr3_-_20329823 | 3.98 |
ENSMUST00000011607.6
|
Cpb1
|
carboxypeptidase B1 (tissue) |
| chr12_-_114793177 | 3.98 |
ENSMUST00000103511.2
ENSMUST00000195735.2 |
Ighv1-31
|
immunoglobulin heavy variable 1-31 |
| chr6_-_69020489 | 3.96 |
ENSMUST00000103342.4
|
Igkv4-79
|
immunoglobulin kappa variable 4-79 |
| chr2_+_31204314 | 3.94 |
ENSMUST00000113532.9
|
Hmcn2
|
hemicentin 2 |
| chr11_+_87684299 | 3.92 |
ENSMUST00000020779.11
|
Mpo
|
myeloperoxidase |
| chr3_-_144514386 | 3.92 |
ENSMUST00000197013.2
|
Clca3a2
|
chloride channel accessory 3A2 |
| chr2_+_57887896 | 3.89 |
ENSMUST00000112616.8
ENSMUST00000166729.2 |
Galnt5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr11_-_117671436 | 3.87 |
ENSMUST00000026659.10
ENSMUST00000127227.2 |
Tmc6
|
transmembrane channel-like gene family 6 |
| chr6_-_69477770 | 3.87 |
ENSMUST00000197448.2
|
Igkv4-58
|
immunoglobulin kappa variable 4-58 |
| chr14_+_79753055 | 3.86 |
ENSMUST00000110835.3
ENSMUST00000227192.2 |
Elf1
|
E74-like factor 1 |
| chr12_-_114443071 | 3.66 |
ENSMUST00000103492.2
|
Ighv10-1
|
immunoglobulin heavy variable 10-1 |
| chr6_-_115569504 | 3.61 |
ENSMUST00000112957.2
|
Mkrn2os
|
makorin, ring finger protein 2, opposite strand |
| chrX_+_139857688 | 3.61 |
ENSMUST00000239541.1
|
Atg4a
|
autophagy related 4A, cysteine peptidase |
| chr4_+_8690398 | 3.60 |
ENSMUST00000127476.8
|
Chd7
|
chromodomain helicase DNA binding protein 7 |
| chr6_+_29853745 | 3.58 |
ENSMUST00000064872.13
ENSMUST00000152581.8 ENSMUST00000176265.8 ENSMUST00000154079.8 |
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr10_-_40018243 | 3.58 |
ENSMUST00000092566.8
ENSMUST00000213488.2 |
Slc16a10
|
solute carrier family 16 (monocarboxylic acid transporters), member 10 |
| chr11_+_58808716 | 3.56 |
ENSMUST00000069941.13
|
Btnl10
|
butyrophilin-like 10 |
| chr3_-_19319123 | 3.48 |
ENSMUST00000121951.2
|
Pde7a
|
phosphodiesterase 7A |
| chr3_-_130524024 | 3.45 |
ENSMUST00000079085.11
|
Rpl34
|
ribosomal protein L34 |
| chr1_-_134883645 | 3.43 |
ENSMUST00000045665.13
ENSMUST00000086444.6 ENSMUST00000112163.2 |
Ppp1r12b
|
protein phosphatase 1, regulatory subunit 12B |
| chrX_+_139857640 | 3.39 |
ENSMUST00000112971.2
|
Atg4a
|
autophagy related 4A, cysteine peptidase |
| chr8_+_120426748 | 3.39 |
ENSMUST00000095171.5
ENSMUST00000212454.2 |
Atp2c2
|
ATPase, Ca++ transporting, type 2C, member 2 |
| chr10_+_115854118 | 3.38 |
ENSMUST00000063470.11
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr9_-_96513529 | 3.37 |
ENSMUST00000034984.8
|
Rasa2
|
RAS p21 protein activator 2 |
| chr10_+_20223516 | 3.36 |
ENSMUST00000169712.3
ENSMUST00000217608.2 |
Mtfr2
|
mitochondrial fission regulator 2 |
| chr1_+_139382485 | 3.36 |
ENSMUST00000200083.5
ENSMUST00000053364.12 |
Aspm
|
abnormal spindle microtubule assembly |
| chr9_-_107483855 | 3.36 |
ENSMUST00000073448.12
ENSMUST00000194606.2 ENSMUST00000195662.6 |
Sema3b
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B |
| chr6_-_69261303 | 3.35 |
ENSMUST00000103349.2
|
Igkv4-69
|
immunoglobulin kappa variable 4-69 |
| chr9_+_55448432 | 3.34 |
ENSMUST00000034869.11
|
Isl2
|
insulin related protein 2 (islet 2) |
| chr5_-_137834444 | 3.31 |
ENSMUST00000197586.2
|
Pilra
|
paired immunoglobin-like type 2 receptor alpha |
| chr10_-_62258195 | 3.31 |
ENSMUST00000020277.9
|
Hkdc1
|
hexokinase domain containing 1 |
| chr19_-_40576782 | 3.30 |
ENSMUST00000176939.8
|
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr6_+_70699533 | 3.29 |
ENSMUST00000103405.2
|
Igkj1
|
immunoglobulin kappa joining 1 |
| chr2_-_113678999 | 3.27 |
ENSMUST00000102545.8
ENSMUST00000110948.8 |
Arhgap11a
|
Rho GTPase activating protein 11A |
| chr5_-_65855511 | 3.27 |
ENSMUST00000201948.4
|
Pds5a
|
PDS5 cohesin associated factor A |
| chr6_-_69609162 | 3.22 |
ENSMUST00000199437.2
|
Igkv4-54
|
immunoglobulin kappa chain variable 4-54 |
| chr6_-_68784692 | 3.19 |
ENSMUST00000103334.4
|
Igkv4-90
|
immunoglobulin kappa chain variable 4-90 |
| chr9_+_120400510 | 3.19 |
ENSMUST00000165532.3
|
Rpl14
|
ribosomal protein L14 |
| chr2_-_119985078 | 3.17 |
ENSMUST00000028755.8
|
Ehd4
|
EH-domain containing 4 |
| chr5_-_108943211 | 3.17 |
ENSMUST00000004943.2
|
Tmed11
|
transmembrane p24 trafficking protein 11 |
| chr2_+_155593030 | 3.15 |
ENSMUST00000029140.12
ENSMUST00000132608.2 |
Procr
|
protein C receptor, endothelial |
| chr4_-_155012643 | 3.10 |
ENSMUST00000123514.8
|
Tnfrsf14
|
tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator) |
| chr7_+_99825886 | 3.08 |
ENSMUST00000178946.9
|
Kcne3
|
potassium voltage-gated channel, Isk-related subfamily, gene 3 |
| chr5_+_4073343 | 3.07 |
ENSMUST00000238634.2
|
Akap9
|
A kinase (PRKA) anchor protein (yotiao) 9 |
| chr1_+_130754413 | 3.06 |
ENSMUST00000027675.14
ENSMUST00000133792.8 |
Pigr
|
polymeric immunoglobulin receptor |
| chr19_-_6065872 | 3.05 |
ENSMUST00000164843.10
|
Capn1
|
calpain 1 |
| chr1_+_107456731 | 3.04 |
ENSMUST00000182198.8
|
Serpinb10
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr10_+_128173603 | 3.04 |
ENSMUST00000005826.9
|
Cs
|
citrate synthase |
| chr5_-_62923463 | 3.01 |
ENSMUST00000076623.8
ENSMUST00000159470.3 |
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr9_-_123507847 | 3.01 |
ENSMUST00000170591.2
ENSMUST00000171647.9 |
Slc6a20a
|
solute carrier family 6 (neurotransmitter transporter), member 20A |
| chr6_+_41107047 | 2.97 |
ENSMUST00000103271.2
|
Trbv13-3
|
T cell receptor beta, variable 13-3 |
| chr2_+_89642395 | 2.95 |
ENSMUST00000214508.2
|
Olfr1255
|
olfactory receptor 1255 |
| chr7_+_27151838 | 2.94 |
ENSMUST00000108357.8
|
Blvrb
|
biliverdin reductase B (flavin reductase (NADPH)) |
| chr2_-_73284262 | 2.94 |
ENSMUST00000102679.8
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr11_+_117700479 | 2.93 |
ENSMUST00000026649.14
ENSMUST00000177131.8 ENSMUST00000120928.2 ENSMUST00000175737.2 ENSMUST00000132298.2 |
Syngr2
Gm20708
|
synaptogyrin 2 predicted gene 20708 |
| chr18_-_67378512 | 2.92 |
ENSMUST00000237631.2
|
Mppe1
|
metallophosphoesterase 1 |
| chr11_-_84058292 | 2.92 |
ENSMUST00000050771.8
|
Gm11437
|
predicted gene 11437 |
| chr6_-_69377328 | 2.90 |
ENSMUST00000198345.2
|
Igkv4-62
|
immunoglobulin kappa variable 4-62 |
| chr13_+_4283729 | 2.90 |
ENSMUST00000081326.7
|
Akr1c19
|
aldo-keto reductase family 1, member C19 |
| chr2_-_144112700 | 2.87 |
ENSMUST00000110030.10
|
Snx5
|
sorting nexin 5 |
| chr10_-_129738595 | 2.86 |
ENSMUST00000071557.2
|
Olfr815
|
olfactory receptor 815 |
| chr8_-_4829473 | 2.85 |
ENSMUST00000207262.2
|
Shcbp1
|
Shc SH2-domain binding protein 1 |
| chr9_+_7502341 | 2.80 |
ENSMUST00000034488.4
|
Mmp10
|
matrix metallopeptidase 10 |
| chr12_-_114955196 | 2.78 |
ENSMUST00000194865.2
|
Ighv1-47
|
immunoglobulin heavy variable 1-47 |
| chr13_-_23735822 | 2.78 |
ENSMUST00000102971.2
|
H4c6
|
H4 clustered histone 6 |
| chr18_-_10182007 | 2.78 |
ENSMUST00000067947.7
|
Rock1
|
Rho-associated coiled-coil containing protein kinase 1 |
| chr13_+_51799268 | 2.73 |
ENSMUST00000075853.6
|
Cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr9_-_103357564 | 2.70 |
ENSMUST00000124310.5
|
Bfsp2
|
beaded filament structural protein 2, phakinin |
| chr3_-_86455575 | 2.70 |
ENSMUST00000077524.4
|
Mab21l2
|
mab-21-like 2 |
| chr14_+_32507920 | 2.70 |
ENSMUST00000039191.8
ENSMUST00000227060.2 ENSMUST00000228481.2 |
Tmem273
|
transmembrane protein 273 |
| chr6_-_129252396 | 2.67 |
ENSMUST00000032259.6
|
Cd69
|
CD69 antigen |
| chr5_-_87716882 | 2.66 |
ENSMUST00000113314.3
|
Sult1d1
|
sulfotransferase family 1D, member 1 |
| chr3_-_16060545 | 2.65 |
ENSMUST00000194367.6
|
Gm5150
|
predicted gene 5150 |
| chr14_+_54701594 | 2.60 |
ENSMUST00000022782.10
|
Lrp10
|
low-density lipoprotein receptor-related protein 10 |
| chr6_+_29859660 | 2.59 |
ENSMUST00000128927.9
|
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr10_+_101994719 | 2.59 |
ENSMUST00000138522.8
ENSMUST00000163753.8 ENSMUST00000138016.8 |
Mgat4c
|
MGAT4 family, member C |
| chr5_+_87148697 | 2.56 |
ENSMUST00000031186.9
|
Ugt2b35
|
UDP glucuronosyltransferase 2 family, polypeptide B35 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 14.0 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 3.3 | 9.8 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 2.6 | 7.9 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 2.4 | 9.8 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 2.3 | 6.8 | GO:0071226 | cellular response to molecule of fungal origin(GO:0071226) |
| 2.2 | 28.0 | GO:1903208 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 2.1 | 6.3 | GO:0002344 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 2.0 | 6.1 | GO:0070949 | positive regulation of eosinophil degranulation(GO:0043311) regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) positive regulation of renin secretion into blood stream(GO:1900135) positive regulation of eosinophil activation(GO:1902568) |
| 1.9 | 5.7 | GO:0006592 | ornithine biosynthetic process(GO:0006592) |
| 1.9 | 7.5 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 1.8 | 7.3 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 1.6 | 4.8 | GO:0008292 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 1.6 | 6.3 | GO:0033382 | protein localization to secretory granule(GO:0033366) protein localization to mast cell secretory granule(GO:0033367) protease localization to mast cell secretory granule(GO:0033368) maintenance of protein location in mast cell secretory granule(GO:0033370) T cell secretory granule organization(GO:0033371) maintenance of protease location in mast cell secretory granule(GO:0033373) protein localization to T cell secretory granule(GO:0033374) protease localization to T cell secretory granule(GO:0033375) maintenance of protein location in T cell secretory granule(GO:0033377) maintenance of protease location in T cell secretory granule(GO:0033379) granzyme B localization to T cell secretory granule(GO:0033380) maintenance of granzyme B location in T cell secretory granule(GO:0033382) |
| 1.5 | 6.1 | GO:0045575 | basophil activation(GO:0045575) |
| 1.5 | 4.4 | GO:0035874 | amiloride transport(GO:0015898) cellular response to copper ion starvation(GO:0035874) response to azide(GO:0097184) cellular response to azide(GO:0097185) |
| 1.4 | 4.3 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 1.4 | 4.2 | GO:1900239 | regulation of phenotypic switching(GO:1900239) negative regulation of vascular associated smooth muscle cell migration(GO:1904753) regulation of cardiac vascular smooth muscle cell differentiation(GO:2000722) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 1.3 | 6.6 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 1.3 | 3.9 | GO:0002149 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 1.2 | 4.7 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 1.1 | 6.8 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 1.1 | 13.7 | GO:0002826 | negative regulation of T-helper 1 type immune response(GO:0002826) |
| 1.1 | 3.3 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 1.1 | 9.9 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 1.1 | 9.9 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 1.0 | 7.0 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.9 | 244.1 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.9 | 2.7 | GO:0010767 | regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) |
| 0.9 | 3.6 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.8 | 3.1 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.7 | 6.7 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.7 | 11.6 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.7 | 2.1 | GO:2000040 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.6 | 2.6 | GO:0001878 | response to yeast(GO:0001878) |
| 0.6 | 4.4 | GO:0019371 | cyclooxygenase pathway(GO:0019371) maintenance of blood-brain barrier(GO:0035633) |
| 0.6 | 1.9 | GO:0015881 | creatine transport(GO:0015881) |
| 0.6 | 2.5 | GO:1905154 | negative regulation of membrane invagination(GO:1905154) |
| 0.6 | 1.2 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.6 | 4.0 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.6 | 83.7 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.6 | 1.7 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.6 | 2.8 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.5 | 4.9 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.5 | 7.1 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.5 | 3.0 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.5 | 1.5 | GO:0006429 | leucyl-tRNA aminoacylation(GO:0006429) |
| 0.5 | 4.8 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.4 | 1.7 | GO:0014732 | skeletal muscle atrophy(GO:0014732) |
| 0.4 | 2.4 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.4 | 6.7 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.4 | 2.7 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.4 | 1.1 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) |
| 0.4 | 2.9 | GO:0046149 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.4 | 1.1 | GO:0035739 | CD4-positive, alpha-beta T cell proliferation(GO:0035739) negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.4 | 2.5 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.4 | 1.1 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.3 | 1.0 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.3 | 8.4 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.3 | 3.0 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.3 | 0.9 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 0.3 | 1.2 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.3 | 1.5 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.3 | 2.0 | GO:0044334 | canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) |
| 0.3 | 1.9 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.3 | 1.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.3 | 1.6 | GO:0031394 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.3 | 1.8 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.3 | 0.8 | GO:0002545 | chronic inflammatory response to non-antigenic stimulus(GO:0002545) regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002880) |
| 0.2 | 1.5 | GO:2000121 | regulation of removal of superoxide radicals(GO:2000121) |
| 0.2 | 15.8 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.2 | 0.7 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) mRNA 3'-splice site recognition(GO:0000389) |
| 0.2 | 4.0 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.2 | 4.5 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.2 | 1.4 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.2 | 2.5 | GO:0002579 | positive regulation of antigen processing and presentation(GO:0002579) |
| 0.2 | 1.6 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.2 | 1.5 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.2 | 2.0 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.2 | 1.1 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.2 | 3.6 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.2 | 1.7 | GO:0002441 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.2 | 3.9 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.2 | 1.8 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.2 | 1.8 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.2 | 1.5 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.2 | 5.3 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.2 | 5.0 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.2 | 2.6 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.2 | 1.4 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.2 | 1.5 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.4 | GO:1903413 | response to bile acid(GO:1903412) cellular response to bile acid(GO:1903413) |
| 0.1 | 7.3 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 4.4 | GO:0002820 | negative regulation of adaptive immune response(GO:0002820) |
| 0.1 | 2.7 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 3.8 | GO:0097435 | fibril organization(GO:0097435) |
| 0.1 | 0.3 | GO:0010899 | regulation of phosphatidylcholine catabolic process(GO:0010899) |
| 0.1 | 2.7 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 1.2 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.1 | 3.3 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.1 | 5.1 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.1 | 8.1 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.1 | 0.9 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.1 | 7.1 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 2.8 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 1.7 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 0.7 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.1 | 1.5 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 0.5 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 | 5.1 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 1.3 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.1 | 2.0 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.1 | 3.1 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.1 | 1.4 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.1 | 5.1 | GO:0050766 | positive regulation of phagocytosis(GO:0050766) |
| 0.1 | 2.3 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 2.5 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.1 | 0.3 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.1 | 0.3 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.1 | 1.8 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.1 | 0.7 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.1 | 3.0 | GO:0001562 | response to protozoan(GO:0001562) |
| 0.1 | 1.8 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 2.7 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 1.9 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 0.6 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.1 | 3.6 | GO:0043276 | anoikis(GO:0043276) |
| 0.1 | 1.1 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.1 | 1.6 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.1 | 4.0 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.1 | 0.3 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.1 | 2.1 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.1 | 4.8 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 0.8 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.1 | 1.0 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.1 | 0.6 | GO:0009137 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.1 | 1.0 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 0.1 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.1 | 0.3 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 0.5 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 0.4 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.1 | 0.1 | GO:0061355 | Wnt protein secretion(GO:0061355) regulation of Wnt protein secretion(GO:0061356) positive regulation of Wnt protein secretion(GO:0061357) |
| 0.1 | 8.9 | GO:0006821 | chloride transport(GO:0006821) |
| 0.1 | 0.7 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.1 | 2.2 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.1 | 0.2 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) late endosomal microautophagy(GO:0061738) |
| 0.1 | 0.3 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.1 | 0.6 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 2.6 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.1 | 0.5 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 1.4 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.1 | 1.4 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 0.9 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.1 | 0.5 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.7 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 5.5 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 1.0 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.2 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.6 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.2 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.0 | 0.3 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.0 | 0.6 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 3.1 | GO:0043407 | negative regulation of MAP kinase activity(GO:0043407) |
| 0.0 | 0.6 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.8 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 3.7 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 | 0.4 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.5 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 0.8 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.6 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.4 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.9 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.5 | GO:0033750 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) |
| 0.0 | 2.7 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 2.0 | GO:0042491 | auditory receptor cell differentiation(GO:0042491) |
| 0.0 | 3.5 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 1.5 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 1.8 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 2.8 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.2 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.6 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.7 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.2 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.5 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.3 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.0 | 0.8 | GO:0090280 | positive regulation of calcium ion import(GO:0090280) |
| 0.0 | 0.4 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.0 | 1.1 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 0.9 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 2.1 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.8 | GO:0035036 | sperm-egg recognition(GO:0035036) |
| 0.0 | 0.9 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 0.5 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.5 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 1.5 | GO:0007605 | sensory perception of sound(GO:0007605) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 16.7 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 1.0 | 4.8 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 0.9 | 36.7 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.8 | 6.6 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.8 | 4.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.7 | 4.5 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.7 | 81.8 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.6 | 3.1 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.5 | 6.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.4 | 3.9 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.4 | 1.9 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.4 | 2.9 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) tubular endosome(GO:0097422) |
| 0.4 | 7.8 | GO:0042581 | specific granule(GO:0042581) |
| 0.4 | 1.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.3 | 1.3 | GO:0008623 | CHRAC(GO:0008623) |
| 0.3 | 1.0 | GO:0090537 | CERF complex(GO:0090537) |
| 0.3 | 1.5 | GO:0034687 | integrin alphaL-beta2 complex(GO:0034687) |
| 0.3 | 4.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.3 | 0.9 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.3 | 1.8 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.3 | 1.5 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.3 | 6.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.3 | 3.4 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.3 | 1.4 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.2 | 3.8 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.2 | 2.4 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.2 | 2.0 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.2 | 2.0 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.2 | 2.3 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.2 | 0.5 | GO:0016014 | dystrobrevin complex(GO:0016014) |
| 0.2 | 1.7 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.2 | 12.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.2 | 3.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 6.5 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 6.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 2.3 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.1 | 267.7 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 0.6 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 1.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 2.4 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 2.9 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 0.8 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 0.3 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.1 | 1.0 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 0.9 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 3.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 0.2 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.1 | 2.8 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 18.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 1.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.7 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 4.4 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 2.2 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 0.6 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 12.9 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 1.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.5 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.1 | 2.9 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 5.0 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 1.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 1.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.4 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.8 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 2.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.6 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 4.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 2.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.5 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.5 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.3 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 4.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.3 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.6 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 1.1 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.6 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 15.5 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.5 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.3 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 1.4 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 6.0 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.5 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 1.1 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 1.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.1 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.3 | GO:0097346 | Ino80 complex(GO:0031011) INO80-type complex(GO:0097346) |
| 0.0 | 0.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 2.8 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 4.6 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 13.2 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 4.0 | 16.0 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 2.2 | 6.7 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 1.9 | 5.7 | GO:0004349 | glutamate 5-kinase activity(GO:0004349) glutamate-5-semialdehyde dehydrogenase activity(GO:0004350) delta1-pyrroline-5-carboxylate synthetase activity(GO:0017084) amino acid kinase activity(GO:0019202) |
| 1.5 | 6.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 1.5 | 4.4 | GO:0052598 | diamine oxidase activity(GO:0052597) histamine oxidase activity(GO:0052598) methylputrescine oxidase activity(GO:0052599) propane-1,3-diamine oxidase activity(GO:0052600) |
| 1.5 | 16.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 1.4 | 9.9 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 1.2 | 4.8 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 1.1 | 6.6 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 1.0 | 2.9 | GO:0042602 | riboflavin reductase (NADPH) activity(GO:0042602) |
| 1.0 | 6.7 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.9 | 3.8 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.9 | 19.6 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.9 | 9.8 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.9 | 28.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.8 | 0.8 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.7 | 6.7 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.7 | 7.3 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.7 | 3.6 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.7 | 2.7 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.6 | 81.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.6 | 7.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.6 | 14.0 | GO:0022840 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.5 | 4.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.5 | 5.5 | GO:1901611 | phosphatidylglycerol binding(GO:1901611) |
| 0.5 | 2.0 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.5 | 1.5 | GO:0004823 | leucine-tRNA ligase activity(GO:0004823) |
| 0.5 | 2.7 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.4 | 3.0 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.4 | 3.8 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.4 | 2.5 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.4 | 1.7 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.4 | 3.3 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.4 | 1.2 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.4 | 6.3 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.4 | 32.7 | GO:0003823 | antigen binding(GO:0003823) |
| 0.4 | 1.2 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.4 | 1.5 | GO:0034597 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.4 | 1.1 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.4 | 1.9 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.3 | 1.4 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.3 | 2.9 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.3 | 1.2 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.3 | 1.5 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.3 | 1.8 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.3 | 2.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.3 | 1.7 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.3 | 3.3 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.3 | 2.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.3 | 1.8 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.3 | 1.0 | GO:0004008 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.2 | 4.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.2 | 1.7 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.2 | 2.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 1.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.2 | 1.5 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.2 | 6.9 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.2 | 1.0 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.2 | 2.9 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.2 | 0.6 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.2 | 0.8 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.2 | 1.6 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.2 | 36.3 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.2 | 1.4 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.2 | 3.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.2 | 0.5 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.2 | 1.8 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 4.0 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.2 | 0.9 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.2 | 0.6 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.2 | 4.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.6 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.1 | 4.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.6 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 3.1 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 2.8 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 3.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 1.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 1.8 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 1.4 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 8.4 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.1 | 2.3 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.7 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 2.7 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 12.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 15.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 0.3 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 5.9 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 4.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 3.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.9 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 2.0 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 5.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.8 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 2.6 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 3.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 4.8 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.6 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 2.3 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.1 | 2.3 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 3.3 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 0.4 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 0.6 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.1 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.1 | 0.2 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.1 | 0.5 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 4.4 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 2.0 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 7.1 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 11.3 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 0.4 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.3 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.4 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 1.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 2.1 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 2.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 8.3 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.6 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 3.1 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 11.4 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 1.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.5 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 1.3 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.7 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.3 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.2 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 6.6 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 2.3 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 3.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.7 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 1.2 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.0 | 1.4 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.2 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.7 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 3.3 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.2 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 1.5 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 1.3 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.0 | 1.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.6 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.4 | 6.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.3 | 2.7 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.2 | 11.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 6.5 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.2 | 6.1 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.2 | 4.7 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 8.1 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 6.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 2.8 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 11.9 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 3.9 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 2.6 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 4.0 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 2.6 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 4.3 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 18.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 5.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 2.8 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 3.4 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 3.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 2.6 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 3.2 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 4.9 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 1.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 3.7 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.9 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 5.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 2.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 3.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.1 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.0 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 1.1 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 2.9 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.1 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 7.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.6 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.1 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 5.5 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 0.2 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.2 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 2.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.3 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 16.1 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.5 | 11.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.4 | 6.5 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.3 | 7.0 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.3 | 6.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.3 | 17.8 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.3 | 10.7 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.2 | 2.0 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.2 | 2.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.2 | 2.5 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.2 | 2.5 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.2 | 5.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 7.8 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.2 | 4.0 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 2.0 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.2 | 3.8 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 8.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 2.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 1.6 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.1 | 10.1 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.1 | 3.1 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.1 | 6.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 1.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 0.3 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.1 | 10.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 1.8 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.1 | 2.8 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 10.6 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 1.7 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 2.8 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 4.4 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 4.7 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 0.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 2.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 0.7 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 11.1 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.1 | 3.1 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 1.7 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.1 | 6.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 7.0 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 1.6 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 1.8 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 2.0 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 1.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.7 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.6 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 4.5 | REACTOME SIGNALING BY ILS | Genes involved in Signaling by Interleukins |
| 0.0 | 0.9 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 1.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.0 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.9 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 1.4 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.3 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |