Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Dmc1
Z-value: 1.45
Transcription factors associated with Dmc1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Dmc1
|
ENSMUSG00000022429.12 | Dmc1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Dmc1 | mm39_v1_chr15_-_79489286_79489315 | -0.09 | 4.8e-01 | Click! |
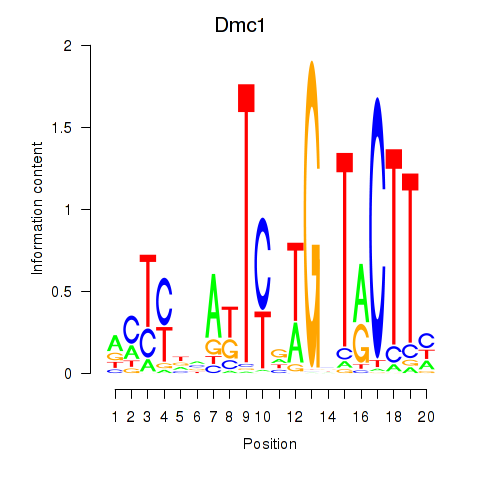
Activity profile of Dmc1 motif
Sorted Z-values of Dmc1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Dmc1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_39729431 | 15.22 |
ENSMUST00000099472.4
|
Cyp2c68
|
cytochrome P450, family 2, subfamily c, polypeptide 68 |
| chr5_-_87485023 | 14.36 |
ENSMUST00000031195.3
|
Ugt2a3
|
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr5_-_87054796 | 12.16 |
ENSMUST00000031181.16
ENSMUST00000113333.2 |
Ugt2b34
|
UDP glucuronosyltransferase 2 family, polypeptide B34 |
| chr6_+_71176811 | 11.68 |
ENSMUST00000067492.8
|
Fabp1
|
fatty acid binding protein 1, liver |
| chr7_-_12731594 | 10.83 |
ENSMUST00000133977.3
|
Slc27a5
|
solute carrier family 27 (fatty acid transporter), member 5 |
| chr7_-_12732067 | 10.60 |
ENSMUST00000032539.14
ENSMUST00000120903.8 |
Slc27a5
|
solute carrier family 27 (fatty acid transporter), member 5 |
| chr13_-_56696310 | 10.47 |
ENSMUST00000062806.6
|
Lect2
|
leukocyte cell-derived chemotaxin 2 |
| chr8_+_105460627 | 10.36 |
ENSMUST00000034346.15
ENSMUST00000164182.3 |
Ces2a
|
carboxylesterase 2A |
| chr5_+_87148697 | 9.87 |
ENSMUST00000031186.9
|
Ugt2b35
|
UDP glucuronosyltransferase 2 family, polypeptide B35 |
| chr16_+_22710134 | 9.59 |
ENSMUST00000231328.2
|
Ahsg
|
alpha-2-HS-glycoprotein |
| chr4_-_49549489 | 8.93 |
ENSMUST00000029987.10
|
Aldob
|
aldolase B, fructose-bisphosphate |
| chr19_-_46661321 | 8.39 |
ENSMUST00000026012.8
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr13_-_56696222 | 8.11 |
ENSMUST00000225183.2
|
Lect2
|
leukocyte cell-derived chemotaxin 2 |
| chr11_+_108271990 | 8.09 |
ENSMUST00000146050.2
ENSMUST00000152958.8 |
Apoh
|
apolipoprotein H |
| chr19_+_40078132 | 7.16 |
ENSMUST00000068094.13
ENSMUST00000080171.3 |
Cyp2c50
|
cytochrome P450, family 2, subfamily c, polypeptide 50 |
| chr8_+_22145796 | 6.83 |
ENSMUST00000079528.6
|
Defa17
|
defensin, alpha, 17 |
| chr10_+_80165961 | 6.75 |
ENSMUST00000186864.7
ENSMUST00000040081.7 |
Reep6
|
receptor accessory protein 6 |
| chr3_+_82933383 | 6.57 |
ENSMUST00000029630.15
ENSMUST00000166581.4 |
Fga
|
fibrinogen alpha chain |
| chr5_-_87074380 | 6.44 |
ENSMUST00000031183.3
|
Ugt2b1
|
UDP glucuronosyltransferase 2 family, polypeptide B1 |
| chr5_+_137568086 | 6.43 |
ENSMUST00000198866.5
|
Tfr2
|
transferrin receptor 2 |
| chr19_-_39801188 | 6.37 |
ENSMUST00000162507.2
ENSMUST00000160476.9 ENSMUST00000239028.2 |
Cyp2c40
|
cytochrome P450, family 2, subfamily c, polypeptide 40 |
| chr14_+_30608433 | 6.08 |
ENSMUST00000120269.11
ENSMUST00000078490.14 ENSMUST00000006703.15 |
Itih4
|
inter alpha-trypsin inhibitor, heavy chain 4 |
| chr8_+_21681630 | 6.04 |
ENSMUST00000098896.5
|
Defa31
|
defensin, alpha, 31 |
| chr12_+_8027767 | 5.84 |
ENSMUST00000037520.14
|
Apob
|
apolipoprotein B |
| chr11_+_108286114 | 5.82 |
ENSMUST00000000049.6
|
Apoh
|
apolipoprotein H |
| chr17_+_64907697 | 5.80 |
ENSMUST00000086723.10
|
Man2a1
|
mannosidase 2, alpha 1 |
| chr8_+_21777425 | 5.79 |
ENSMUST00000098893.4
|
Defa3
|
defensin, alpha, 3 |
| chr14_+_30608478 | 5.75 |
ENSMUST00000168782.4
|
Itih4
|
inter alpha-trypsin inhibitor, heavy chain 4 |
| chr4_-_107164315 | 5.72 |
ENSMUST00000126291.2
ENSMUST00000106748.2 ENSMUST00000129138.2 |
Dio1
|
deiodinase, iodothyronine, type I |
| chr19_-_40062174 | 5.50 |
ENSMUST00000048959.5
|
Cyp2c54
|
cytochrome P450, family 2, subfamily c, polypeptide 54 |
| chr16_+_22710027 | 5.49 |
ENSMUST00000231848.2
|
Ahsg
|
alpha-2-HS-glycoprotein |
| chr15_-_82678490 | 5.47 |
ENSMUST00000006094.6
|
Cyp2d26
|
cytochrome P450, family 2, subfamily d, polypeptide 26 |
| chr8_+_22224506 | 5.44 |
ENSMUST00000080533.6
|
Defa24
|
defensin, alpha, 24 |
| chr5_+_90608751 | 5.40 |
ENSMUST00000031314.10
|
Alb
|
albumin |
| chr4_-_96552349 | 5.38 |
ENSMUST00000030299.8
|
Cyp2j5
|
cytochrome P450, family 2, subfamily j, polypeptide 5 |
| chr5_+_137568113 | 5.21 |
ENSMUST00000031729.13
ENSMUST00000199054.5 |
Tfr2
|
transferrin receptor 2 |
| chr16_+_22710785 | 5.20 |
ENSMUST00000023583.7
ENSMUST00000232098.2 |
Ahsg
|
alpha-2-HS-glycoprotein |
| chr13_-_24098981 | 5.15 |
ENSMUST00000110407.4
|
Slc17a4
|
solute carrier family 17 (sodium phosphate), member 4 |
| chr6_+_41098273 | 5.11 |
ENSMUST00000103270.4
|
Trbv13-2
|
T cell receptor beta, variable 13-2 |
| chr6_-_128503666 | 5.07 |
ENSMUST00000143664.2
ENSMUST00000112132.8 |
Pzp
|
PZP, alpha-2-macroglobulin like |
| chr7_+_140500808 | 5.05 |
ENSMUST00000106045.8
ENSMUST00000183845.8 |
Nlrp6
|
NLR family, pyrin domain containing 6 |
| chr8_+_21545063 | 4.64 |
ENSMUST00000098899.4
|
Defa23
|
defensin, alpha, 23 |
| chr19_-_46661501 | 4.50 |
ENSMUST00000236174.2
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr17_-_33136021 | 4.48 |
ENSMUST00000054174.9
|
Cyp4f14
|
cytochrome P450, family 4, subfamily f, polypeptide 14 |
| chr1_+_58152295 | 4.45 |
ENSMUST00000040999.14
ENSMUST00000162011.3 |
Aox3
|
aldehyde oxidase 3 |
| chr6_+_121983720 | 4.41 |
ENSMUST00000081777.8
|
Mug2
|
murinoglobulin 2 |
| chr4_-_133822389 | 4.34 |
ENSMUST00000000696.7
|
Cd52
|
CD52 antigen |
| chr4_+_133280680 | 4.33 |
ENSMUST00000042706.3
|
Nr0b2
|
nuclear receptor subfamily 0, group B, member 2 |
| chr15_-_78280099 | 4.28 |
ENSMUST00000229878.2
ENSMUST00000165170.8 ENSMUST00000074380.14 |
Tex33
|
testis expressed 33 |
| chr19_-_40175709 | 4.24 |
ENSMUST00000051846.13
|
Cyp2c70
|
cytochrome P450, family 2, subfamily c, polypeptide 70 |
| chr10_-_89369432 | 4.20 |
ENSMUST00000105297.2
|
Nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr19_+_39980868 | 4.11 |
ENSMUST00000049178.3
|
Cyp2c37
|
cytochrome P450, family 2. subfamily c, polypeptide 37 |
| chr5_-_89583469 | 4.11 |
ENSMUST00000200534.2
|
Gc
|
vitamin D binding protein |
| chr13_-_24098951 | 4.08 |
ENSMUST00000021769.16
|
Slc17a4
|
solute carrier family 17 (sodium phosphate), member 4 |
| chr7_+_140500848 | 4.07 |
ENSMUST00000184560.2
|
Nlrp6
|
NLR family, pyrin domain containing 6 |
| chr12_+_79344056 | 3.97 |
ENSMUST00000171210.3
|
Rad51b
|
RAD51 paralog B |
| chr5_-_87572060 | 3.97 |
ENSMUST00000072818.6
|
Ugt2b38
|
UDP glucuronosyltransferase 2 family, polypeptide B38 |
| chr1_+_21310821 | 3.96 |
ENSMUST00000121676.8
ENSMUST00000124990.3 |
Gsta3
|
glutathione S-transferase, alpha 3 |
| chr19_-_44017637 | 3.94 |
ENSMUST00000026211.10
ENSMUST00000211830.2 |
Cyp2c23
|
cytochrome P450, family 2, subfamily c, polypeptide 23 |
| chr10_+_87697155 | 3.94 |
ENSMUST00000122100.3
|
Igf1
|
insulin-like growth factor 1 |
| chr3_-_144638284 | 3.80 |
ENSMUST00000098549.4
|
Clca4b
|
chloride channel accessory 4B |
| chr2_+_163348728 | 3.74 |
ENSMUST00000143911.8
|
Hnf4a
|
hepatic nuclear factor 4, alpha |
| chr7_+_19699291 | 3.71 |
ENSMUST00000094753.6
|
Ceacam20
|
carcinoembryonic antigen-related cell adhesion molecule 20 |
| chr6_-_68713748 | 3.71 |
ENSMUST00000183936.2
ENSMUST00000196863.2 |
Igkv19-93
|
immunoglobulin kappa chain variable 19-93 |
| chrX_-_90619517 | 3.66 |
ENSMUST00000078832.5
|
1700084M14Rik
|
RIKEN cDNA 1700084M14 gene |
| chr17_-_57535003 | 3.60 |
ENSMUST00000177046.2
ENSMUST00000024988.15 |
C3
|
complement component 3 |
| chr1_+_21310803 | 3.58 |
ENSMUST00000027067.15
|
Gsta3
|
glutathione S-transferase, alpha 3 |
| chr4_-_117039809 | 3.57 |
ENSMUST00000065896.9
|
Kif2c
|
kinesin family member 2C |
| chr6_-_136758716 | 3.53 |
ENSMUST00000078095.11
ENSMUST00000032338.10 |
Gucy2c
|
guanylate cyclase 2c |
| chr12_-_115258124 | 3.52 |
ENSMUST00000192591.2
|
Ighv8-8
|
immunoglobulin heavy variable 8-8 |
| chr12_-_113928438 | 3.48 |
ENSMUST00000103478.4
|
Ighv3-1
|
immunoglobulin heavy variable 3-1 |
| chr7_+_43284131 | 3.40 |
ENSMUST00000032663.10
|
Ceacam18
|
carcinoembryonic antigen-related cell adhesion molecule 18 |
| chr5_-_115257336 | 3.39 |
ENSMUST00000031524.11
|
Acads
|
acyl-Coenzyme A dehydrogenase, short chain |
| chrX_+_149371219 | 3.38 |
ENSMUST00000153221.8
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr3_-_14843512 | 3.36 |
ENSMUST00000094365.11
|
Car1
|
carbonic anhydrase 1 |
| chr14_-_75829389 | 3.33 |
ENSMUST00000165569.3
ENSMUST00000035243.5 |
Cby2
|
chibby family member 2 |
| chr5_-_87402659 | 3.28 |
ENSMUST00000075858.4
|
Ugt2b37
|
UDP glucuronosyltransferase 2 family, polypeptide B37 |
| chr3_-_73615732 | 3.26 |
ENSMUST00000029367.6
|
Bche
|
butyrylcholinesterase |
| chr6_-_69704122 | 3.22 |
ENSMUST00000103364.3
|
Igkv5-48
|
immunoglobulin kappa variable 5-48 |
| chr13_+_92491234 | 3.22 |
ENSMUST00000022218.6
|
Dhfr
|
dihydrofolate reductase |
| chr4_-_107928567 | 3.22 |
ENSMUST00000106701.2
|
Scp2
|
sterol carrier protein 2, liver |
| chr7_-_119122681 | 3.20 |
ENSMUST00000033267.4
|
Pdilt
|
protein disulfide isomerase-like, testis expressed |
| chr8_+_105048856 | 3.18 |
ENSMUST00000041973.7
|
Cmtm2b
|
CKLF-like MARVEL transmembrane domain containing 2B |
| chr1_+_165957909 | 3.14 |
ENSMUST00000166159.2
|
Gpa33
|
glycoprotein A33 (transmembrane) |
| chr8_-_22193658 | 3.11 |
ENSMUST00000071886.7
|
Defa39
|
defensin, alpha, 39 |
| chr19_-_39875192 | 3.06 |
ENSMUST00000168838.3
|
Cyp2c69
|
cytochrome P450, family 2, subfamily c, polypeptide 69 |
| chr3_+_94284739 | 3.04 |
ENSMUST00000197040.5
|
Rorc
|
RAR-related orphan receptor gamma |
| chr12_-_84923252 | 3.02 |
ENSMUST00000163189.8
ENSMUST00000110254.9 ENSMUST00000002073.13 |
Ltbp2
|
latent transforming growth factor beta binding protein 2 |
| chr1_-_73055043 | 3.00 |
ENSMUST00000027374.7
|
Tnp1
|
transition protein 1 |
| chr1_-_72323464 | 2.99 |
ENSMUST00000027381.13
|
Pecr
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr17_-_74354844 | 2.97 |
ENSMUST00000043458.9
|
Srd5a2
|
steroid 5 alpha-reductase 2 |
| chr3_-_10400710 | 2.95 |
ENSMUST00000078748.4
|
Slc10a5
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 5 |
| chr6_-_6217021 | 2.94 |
ENSMUST00000015256.15
|
Slc25a13
|
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 13 |
| chr17_-_65946817 | 2.89 |
ENSMUST00000233702.2
|
Txndc2
|
thioredoxin domain containing 2 (spermatozoa) |
| chr1_+_88093726 | 2.87 |
ENSMUST00000097659.5
|
Ugt1a5
|
UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr2_-_26096547 | 2.87 |
ENSMUST00000028302.8
|
Lhx3
|
LIM homeobox protein 3 |
| chr4_-_107780716 | 2.87 |
ENSMUST00000106719.8
ENSMUST00000106720.9 ENSMUST00000131644.2 ENSMUST00000030345.15 |
Cpt2
|
carnitine palmitoyltransferase 2 |
| chr6_+_68233361 | 2.86 |
ENSMUST00000103320.3
|
Igkv14-111
|
immunoglobulin kappa variable 14-111 |
| chr6_-_6217126 | 2.85 |
ENSMUST00000188414.4
|
Slc25a13
|
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 13 |
| chr6_-_40877303 | 2.83 |
ENSMUST00000063523.5
|
Prss58
|
protease, serine 58 |
| chr17_-_66901568 | 2.80 |
ENSMUST00000024914.4
|
Themis3
|
thymocyte selection associated family member 3 |
| chr1_-_72323407 | 2.79 |
ENSMUST00000097698.5
|
Pecr
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr10_-_75673175 | 2.77 |
ENSMUST00000220440.2
|
Gstt2
|
glutathione S-transferase, theta 2 |
| chr18_+_52779281 | 2.76 |
ENSMUST00000118724.8
ENSMUST00000091904.6 |
1700034E13Rik
|
RIKEN cDNA 1700034E13 gene |
| chr5_+_122239030 | 2.70 |
ENSMUST00000139213.8
ENSMUST00000111751.8 ENSMUST00000155612.8 |
Myl2
|
myosin, light polypeptide 2, regulatory, cardiac, slow |
| chr3_+_94284812 | 2.69 |
ENSMUST00000200009.2
|
Rorc
|
RAR-related orphan receptor gamma |
| chr5_+_77413282 | 2.68 |
ENSMUST00000080359.12
|
Rest
|
RE1-silencing transcription factor |
| chrX_-_94521712 | 2.67 |
ENSMUST00000033549.3
|
Asb12
|
ankyrin repeat and SOCS box-containing 12 |
| chr1_-_23141324 | 2.65 |
ENSMUST00000073179.6
|
Ppp1r14bl
|
protein phosphatase 1, regulatory inhibitor subunit 14B like |
| chr6_-_69626340 | 2.65 |
ENSMUST00000198328.2
|
Igkv4-53
|
immunoglobulin kappa variable 4-53 |
| chr6_+_137731526 | 2.64 |
ENSMUST00000203216.3
ENSMUST00000087675.9 ENSMUST00000203693.3 |
Dera
|
deoxyribose-phosphate aldolase (putative) |
| chr14_-_68819544 | 2.64 |
ENSMUST00000022641.9
|
Adamdec1
|
ADAM-like, decysin 1 |
| chr14_+_75373766 | 2.62 |
ENSMUST00000145303.8
|
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr15_-_75881289 | 2.57 |
ENSMUST00000170153.2
|
Fam83h
|
family with sequence similarity 83, member H |
| chr11_+_5519677 | 2.55 |
ENSMUST00000109856.8
ENSMUST00000109855.8 ENSMUST00000118112.9 |
Ankrd36
|
ankyrin repeat domain 36 |
| chr5_-_72716942 | 2.55 |
ENSMUST00000074948.5
ENSMUST00000087216.12 |
Nfxl1
|
nuclear transcription factor, X-box binding-like 1 |
| chr19_-_7780025 | 2.53 |
ENSMUST00000065634.8
|
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr10_-_43880353 | 2.53 |
ENSMUST00000020017.14
|
Crybg1
|
crystallin beta-gamma domain containing 1 |
| chr6_+_40763875 | 2.50 |
ENSMUST00000195870.3
|
Mgam2-ps
|
maltase-glucoamylase 2, pseudogene |
| chr11_+_29497950 | 2.49 |
ENSMUST00000020753.4
ENSMUST00000208530.2 |
Clhc1
|
clathrin heavy chain linker domain containing 1 |
| chr3_-_85909798 | 2.48 |
ENSMUST00000061343.4
|
Prss48
|
protease, serine 48 |
| chr10_+_127637015 | 2.46 |
ENSMUST00000071646.2
|
Rdh16
|
retinol dehydrogenase 16 |
| chr1_+_170846482 | 2.45 |
ENSMUST00000078825.5
|
Fcgr4
|
Fc receptor, IgG, low affinity IV |
| chr1_-_58735106 | 2.45 |
ENSMUST00000055313.14
ENSMUST00000188772.7 |
Flacc1
|
flagellum associated containing coiled-coil domains 1 |
| chr1_-_26726539 | 2.44 |
ENSMUST00000097801.4
|
4931408C20Rik
|
RIKEN cDNA 4931408C20 gene |
| chr8_+_71171183 | 2.44 |
ENSMUST00000179347.3
ENSMUST00000213065.2 |
Gm3336
|
predicted gene 3336 |
| chr6_+_121709891 | 2.44 |
ENSMUST00000204124.2
|
Gm7298
|
predicted gene 7298 |
| chr7_-_126651847 | 2.41 |
ENSMUST00000205424.2
|
Zg16
|
zymogen granule protein 16 |
| chr9_-_121745354 | 2.41 |
ENSMUST00000062474.5
|
Cyp8b1
|
cytochrome P450, family 8, subfamily b, polypeptide 1 |
| chr2_+_144665576 | 2.38 |
ENSMUST00000028918.4
|
Scp2d1
|
SCP2 sterol-binding domain containing 1 |
| chr4_-_92035446 | 2.38 |
ENSMUST00000107108.8
ENSMUST00000143542.2 |
Izumo3
|
IZUMO family member 3 |
| chr4_+_148686985 | 2.33 |
ENSMUST00000105701.9
ENSMUST00000052060.7 |
Masp2
|
mannan-binding lectin serine peptidase 2 |
| chr19_+_8906916 | 2.31 |
ENSMUST00000096241.6
|
Eml3
|
echinoderm microtubule associated protein like 3 |
| chrX_+_91533555 | 2.30 |
ENSMUST00000096371.3
|
Gm5941
|
predicted gene 5941 |
| chr1_-_171359228 | 2.29 |
ENSMUST00000168184.2
|
Itln1
|
intelectin 1 (galactofuranose binding) |
| chr5_-_24745436 | 2.26 |
ENSMUST00000048302.13
ENSMUST00000119657.2 |
Asb10
|
ankyrin repeat and SOCS box-containing 10 |
| chr4_-_133615075 | 2.26 |
ENSMUST00000003741.16
ENSMUST00000105894.11 |
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr17_+_80514889 | 2.25 |
ENSMUST00000134652.2
|
Ttc39d
|
tetratricopeptide repeat domain 39D |
| chr10_-_86843878 | 2.24 |
ENSMUST00000035288.17
|
Stab2
|
stabilin 2 |
| chr1_+_163942833 | 2.24 |
ENSMUST00000162746.2
|
Selp
|
selectin, platelet |
| chr6_-_69245427 | 2.23 |
ENSMUST00000103348.3
|
Igkv4-70
|
immunoglobulin kappa chain variable 4-70 |
| chr19_+_38121214 | 2.22 |
ENSMUST00000112329.3
|
Pde6c
|
phosphodiesterase 6C, cGMP specific, cone, alpha prime |
| chr4_+_117109148 | 2.19 |
ENSMUST00000062824.12
|
Tmem53
|
transmembrane protein 53 |
| chr7_+_139695707 | 2.18 |
ENSMUST00000211757.2
|
Paox
|
polyamine oxidase (exo-N4-amino) |
| chr9_-_35010357 | 2.17 |
ENSMUST00000214526.2
ENSMUST00000217149.2 |
St3gal4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr1_+_74414354 | 2.16 |
ENSMUST00000187516.7
ENSMUST00000027368.6 |
Slc11a1
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 1 |
| chr17_-_31496352 | 2.16 |
ENSMUST00000024832.9
|
Rsph1
|
radial spoke head 1 homolog (Chlamydomonas) |
| chr5_+_104447037 | 2.16 |
ENSMUST00000031246.9
|
Ibsp
|
integrin binding sialoprotein |
| chr11_+_80749184 | 2.15 |
ENSMUST00000103223.8
ENSMUST00000103222.4 |
Spaca3
|
sperm acrosome associated 3 |
| chr6_+_41092928 | 2.15 |
ENSMUST00000194399.2
|
Trbv13-1
|
T cell receptor beta, variable 13-1 |
| chr1_-_156766381 | 2.15 |
ENSMUST00000188656.7
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr15_+_75881712 | 2.15 |
ENSMUST00000187868.3
|
Iqank1
|
IQ motif and ankyrin repeat containing 1 |
| chr2_+_164131181 | 2.15 |
ENSMUST00000017147.8
ENSMUST00000109370.2 |
Svs3a
|
seminal vesicle secretory protein 3A |
| chr5_+_115003526 | 2.15 |
ENSMUST00000134389.3
|
4930519G04Rik
|
RIKEN cDNA 4930519G04 gene |
| chr2_+_148631276 | 2.14 |
ENSMUST00000168443.8
ENSMUST00000028932.4 |
Cst12
|
cystatin 12 |
| chr11_-_116089866 | 2.13 |
ENSMUST00000066587.12
|
Acox1
|
acyl-Coenzyme A oxidase 1, palmitoyl |
| chr18_-_66072119 | 2.13 |
ENSMUST00000025396.5
|
Rax
|
retina and anterior neural fold homeobox |
| chr5_-_104169785 | 2.12 |
ENSMUST00000031251.16
|
Hsd17b11
|
hydroxysteroid (17-beta) dehydrogenase 11 |
| chr4_+_63133639 | 2.11 |
ENSMUST00000036300.13
|
Col27a1
|
collagen, type XXVII, alpha 1 |
| chr5_-_92496730 | 2.11 |
ENSMUST00000038816.13
ENSMUST00000118006.3 |
Cxcl10
|
chemokine (C-X-C motif) ligand 10 |
| chr1_-_156766351 | 2.09 |
ENSMUST00000189648.2
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr17_+_47083561 | 2.08 |
ENSMUST00000071430.7
|
2310039H08Rik
|
RIKEN cDNA 2310039H08 gene |
| chr12_-_115611981 | 2.08 |
ENSMUST00000103540.3
ENSMUST00000199266.2 |
Ighv8-12
|
immunoglobulin heavy variable V8-12 |
| chr19_-_7779943 | 2.05 |
ENSMUST00000120522.8
|
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr2_+_14234198 | 2.04 |
ENSMUST00000028045.4
|
Mrc1
|
mannose receptor, C type 1 |
| chr5_-_82271183 | 2.04 |
ENSMUST00000186079.2
ENSMUST00000185607.2 |
1700031L13Rik
|
RIKEN cDNA 1700031L13 gene |
| chr3_-_146357059 | 2.03 |
ENSMUST00000149825.2
ENSMUST00000049703.6 |
4930503B20Rik
|
RIKEN cDNA 4930503B20 gene |
| chr1_+_131725119 | 2.03 |
ENSMUST00000112393.9
ENSMUST00000048660.12 |
Pm20d1
|
peptidase M20 domain containing 1 |
| chr18_-_66072168 | 2.03 |
ENSMUST00000238035.2
|
Rax
|
retina and anterior neural fold homeobox |
| chr3_-_73615535 | 2.02 |
ENSMUST00000138216.8
|
Bche
|
butyrylcholinesterase |
| chr9_-_103099262 | 2.02 |
ENSMUST00000170904.2
|
Trf
|
transferrin |
| chr4_-_49473904 | 2.01 |
ENSMUST00000135976.2
|
Acnat1
|
acyl-coenzyme A amino acid N-acyltransferase 1 |
| chr16_+_32555015 | 2.01 |
ENSMUST00000239554.1
|
Muc4
|
mucin 4 |
| chr14_-_56026266 | 2.01 |
ENSMUST00000168716.8
ENSMUST00000178399.3 ENSMUST00000022830.14 |
Ripk3
|
receptor-interacting serine-threonine kinase 3 |
| chr1_-_192946359 | 2.01 |
ENSMUST00000161737.8
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr1_+_163889551 | 2.00 |
ENSMUST00000192047.6
ENSMUST00000027871.13 |
Sell
|
selectin, lymphocyte |
| chr11_+_104077153 | 2.00 |
ENSMUST00000107000.2
ENSMUST00000059448.8 |
Sppl2c
|
signal peptide peptidase 2C |
| chr2_-_164098558 | 1.99 |
ENSMUST00000063132.6
|
Svs3b
|
seminal vesicle secretory protein 3B |
| chr1_+_136059101 | 1.99 |
ENSMUST00000075164.11
|
Kif21b
|
kinesin family member 21B |
| chr6_-_69204417 | 1.99 |
ENSMUST00000103346.3
|
Igkv4-72
|
immunoglobulin kappa chain variable 4-72 |
| chr1_-_184578057 | 1.97 |
ENSMUST00000068725.10
|
Mtarc2
|
mitochondrial amidoxime reducing component 2 |
| chr7_+_119803181 | 1.94 |
ENSMUST00000084640.3
|
Abca14
|
ATP-binding cassette, sub-family A (ABC1), member 14 |
| chr6_-_70313491 | 1.93 |
ENSMUST00000103388.4
|
Igkv6-20
|
immunoglobulin kappa variable 6-20 |
| chr5_-_38649291 | 1.92 |
ENSMUST00000129099.8
|
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr4_+_117109204 | 1.92 |
ENSMUST00000125943.8
ENSMUST00000106434.8 |
Tmem53
|
transmembrane protein 53 |
| chr16_-_16950241 | 1.91 |
ENSMUST00000023453.10
|
Sdf2l1
|
stromal cell-derived factor 2-like 1 |
| chr6_+_70192384 | 1.89 |
ENSMUST00000103383.3
|
Igkv6-25
|
immunoglobulin kappa chain variable 6-25 |
| chr13_-_26954110 | 1.89 |
ENSMUST00000055915.6
|
Hdgfl1
|
HDGF like 1 |
| chr13_+_59860096 | 1.89 |
ENSMUST00000165133.3
|
Spata31d1b
|
spermatogenesis associated 31 subfamily D, member 1B |
| chr6_+_48653047 | 1.88 |
ENSMUST00000054050.5
|
Gimap9
|
GTPase, IMAP family member 9 |
| chr4_-_63540653 | 1.88 |
ENSMUST00000102861.8
ENSMUST00000102862.4 |
Tex48
|
testis expressed 48 |
| chr7_-_48494959 | 1.87 |
ENSMUST00000208050.2
|
Csrp3
|
cysteine and glycine-rich protein 3 |
| chr9_-_106438798 | 1.87 |
ENSMUST00010126732.2
ENSMUST00010126033.2 ENSMUST00010181659.1 ENSMUST00010126065.2 ENSMUST00010126032.3 ENSMUST00000062917.16 |
ENSMUSG00001074846.1
ENSMUSG00000118396.3
|
IQ motif containing F3 IQ motif containing F3 |
| chr8_+_46984016 | 1.87 |
ENSMUST00000152423.2
|
Acsl1
|
acyl-CoA synthetase long-chain family member 1 |
| chrX_+_90436106 | 1.86 |
ENSMUST00000063726.5
|
Mageb6b1
|
MAGE family member B6B1 |
| chr7_+_106786300 | 1.85 |
ENSMUST00000142623.3
|
Nlrp14
|
NLR family, pyrin domain containing 14 |
| chr14_+_53157900 | 1.85 |
ENSMUST00000178252.3
|
Trav9d-3
|
T cell receptor alpha variable 9D-3 |
| chr19_+_6144449 | 1.85 |
ENSMUST00000235513.2
|
Gm550
|
predicted gene 550 |
| chr10_-_23226684 | 1.85 |
ENSMUST00000220299.2
|
Eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr10_+_22034460 | 1.85 |
ENSMUST00000181645.8
ENSMUST00000105522.9 |
Raet1e
H60b
|
retinoic acid early transcript 1E histocompatibility 60b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 21.4 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 2.5 | 7.5 | GO:0043387 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) |
| 2.2 | 6.6 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 2.1 | 6.4 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 1.9 | 5.8 | GO:0033306 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 1.8 | 14.7 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 1.8 | 5.4 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) |
| 1.6 | 6.6 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 1.5 | 4.5 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 1.5 | 8.9 | GO:0006116 | NADH oxidation(GO:0006116) |
| 1.4 | 15.4 | GO:0097460 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 1.4 | 45.3 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 1.2 | 3.7 | GO:0010534 | regulation of activation of JAK2 kinase activity(GO:0010534) activation of JAK2 kinase activity(GO:0042977) negative regulation of activation of JAK2 kinase activity(GO:1902569) |
| 1.2 | 17.2 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 1.2 | 5.8 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 1.1 | 3.4 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 1.1 | 2.1 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 1.0 | 4.2 | GO:0034971 | histone H3-R17 methylation(GO:0034971) bile acid signaling pathway(GO:0038183) |
| 1.0 | 19.1 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.9 | 3.6 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.9 | 4.4 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.9 | 5.3 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.8 | 0.8 | GO:0002590 | negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) |
| 0.7 | 2.2 | GO:1904156 | DN2 thymocyte differentiation(GO:1904155) DN3 thymocyte differentiation(GO:1904156) |
| 0.7 | 11.7 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.7 | 21.9 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.7 | 2.2 | GO:0046203 | spermidine catabolic process(GO:0046203) |
| 0.7 | 2.2 | GO:0070839 | divalent metal ion export(GO:0070839) |
| 0.7 | 2.9 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.7 | 2.0 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.6 | 5.7 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 0.6 | 1.9 | GO:1903920 | positive regulation of actin filament severing(GO:1903920) |
| 0.6 | 3.7 | GO:0009624 | response to nematode(GO:0009624) |
| 0.6 | 3.0 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.6 | 4.8 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.6 | 3.6 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.6 | 0.6 | GO:1904464 | regulation of matrix metallopeptidase secretion(GO:1904464) matrix metallopeptidase secretion(GO:1990773) |
| 0.6 | 3.9 | GO:1904073 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.6 | 2.8 | GO:1903677 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.6 | 1.7 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.5 | 2.1 | GO:0044704 | mating plug formation(GO:0042628) single-organism reproductive behavior(GO:0044704) post-mating behavior(GO:0045297) |
| 0.5 | 2.7 | GO:2000065 | negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.5 | 2.6 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.5 | 5.8 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.5 | 2.1 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.5 | 0.5 | GO:0090264 | immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) |
| 0.5 | 2.0 | GO:2000449 | regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000449) |
| 0.5 | 5.8 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.5 | 1.5 | GO:0042197 | dichloromethane metabolic process(GO:0018900) chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 0.5 | 9.1 | GO:0070255 | regulation of mucus secretion(GO:0070255) |
| 0.4 | 2.7 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.4 | 1.7 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.4 | 1.7 | GO:0018317 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.4 | 2.6 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.4 | 4.2 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.4 | 1.2 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.4 | 3.2 | GO:0006545 | glycine biosynthetic process(GO:0006545) tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.4 | 1.2 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.4 | 1.6 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.4 | 1.1 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) |
| 0.4 | 0.4 | GO:0018307 | enzyme active site formation(GO:0018307) |
| 0.4 | 2.1 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.3 | 1.4 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.3 | 3.4 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.3 | 1.3 | GO:1901594 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.3 | 1.3 | GO:0019230 | proprioception(GO:0019230) |
| 0.3 | 1.0 | GO:0045083 | negative regulation of interleukin-12 biosynthetic process(GO:0045083) response to diterpene(GO:1904629) cellular response to diterpene(GO:1904630) response to glucoside(GO:1904631) cellular response to glucoside(GO:1904632) |
| 0.3 | 1.0 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.3 | 1.3 | GO:0006772 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.3 | 1.2 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.3 | 2.5 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.3 | 0.9 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.3 | 2.0 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.3 | 0.9 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.3 | 2.3 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.3 | 4.1 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.3 | 1.6 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.3 | 11.3 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.3 | 4.0 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.3 | 2.1 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.3 | 1.8 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.3 | 0.8 | GO:1903898 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.2 | 0.7 | GO:0060722 | spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) |
| 0.2 | 0.7 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.2 | 0.7 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.2 | 1.7 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.2 | 6.7 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.2 | 0.9 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.2 | 2.3 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.2 | 4.1 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) |
| 0.2 | 0.5 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 0.2 | 3.4 | GO:0015747 | urate transport(GO:0015747) |
| 0.2 | 1.8 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.2 | 0.6 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.2 | 0.8 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.2 | 1.0 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.2 | 8.1 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.2 | 0.4 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.2 | 0.8 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.2 | 3.6 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.2 | 1.4 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.2 | 2.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.2 | 0.7 | GO:0035795 | negative regulation of mitochondrial membrane permeability(GO:0035795) |
| 0.2 | 5.1 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.2 | 0.8 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.2 | 5.1 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.2 | 1.5 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.2 | 0.6 | GO:0007522 | visceral muscle development(GO:0007522) |
| 0.2 | 2.9 | GO:0006662 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.2 | 0.6 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.1 | 2.4 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.1 | 0.4 | GO:0000451 | rRNA 2'-O-methylation(GO:0000451) |
| 0.1 | 1.3 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 1.7 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 3.0 | GO:0097435 | fibril organization(GO:0097435) |
| 0.1 | 0.8 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.1 | 4.3 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.1 | 0.8 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.1 | 1.3 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 0.8 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.1 | 7.7 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.1 | 31.6 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 1.8 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 1.9 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 0.7 | GO:0006544 | glycine metabolic process(GO:0006544) |
| 0.1 | 0.5 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.1 | 1.3 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.1 | 1.3 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.1 | 1.6 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.1 | 1.6 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.1 | 1.9 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 1.0 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 | 1.8 | GO:0001913 | T cell mediated cytotoxicity(GO:0001913) |
| 0.1 | 1.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 0.8 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.1 | 1.0 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.1 | 3.2 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) |
| 0.1 | 1.7 | GO:0015816 | glycine transport(GO:0015816) |
| 0.1 | 1.3 | GO:2001028 | positive regulation of endothelial cell chemotaxis(GO:2001028) |
| 0.1 | 0.4 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 3.1 | GO:0001562 | response to protozoan(GO:0001562) |
| 0.1 | 1.4 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 | 1.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 1.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.3 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.1 | 0.9 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 1.0 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 0.5 | GO:2000327 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.1 | 0.2 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.1 | 0.3 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.1 | 1.1 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 0.1 | 0.3 | GO:0090135 | actin filament branching(GO:0090135) |
| 0.1 | 2.0 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 1.3 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.1 | 0.5 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.1 | 11.3 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 5.7 | GO:0042446 | hormone biosynthetic process(GO:0042446) |
| 0.1 | 1.0 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 1.7 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.1 | 0.4 | GO:0019660 | glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.1 | 0.1 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.1 | 0.2 | GO:0032066 | nucleolus to nucleoplasm transport(GO:0032066) |
| 0.1 | 0.3 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.1 | 3.1 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 | 1.3 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 1.8 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 1.5 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 0.3 | GO:0043091 | L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.1 | 3.6 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.1 | 1.6 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.1 | 5.7 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.1 | 2.0 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.1 | 0.9 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) |
| 0.1 | 2.8 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 0.7 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.1 | 1.7 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.1 | 1.2 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.1 | 0.2 | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.1 | 9.3 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.1 | 1.3 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 0.3 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.1 | 0.6 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.3 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 1.7 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.1 | 1.2 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.1 | 0.2 | GO:0072347 | response to anesthetic(GO:0072347) |
| 0.0 | 1.6 | GO:0072678 | T cell migration(GO:0072678) |
| 0.0 | 3.7 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 9.4 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 0.2 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.4 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.0 | 1.0 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.7 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.0 | 2.6 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) |
| 0.0 | 0.9 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.3 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.1 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.3 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.6 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.6 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 2.0 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.3 | GO:0002553 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.0 | 0.5 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.4 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 1.0 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.0 | 0.8 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.7 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 2.8 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.1 | GO:0042148 | strand invasion(GO:0042148) |
| 0.0 | 1.0 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.2 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.0 | 1.6 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.4 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.8 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.0 | 1.3 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.8 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.3 | GO:0032350 | regulation of hormone metabolic process(GO:0032350) |
| 0.0 | 0.6 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 1.1 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.2 | GO:0015817 | histidine transport(GO:0015817) |
| 0.0 | 7.2 | GO:0043413 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 0.5 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) |
| 0.0 | 0.3 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 1.1 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.5 | GO:0010529 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.1 | GO:0097531 | mast cell chemotaxis(GO:0002551) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.7 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.2 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 | 0.6 | GO:0009072 | aromatic amino acid family metabolic process(GO:0009072) |
| 0.0 | 0.4 | GO:0070233 | negative regulation of T cell apoptotic process(GO:0070233) |
| 0.0 | 0.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.7 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.6 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.1 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.6 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.4 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 1.0 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.8 | GO:0043407 | negative regulation of MAP kinase activity(GO:0043407) |
| 0.0 | 0.2 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.1 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 2.5 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.4 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.0 | 0.9 | GO:0030301 | cholesterol transport(GO:0030301) |
| 0.0 | 0.3 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.5 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.3 | GO:0007635 | chemosensory behavior(GO:0007635) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.7 | GO:0044317 | rod spherule(GO:0044317) |
| 1.9 | 5.8 | GO:0034359 | mature chylomicron(GO:0034359) |
| 1.8 | 8.9 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 1.2 | 15.4 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 1.1 | 3.4 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.9 | 10.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.7 | 6.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.7 | 13.9 | GO:0042627 | chylomicron(GO:0042627) |
| 0.7 | 4.8 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.6 | 2.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.5 | 6.9 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.5 | 9.1 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.4 | 1.3 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.4 | 2.2 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.4 | 3.9 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.4 | 2.7 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.3 | 1.0 | GO:0002945 | cyclin K-CDK12 complex(GO:0002944) cyclin K-CDK13 complex(GO:0002945) |
| 0.3 | 9.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.3 | 5.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.3 | 2.0 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.3 | 1.6 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.3 | 0.8 | GO:0034665 | integrin alpha1-beta1 complex(GO:0034665) |
| 0.3 | 0.8 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.3 | 0.8 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.3 | 3.9 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 1.0 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.2 | 2.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.2 | 4.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.2 | 58.4 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.2 | 1.8 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.2 | 3.0 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.2 | 1.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.2 | 2.0 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.2 | 6.6 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.2 | 1.1 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.2 | 5.4 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.2 | 1.8 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.2 | 15.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 1.2 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.2 | 3.0 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.2 | 3.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.2 | 2.0 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 2.0 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 0.6 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 1.2 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 1.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 0.5 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.1 | 1.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 1.9 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 3.6 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 2.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 2.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 1.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 9.8 | GO:0044439 | microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.1 | 1.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 1.2 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.8 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.1 | 5.2 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 1.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 2.0 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 4.2 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 0.5 | GO:0044299 | C-fiber(GO:0044299) |
| 0.1 | 0.8 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.1 | 1.6 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 1.0 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 0.8 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.1 | 0.5 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 1.4 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 3.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.7 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 0.5 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.3 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 1.6 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 0.8 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 2.5 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 2.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 1.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 1.7 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 1.0 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.3 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 0.0 | 0.4 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 1.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 2.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.4 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.5 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.6 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.3 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.6 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.6 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 2.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.3 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 1.0 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 1.0 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.6 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 1.5 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 1.4 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.6 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.8 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.4 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.3 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 2.0 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.7 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 1.6 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 29.5 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 1.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 24.0 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.7 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.6 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.0 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 20.3 | GO:0030294 | receptor signaling protein tyrosine kinase inhibitor activity(GO:0030294) |
| 3.2 | 12.7 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 2.3 | 11.6 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 2.3 | 13.9 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 1.9 | 5.8 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 1.8 | 5.3 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 1.7 | 31.9 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 1.5 | 4.5 | GO:0052871 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) |
| 1.4 | 5.8 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 1.4 | 5.7 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 1.4 | 4.2 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 1.3 | 53.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 1.2 | 3.7 | GO:0070540 | stearic acid binding(GO:0070540) |
| 1.2 | 5.8 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 1.0 | 4.1 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 1.0 | 32.7 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 1.0 | 5.7 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.9 | 9.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.9 | 18.7 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.9 | 4.4 | GO:0016623 | aldehyde oxidase activity(GO:0004031) xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) molybdenum ion binding(GO:0030151) |
| 0.8 | 3.2 | GO:1904121 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.7 | 3.0 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.7 | 2.0 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.6 | 10.3 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.6 | 2.2 | GO:0042806 | fucose binding(GO:0042806) |
| 0.5 | 10.8 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) aromatase activity(GO:0070330) |
| 0.5 | 4.8 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.5 | 5.8 | GO:0035473 | lipase binding(GO:0035473) |
| 0.5 | 13.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.5 | 2.5 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.5 | 1.5 | GO:0019120 | hydrolase activity, acting on acid halide bonds(GO:0016824) hydrolase activity, acting on acid halide bonds, in C-halide compounds(GO:0019120) alkylhalidase activity(GO:0047651) |
| 0.5 | 2.9 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.5 | 5.6 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.4 | 3.5 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.4 | 8.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.4 | 2.2 | GO:0046592 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.4 | 4.3 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.4 | 3.0 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.4 | 2.9 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.4 | 1.2 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.4 | 2.0 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.4 | 5.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.4 | 3.4 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.4 | 1.9 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.4 | 2.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.3 | 1.3 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.3 | 1.0 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.3 | 2.0 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.3 | 2.9 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.3 | 1.3 | GO:0047874 | dolichyldiphosphatase activity(GO:0047874) |
| 0.3 | 2.2 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.3 | 0.9 | GO:0019863 | IgE binding(GO:0019863) |
| 0.3 | 0.9 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.3 | 2.1 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.3 | 2.0 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.3 | 0.8 | GO:0052593 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.3 | 4.2 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.3 | 0.8 | GO:0045142 | triplex DNA binding(GO:0045142) |
| 0.3 | 0.8 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.3 | 1.9 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.3 | 3.2 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.3 | 1.3 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.3 | 2.8 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.2 | 3.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.2 | 4.5 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.2 | 0.7 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.2 | 4.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.2 | 0.6 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.2 | 0.8 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.2 | 1.4 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.2 | 2.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.2 | 2.7 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.2 | 3.3 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.2 | 2.3 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.2 | 10.3 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.2 | 2.0 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.2 | 0.7 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.2 | 1.6 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.2 | 5.6 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.2 | 1.2 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.2 | 0.5 | GO:0080023 | 3R-hydroxyacyl-CoA dehydratase activity(GO:0080023) |
| 0.2 | 1.2 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.2 | 1.3 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.2 | 1.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.2 | 1.4 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 0.7 | GO:0004904 | interferon receptor activity(GO:0004904) type I interferon receptor activity(GO:0004905) type I interferon binding(GO:0019962) |
| 0.1 | 0.4 | GO:0070039 | rRNA (guanosine-2'-O-)-methyltransferase activity(GO:0070039) |
| 0.1 | 0.6 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.1 | 1.0 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 0.8 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.1 | 4.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.4 | GO:0050253 | sterol esterase activity(GO:0004771) retinyl-palmitate esterase activity(GO:0050253) |
| 0.1 | 0.8 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.1 | 2.8 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 0.8 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 3.7 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.1 | 2.2 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.1 | 2.1 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.1 | 1.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.1 | 1.7 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 1.7 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 0.3 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.1 | 0.7 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.1 | 1.8 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 2.5 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 1.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 1.7 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 0.3 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.1 | 2.0 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.5 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 3.9 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 1.8 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.1 | 2.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 1.3 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 0.3 | GO:0030622 | U4atac snRNA binding(GO:0030622) |
| 0.1 | 0.6 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 1.2 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.1 | 4.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.7 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 2.0 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.3 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 1.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 1.0 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.1 | 0.5 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.1 | 0.3 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.1 | 2.0 | GO:0051861 | glycolipid binding(GO:0051861) |
| 0.1 | 2.0 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 17.2 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 0.3 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.1 | 2.0 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 4.1 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 1.3 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 0.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.3 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.1 | 0.9 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 0.2 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.1 | 0.4 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.1 | 1.0 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.1 | 0.6 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 0.2 | GO:0086077 | gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.1 | 0.4 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 1.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 1.8 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.1 | 0.2 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 11.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 0.0 | 0.9 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 1.4 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.3 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 1.0 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 1.2 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.2 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 1.0 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 0.0 | 1.4 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 1.0 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.4 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.9 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 1.7 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 1.0 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.5 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 4.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 2.3 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 1.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 5.2 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 2.2 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.4 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.3 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.3 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 1.0 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 1.7 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.2 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.2 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 4.1 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.4 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.0 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 1.6 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 1.4 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.8 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 2.2 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.9 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.2 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 1.7 | GO:0008201 | heparin binding(GO:0008201) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 7.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.3 | 17.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.2 | 17.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 6.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 4.5 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 9.1 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 2.0 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 3.1 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.1 | 14.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 0.7 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 3.2 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 3.9 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 4.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 5.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 2.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 16.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 1.6 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 3.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 1.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 2.1 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 2.0 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.3 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 2.0 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 1.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.8 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 1.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.4 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 2.1 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.7 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 5.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.9 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 1.4 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 2.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.9 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.7 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.0 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 26.8 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 1.1 | 15.9 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.6 | 1.7 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.6 | 8.0 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.4 | 6.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.4 | 5.9 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.3 | 7.1 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.3 | 5.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.3 | 5.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.3 | 2.9 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 6.1 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.2 | 12.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.2 | 4.0 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.2 | 5.8 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 3.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.2 | 2.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 7.7 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.2 | 4.4 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.2 | 7.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 2.0 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.1 | 2.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 2.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 4.1 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.1 | 2.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 3.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 1.3 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 6.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 14.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 2.0 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 5.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 0.6 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 1.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 0.6 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 1.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 2.3 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 1.2 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 2.7 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 1.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.8 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 1.1 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.3 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 1.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 2.0 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 2.2 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.3 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.7 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 1.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 2.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.8 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.3 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 3.0 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.0 | 0.8 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.9 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 1.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.4 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.5 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.6 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 1.1 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.2 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.7 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |