Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
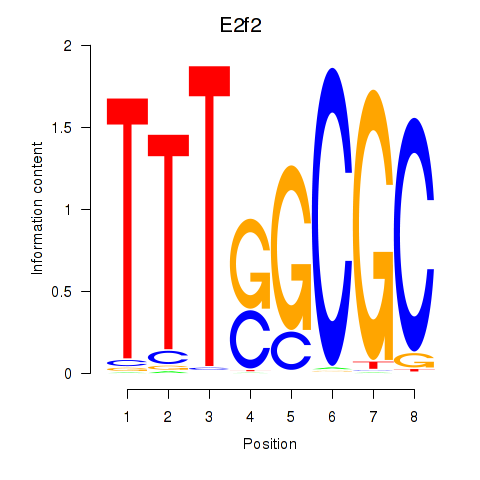
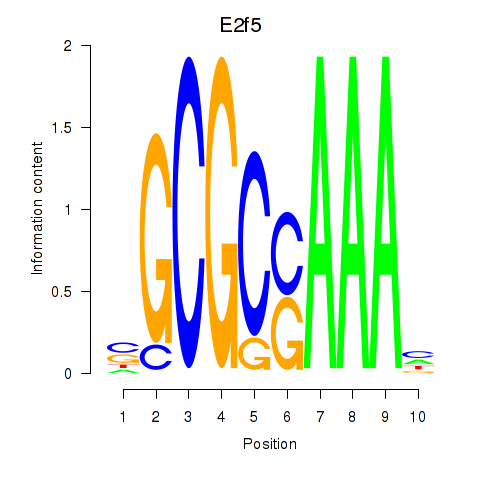
Results for E2f2_E2f5
Z-value: 3.86
Transcription factors associated with E2f2_E2f5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2f2
|
ENSMUSG00000018983.10 | E2f2 |
|
E2f5
|
ENSMUSG00000027552.15 | E2f5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2f2 | mm39_v1_chr4_+_135899678_135899738 | 0.92 | 3.9e-30 | Click! |
| E2f5 | mm39_v1_chr3_+_14643669_14643756 | -0.01 | 9.1e-01 | Click! |
Activity profile of E2f2_E2f5 motif
Sorted Z-values of E2f2_E2f5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of E2f2_E2f5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_135899678 | 64.78 |
ENSMUST00000061721.6
|
E2f2
|
E2F transcription factor 2 |
| chr12_+_24758240 | 60.09 |
ENSMUST00000020980.12
|
Rrm2
|
ribonucleotide reductase M2 |
| chr13_-_55477535 | 54.35 |
ENSMUST00000021941.8
|
Mxd3
|
Max dimerization protein 3 |
| chr10_-_69188716 | 52.36 |
ENSMUST00000119827.8
ENSMUST00000020099.13 |
Cdk1
|
cyclin-dependent kinase 1 |
| chr17_+_56611313 | 51.17 |
ENSMUST00000113035.8
ENSMUST00000113039.9 ENSMUST00000142387.2 |
Uhrf1
|
ubiquitin-like, containing PHD and RING finger domains, 1 |
| chr2_+_72306503 | 48.11 |
ENSMUST00000102691.11
ENSMUST00000157019.2 |
Cdca7
|
cell division cycle associated 7 |
| chr2_-_157046386 | 42.91 |
ENSMUST00000029170.8
|
Rbl1
|
RB transcriptional corepressor like 1 |
| chr8_+_75836187 | 42.49 |
ENSMUST00000164309.3
ENSMUST00000212426.2 ENSMUST00000212811.2 |
Mcm5
|
minichromosome maintenance complex component 5 |
| chr12_+_24758968 | 38.30 |
ENSMUST00000154588.2
|
Rrm2
|
ribonucleotide reductase M2 |
| chr9_+_122780111 | 38.09 |
ENSMUST00000040717.7
ENSMUST00000214652.2 ENSMUST00000217401.2 |
Kif15
|
kinesin family member 15 |
| chr10_-_5755412 | 34.62 |
ENSMUST00000019907.8
|
Fbxo5
|
F-box protein 5 |
| chr12_+_24758724 | 34.46 |
ENSMUST00000153058.8
|
Rrm2
|
ribonucleotide reductase M2 |
| chr5_-_138170077 | 34.00 |
ENSMUST00000155902.8
ENSMUST00000148879.8 |
Mcm7
|
minichromosome maintenance complex component 7 |
| chrX_-_50294652 | 32.27 |
ENSMUST00000114875.8
|
Mbnl3
|
muscleblind like splicing factor 3 |
| chr4_+_126450728 | 31.12 |
ENSMUST00000048391.15
|
Clspn
|
claspin |
| chr10_-_21036792 | 30.71 |
ENSMUST00000188495.8
|
Myb
|
myeloblastosis oncogene |
| chr2_-_113678999 | 29.46 |
ENSMUST00000102545.8
ENSMUST00000110948.8 |
Arhgap11a
|
Rho GTPase activating protein 11A |
| chr15_-_57998443 | 29.45 |
ENSMUST00000038194.5
|
Atad2
|
ATPase family, AAA domain containing 2 |
| chr1_-_128287347 | 28.57 |
ENSMUST00000190495.2
ENSMUST00000027601.11 |
Mcm6
|
minichromosome maintenance complex component 6 |
| chr10_-_21036824 | 28.21 |
ENSMUST00000020158.9
|
Myb
|
myeloblastosis oncogene |
| chr5_+_110434172 | 27.49 |
ENSMUST00000007296.12
ENSMUST00000112482.2 |
Pole
|
polymerase (DNA directed), epsilon |
| chr6_-_88875646 | 26.78 |
ENSMUST00000058011.8
|
Mcm2
|
minichromosome maintenance complex component 2 |
| chr1_-_20890437 | 26.73 |
ENSMUST00000053266.11
|
Mcm3
|
minichromosome maintenance complex component 3 |
| chr4_+_134591847 | 26.15 |
ENSMUST00000030627.8
|
Rhd
|
Rh blood group, D antigen |
| chr2_-_113678945 | 25.93 |
ENSMUST00000110949.9
|
Arhgap11a
|
Rho GTPase activating protein 11A |
| chr6_-_47571901 | 25.35 |
ENSMUST00000081721.13
ENSMUST00000114618.8 ENSMUST00000114616.8 |
Ezh2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr2_+_118943274 | 24.28 |
ENSMUST00000140939.8
ENSMUST00000028795.10 |
Rad51
|
RAD51 recombinase |
| chr7_-_48530777 | 23.15 |
ENSMUST00000058745.15
|
E2f8
|
E2F transcription factor 8 |
| chr11_+_79980210 | 22.68 |
ENSMUST00000017694.7
ENSMUST00000108239.7 |
Atad5
|
ATPase family, AAA domain containing 5 |
| chr7_-_48531344 | 22.68 |
ENSMUST00000119223.2
|
E2f8
|
E2F transcription factor 8 |
| chr17_+_56610396 | 22.35 |
ENSMUST00000113038.8
|
Uhrf1
|
ubiquitin-like, containing PHD and RING finger domains, 1 |
| chr5_-_138170644 | 22.15 |
ENSMUST00000000505.16
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr12_-_69274936 | 22.12 |
ENSMUST00000221411.2
ENSMUST00000021359.7 |
Pole2
|
polymerase (DNA directed), epsilon 2 (p59 subunit) |
| chr16_-_15455141 | 21.61 |
ENSMUST00000023353.4
|
Mcm4
|
minichromosome maintenance complex component 4 |
| chr2_+_150751475 | 21.13 |
ENSMUST00000028948.5
|
Gins1
|
GINS complex subunit 1 (Psf1 homolog) |
| chr2_-_113678616 | 20.74 |
ENSMUST00000110947.2
|
Arhgap11a
|
Rho GTPase activating protein 11A |
| chr13_-_24945844 | 20.51 |
ENSMUST00000006898.10
ENSMUST00000110382.9 |
Gmnn
|
geminin |
| chrX_-_50294867 | 20.47 |
ENSMUST00000114876.9
|
Mbnl3
|
muscleblind like splicing factor 3 |
| chr9_+_65797519 | 20.08 |
ENSMUST00000045802.7
|
Pclaf
|
PCNA clamp associated factor |
| chr4_+_114857348 | 19.16 |
ENSMUST00000030490.13
|
Stil
|
Scl/Tal1 interrupting locus |
| chr12_-_11315755 | 19.09 |
ENSMUST00000166117.4
ENSMUST00000219600.2 ENSMUST00000218487.2 |
Gen1
|
GEN1, Holliday junction 5' flap endonuclease |
| chr4_+_134195631 | 18.81 |
ENSMUST00000030636.11
ENSMUST00000127279.8 ENSMUST00000105867.8 |
Stmn1
|
stathmin 1 |
| chr14_-_20438890 | 18.71 |
ENSMUST00000022345.7
|
Dnajc9
|
DnaJ heat shock protein family (Hsp40) member C9 |
| chr5_+_88912855 | 18.44 |
ENSMUST00000031311.10
|
Dck
|
deoxycytidine kinase |
| chr13_-_24945423 | 17.60 |
ENSMUST00000176890.8
ENSMUST00000175689.8 |
Gmnn
|
geminin |
| chr9_+_64188857 | 17.02 |
ENSMUST00000215031.2
ENSMUST00000213165.2 ENSMUST00000213289.2 ENSMUST00000216594.2 ENSMUST00000034964.7 |
Tipin
|
timeless interacting protein |
| chr9_+_44245981 | 16.82 |
ENSMUST00000052686.4
|
H2ax
|
H2A.X variant histone |
| chr4_-_133695264 | 16.30 |
ENSMUST00000102553.11
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr12_+_116369017 | 16.22 |
ENSMUST00000084828.5
ENSMUST00000222469.2 ENSMUST00000221114.2 ENSMUST00000221970.2 |
Ncapg2
|
non-SMC condensin II complex, subunit G2 |
| chr9_+_109704609 | 16.20 |
ENSMUST00000094324.8
|
Cdc25a
|
cell division cycle 25A |
| chr15_-_8473918 | 15.81 |
ENSMUST00000052965.8
|
Nipbl
|
NIPBL cohesin loading factor |
| chr6_+_51447613 | 15.63 |
ENSMUST00000114445.8
ENSMUST00000114446.8 ENSMUST00000141711.3 |
Cbx3
|
chromobox 3 |
| chrX_-_92675719 | 15.58 |
ENSMUST00000006856.3
|
Pola1
|
polymerase (DNA directed), alpha 1 |
| chr1_-_191307648 | 15.47 |
ENSMUST00000027933.11
|
Dtl
|
denticleless E3 ubiquitin protein ligase |
| chr9_-_20864096 | 15.29 |
ENSMUST00000004202.17
|
Dnmt1
|
DNA methyltransferase (cytosine-5) 1 |
| chr10_+_110581293 | 14.85 |
ENSMUST00000174857.8
ENSMUST00000073781.12 ENSMUST00000173471.8 ENSMUST00000173634.2 |
E2f7
|
E2F transcription factor 7 |
| chr1_-_88629843 | 14.78 |
ENSMUST00000159814.2
|
Arl4c
|
ADP-ribosylation factor-like 4C |
| chr15_-_9140403 | 14.77 |
ENSMUST00000096482.10
|
Skp2
|
S-phase kinase-associated protein 2 |
| chr2_-_154411765 | 14.66 |
ENSMUST00000103145.11
|
E2f1
|
E2F transcription factor 1 |
| chr13_-_47259652 | 14.37 |
ENSMUST00000021807.13
ENSMUST00000135278.8 |
Dek
|
DEK proto-oncogene (DNA binding) |
| chr17_+_29020064 | 14.35 |
ENSMUST00000004985.11
|
Brpf3
|
bromodomain and PHD finger containing, 3 |
| chr7_-_125995884 | 14.07 |
ENSMUST00000075671.5
|
Nfatc2ip
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 2 interacting protein |
| chr19_+_38919477 | 14.03 |
ENSMUST00000145051.2
|
Hells
|
helicase, lymphoid specific |
| chr17_+_23945310 | 13.74 |
ENSMUST00000024701.9
|
Pkmyt1
|
protein kinase, membrane associated tyrosine/threonine 1 |
| chr3_+_97920819 | 13.59 |
ENSMUST00000079812.8
|
Notch2
|
notch 2 |
| chr17_+_56610321 | 13.45 |
ENSMUST00000001258.15
|
Uhrf1
|
ubiquitin-like, containing PHD and RING finger domains, 1 |
| chrX_-_7940959 | 13.28 |
ENSMUST00000115636.4
ENSMUST00000115638.10 |
Suv39h1
|
suppressor of variegation 3-9 1 |
| chr8_-_54091980 | 13.21 |
ENSMUST00000047768.11
|
Neil3
|
nei like 3 (E. coli) |
| chr2_-_154411640 | 13.05 |
ENSMUST00000000894.6
|
E2f1
|
E2F transcription factor 1 |
| chr8_+_13388745 | 12.79 |
ENSMUST00000209885.2
ENSMUST00000209396.2 |
Tfdp1
|
transcription factor Dp 1 |
| chr2_-_169984593 | 12.65 |
ENSMUST00000109155.8
|
Zfp217
|
zinc finger protein 217 |
| chr6_+_51447490 | 12.63 |
ENSMUST00000031862.14
|
Cbx3
|
chromobox 3 |
| chr10_-_60055082 | 12.56 |
ENSMUST00000135158.9
|
Chst3
|
carbohydrate sulfotransferase 3 |
| chr4_+_114857370 | 12.56 |
ENSMUST00000129957.8
|
Stil
|
Scl/Tal1 interrupting locus |
| chr12_+_71063431 | 12.53 |
ENSMUST00000125125.2
|
Arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr8_-_34578880 | 12.23 |
ENSMUST00000080152.5
|
Gm10131
|
predicted pseudogene 10131 |
| chr14_-_73563212 | 12.05 |
ENSMUST00000022701.7
|
Rb1
|
RB transcriptional corepressor 1 |
| chr19_+_53588808 | 11.71 |
ENSMUST00000025930.10
|
Smc3
|
structural maintenance of chromosomes 3 |
| chr11_-_6394352 | 11.56 |
ENSMUST00000093346.6
|
H2az2
|
H2A.Z histone variant 2 |
| chr3_-_95125002 | 11.51 |
ENSMUST00000107209.8
|
Gabpb2
|
GA repeat binding protein, beta 2 |
| chr3_-_95125190 | 11.26 |
ENSMUST00000136139.8
|
Gabpb2
|
GA repeat binding protein, beta 2 |
| chr1_+_86454431 | 10.93 |
ENSMUST00000045897.15
ENSMUST00000186255.7 ENSMUST00000188699.7 |
Ptma
|
prothymosin alpha |
| chr15_-_9140460 | 10.91 |
ENSMUST00000110585.10
|
Skp2
|
S-phase kinase-associated protein 2 |
| chr1_+_183766572 | 10.83 |
ENSMUST00000048655.8
|
Dusp10
|
dual specificity phosphatase 10 |
| chr5_-_136596299 | 10.83 |
ENSMUST00000004097.16
|
Cux1
|
cut-like homeobox 1 |
| chrX_+_41239872 | 10.63 |
ENSMUST00000123245.8
|
Stag2
|
stromal antigen 2 |
| chr17_+_29709723 | 10.23 |
ENSMUST00000024811.9
|
Pim1
|
proviral integration site 1 |
| chr19_+_38919353 | 10.14 |
ENSMUST00000025965.12
|
Hells
|
helicase, lymphoid specific |
| chrX_+_49930311 | 9.82 |
ENSMUST00000114887.9
|
Stk26
|
serine/threonine kinase 26 |
| chr1_+_86454511 | 9.46 |
ENSMUST00000188533.2
|
Ptma
|
prothymosin alpha |
| chr9_+_103182352 | 9.43 |
ENSMUST00000035164.10
|
Topbp1
|
topoisomerase (DNA) II binding protein 1 |
| chr17_-_71782296 | 9.32 |
ENSMUST00000127430.2
|
Smchd1
|
SMC hinge domain containing 1 |
| chr5_+_45827249 | 9.20 |
ENSMUST00000117396.3
|
Ncapg
|
non-SMC condensin I complex, subunit G |
| chr11_+_98798627 | 9.12 |
ENSMUST00000092706.13
|
Cdc6
|
cell division cycle 6 |
| chr14_+_79689230 | 9.12 |
ENSMUST00000100359.3
ENSMUST00000226192.2 |
Kbtbd6
|
kelch repeat and BTB (POZ) domain containing 6 |
| chr11_-_87295292 | 9.00 |
ENSMUST00000067692.13
|
Rad51c
|
RAD51 paralog C |
| chr18_+_56840813 | 8.94 |
ENSMUST00000025486.9
|
Lmnb1
|
lamin B1 |
| chr9_-_21202545 | 8.72 |
ENSMUST00000215619.2
|
Cdkn2d
|
cyclin dependent kinase inhibitor 2D |
| chr15_+_61857390 | 8.70 |
ENSMUST00000159327.2
ENSMUST00000167731.8 |
Myc
|
myelocytomatosis oncogene |
| chr6_-_148847854 | 8.58 |
ENSMUST00000139355.8
ENSMUST00000146457.2 ENSMUST00000054080.15 |
Sinhcaf
|
SIN3-HDAC complex associated factor |
| chr7_+_126380655 | 8.56 |
ENSMUST00000172352.8
ENSMUST00000094037.5 |
Tbx6
|
T-box 6 |
| chr15_+_61857226 | 8.54 |
ENSMUST00000161976.8
ENSMUST00000022971.8 |
Myc
|
myelocytomatosis oncogene |
| chr13_-_22017677 | 8.53 |
ENSMUST00000081342.7
|
H2ac24
|
H2A clustered histone 24 |
| chr4_+_126450762 | 8.42 |
ENSMUST00000147675.2
|
Clspn
|
claspin |
| chr2_-_28511941 | 8.38 |
ENSMUST00000028156.8
ENSMUST00000164290.8 |
Gfi1b
|
growth factor independent 1B |
| chr19_-_10181243 | 8.34 |
ENSMUST00000142241.2
ENSMUST00000116542.9 ENSMUST00000025651.6 ENSMUST00000156291.2 |
Fen1
|
flap structure specific endonuclease 1 |
| chr1_-_176641607 | 8.29 |
ENSMUST00000195717.6
ENSMUST00000192961.6 |
Cep170
|
centrosomal protein 170 |
| chr15_-_54953819 | 8.17 |
ENSMUST00000110231.2
ENSMUST00000023059.13 |
Dscc1
|
DNA replication and sister chromatid cohesion 1 |
| chr3_-_127346882 | 8.12 |
ENSMUST00000197668.2
ENSMUST00000029588.10 |
Larp7
|
La ribonucleoprotein domain family, member 7 |
| chr8_-_123404811 | 8.08 |
ENSMUST00000006525.14
ENSMUST00000064674.13 |
Cbfa2t3
|
CBFA2/RUNX1 translocation partner 3 |
| chr18_+_10617773 | 8.00 |
ENSMUST00000002551.5
ENSMUST00000234207.2 |
Snrpd1
|
small nuclear ribonucleoprotein D1 |
| chr1_+_175708341 | 7.95 |
ENSMUST00000195196.6
ENSMUST00000194306.6 ENSMUST00000193822.6 |
Exo1
|
exonuclease 1 |
| chr18_-_80756273 | 7.79 |
ENSMUST00000078049.12
ENSMUST00000236711.2 |
Nfatc1
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 1 |
| chr15_-_79489286 | 7.77 |
ENSMUST00000229408.2
ENSMUST00000023065.8 |
Dmc1
|
DNA meiotic recombinase 1 |
| chr9_-_21202353 | 7.71 |
ENSMUST00000086374.8
|
Cdkn2d
|
cyclin dependent kinase inhibitor 2D |
| chr8_-_106074585 | 7.67 |
ENSMUST00000014922.5
|
Fhod1
|
formin homology 2 domain containing 1 |
| chr4_+_32615428 | 7.64 |
ENSMUST00000178925.8
ENSMUST00000029950.10 |
Casp8ap2
|
caspase 8 associated protein 2 |
| chr13_-_22227114 | 7.61 |
ENSMUST00000091741.6
|
H2ac11
|
H2A clustered histone 11 |
| chr9_+_61280501 | 7.49 |
ENSMUST00000162583.8
ENSMUST00000161993.8 ENSMUST00000160882.8 ENSMUST00000160724.8 ENSMUST00000162973.8 ENSMUST00000159050.8 |
Tle3
|
transducin-like enhancer of split 3 |
| chr2_-_118380149 | 7.35 |
ENSMUST00000090219.13
|
Bmf
|
BCL2 modifying factor |
| chr6_+_113508636 | 7.34 |
ENSMUST00000036340.12
ENSMUST00000204827.3 |
Fancd2
|
Fanconi anemia, complementation group D2 |
| chr4_-_43010225 | 7.30 |
ENSMUST00000030165.5
|
Fancg
|
Fanconi anemia, complementation group G |
| chr6_+_21949570 | 7.28 |
ENSMUST00000031680.10
ENSMUST00000115389.8 ENSMUST00000151473.8 |
Ing3
|
inhibitor of growth family, member 3 |
| chr4_-_133694543 | 7.26 |
ENSMUST00000123234.8
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr1_-_93729562 | 7.23 |
ENSMUST00000112890.3
|
Dtymk
|
deoxythymidylate kinase |
| chr9_+_61280764 | 7.19 |
ENSMUST00000160541.8
ENSMUST00000161207.8 ENSMUST00000159630.8 |
Tle3
|
transducin-like enhancer of split 3 |
| chr12_+_11315868 | 7.18 |
ENSMUST00000020931.6
|
Smc6
|
structural maintenance of chromosomes 6 |
| chr9_-_13738304 | 7.14 |
ENSMUST00000148086.8
ENSMUST00000034398.12 |
Cep57
|
centrosomal protein 57 |
| chr4_+_140428777 | 7.13 |
ENSMUST00000138808.8
ENSMUST00000038893.6 |
Rcc2
|
regulator of chromosome condensation 2 |
| chrX_-_72502595 | 7.13 |
ENSMUST00000033737.15
ENSMUST00000077243.5 |
Haus7
|
HAUS augmin-like complex, subunit 7 |
| chr10_+_128067934 | 7.05 |
ENSMUST00000055539.11
ENSMUST00000105244.8 ENSMUST00000105243.9 |
Timeless
|
timeless circadian clock 1 |
| chr7_-_44198157 | 6.97 |
ENSMUST00000145956.2
ENSMUST00000049343.15 |
Pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr1_-_74544946 | 6.97 |
ENSMUST00000044260.11
ENSMUST00000186282.7 |
Usp37
|
ubiquitin specific peptidase 37 |
| chr17_+_50816296 | 6.95 |
ENSMUST00000043938.8
|
Plcl2
|
phospholipase C-like 2 |
| chr8_-_106434565 | 6.94 |
ENSMUST00000013299.11
|
Enkd1
|
enkurin domain containing 1 |
| chr16_-_18630365 | 6.93 |
ENSMUST00000096990.10
|
Cdc45
|
cell division cycle 45 |
| chr10_-_128361731 | 6.89 |
ENSMUST00000026427.8
|
Esyt1
|
extended synaptotagmin-like protein 1 |
| chr3_+_116388600 | 6.88 |
ENSMUST00000198386.5
ENSMUST00000198311.5 ENSMUST00000197335.2 |
Sass6
|
SAS-6 centriolar assembly protein |
| chr9_-_21202693 | 6.88 |
ENSMUST00000213407.2
|
Cdkn2d
|
cyclin dependent kinase inhibitor 2D |
| chr1_+_131838294 | 6.84 |
ENSMUST00000062264.8
|
Nucks1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr2_-_34803988 | 6.82 |
ENSMUST00000028232.7
ENSMUST00000202907.2 |
Phf19
|
PHD finger protein 19 |
| chr5_+_114268425 | 6.79 |
ENSMUST00000031587.13
|
Ung
|
uracil DNA glycosylase |
| chr13_+_21900554 | 6.67 |
ENSMUST00000070124.5
|
H2ac13
|
H2A clustered histone 13 |
| chr19_-_6014210 | 6.60 |
ENSMUST00000025752.15
ENSMUST00000165143.3 |
Pola2
|
polymerase (DNA directed), alpha 2 |
| chr11_-_6394385 | 6.50 |
ENSMUST00000109737.9
|
H2az2
|
H2A.Z histone variant 2 |
| chr3_+_127347099 | 6.48 |
ENSMUST00000043108.9
ENSMUST00000196141.5 ENSMUST00000195955.2 |
Zgrf1
|
zinc finger, GRF-type containing 1 |
| chr7_+_102090892 | 6.33 |
ENSMUST00000033283.10
|
Rrm1
|
ribonucleotide reductase M1 |
| chr8_-_121316043 | 6.28 |
ENSMUST00000034278.6
|
Gins2
|
GINS complex subunit 2 (Psf2 homolog) |
| chrX_+_41239548 | 6.18 |
ENSMUST00000069619.14
|
Stag2
|
stromal antigen 2 |
| chr6_-_8259098 | 6.15 |
ENSMUST00000012627.5
|
Rpa3
|
replication protein A3 |
| chr9_-_36637670 | 6.13 |
ENSMUST00000172702.9
ENSMUST00000172742.2 |
Chek1
|
checkpoint kinase 1 |
| chr8_+_85598734 | 6.12 |
ENSMUST00000170296.2
ENSMUST00000136026.8 |
Syce2
|
synaptonemal complex central element protein 2 |
| chr9_-_36637923 | 6.03 |
ENSMUST00000034625.12
|
Chek1
|
checkpoint kinase 1 |
| chr2_-_132095146 | 5.96 |
ENSMUST00000028817.7
|
Pcna
|
proliferating cell nuclear antigen |
| chr11_+_79883885 | 5.86 |
ENSMUST00000163272.2
ENSMUST00000017692.15 |
Suz12
|
SUZ12 polycomb repressive complex 2 subunit |
| chr2_-_91067212 | 5.84 |
ENSMUST00000111352.8
|
Ddb2
|
damage specific DNA binding protein 2 |
| chr9_+_13739003 | 5.79 |
ENSMUST00000059579.12
|
Fam76b
|
family with sequence similarity 76, member B |
| chr6_+_92068361 | 5.77 |
ENSMUST00000113460.8
|
Nr2c2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr4_-_132570421 | 5.65 |
ENSMUST00000105919.2
ENSMUST00000030702.14 |
Ppp1r8
|
protein phosphatase 1, regulatory subunit 8 |
| chr19_-_6014159 | 5.61 |
ENSMUST00000235224.2
|
Pola2
|
polymerase (DNA directed), alpha 2 |
| chr7_+_82516491 | 5.61 |
ENSMUST00000082237.7
|
Mex3b
|
mex3 RNA binding family member B |
| chr13_-_23929490 | 5.54 |
ENSMUST00000091752.5
|
H3c3
|
H3 clustered histone 3 |
| chr5_-_136596094 | 5.51 |
ENSMUST00000175975.9
ENSMUST00000176216.9 ENSMUST00000176745.8 |
Cux1
|
cut-like homeobox 1 |
| chr8_+_96360081 | 5.43 |
ENSMUST00000034094.11
ENSMUST00000212434.2 |
Gins3
|
GINS complex subunit 3 (Psf3 homolog) |
| chr7_-_67022520 | 5.38 |
ENSMUST00000156690.8
ENSMUST00000107476.8 ENSMUST00000076325.12 ENSMUST00000032776.15 ENSMUST00000133074.2 |
Mef2a
|
myocyte enhancer factor 2A |
| chr12_+_33197753 | 5.15 |
ENSMUST00000146040.9
|
Atxn7l1
|
ataxin 7-like 1 |
| chr12_+_33197776 | 5.14 |
ENSMUST00000125192.9
ENSMUST00000136644.8 |
Atxn7l1
|
ataxin 7-like 1 |
| chr13_-_21937997 | 5.08 |
ENSMUST00000074752.4
|
H2ac15
|
H2A clustered histone 15 |
| chr7_+_112806672 | 5.08 |
ENSMUST00000047321.9
ENSMUST00000210074.2 ENSMUST00000210238.2 |
Arntl
|
aryl hydrocarbon receptor nuclear translocator-like |
| chr11_+_98441923 | 5.05 |
ENSMUST00000081033.13
ENSMUST00000107511.8 ENSMUST00000107509.8 ENSMUST00000017339.12 |
Zpbp2
|
zona pellucida binding protein 2 |
| chr9_+_61280890 | 5.00 |
ENSMUST00000161689.8
|
Tle3
|
transducin-like enhancer of split 3 |
| chr14_+_54443198 | 4.83 |
ENSMUST00000103728.2
|
Traj13
|
T cell receptor alpha joining 13 |
| chr11_+_85202058 | 4.83 |
ENSMUST00000020835.16
|
Ppm1d
|
protein phosphatase 1D magnesium-dependent, delta isoform |
| chr15_-_97991114 | 4.81 |
ENSMUST00000180657.2
|
Senp1
|
SUMO1/sentrin specific peptidase 1 |
| chr5_+_137743992 | 4.78 |
ENSMUST00000100540.10
|
Tsc22d4
|
TSC22 domain family, member 4 |
| chr4_-_133695204 | 4.77 |
ENSMUST00000100472.10
ENSMUST00000136327.2 |
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr2_+_164721277 | 4.77 |
ENSMUST00000041643.5
|
Pcif1
|
phosphorylated CTD interacting factor 1 |
| chr12_-_4924341 | 4.76 |
ENSMUST00000137337.8
ENSMUST00000045921.14 |
Mfsd2b
|
major facilitator superfamily domain containing 2B |
| chr7_-_99770653 | 4.76 |
ENSMUST00000208670.2
ENSMUST00000032969.14 |
Pold3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chr16_-_4377640 | 4.72 |
ENSMUST00000005862.9
|
Tfap4
|
transcription factor AP4 |
| chr3_+_41517790 | 4.72 |
ENSMUST00000163764.8
|
Jade1
|
jade family PHD finger 1 |
| chr10_-_4337435 | 4.71 |
ENSMUST00000100077.5
|
Zbtb2
|
zinc finger and BTB domain containing 2 |
| chr12_-_75678092 | 4.66 |
ENSMUST00000238938.2
|
Rplp2-ps1
|
ribosomal protein, large P2, pseudogene 1 |
| chr6_+_51447317 | 4.62 |
ENSMUST00000094623.10
|
Cbx3
|
chromobox 3 |
| chr5_+_143846782 | 4.61 |
ENSMUST00000148011.8
ENSMUST00000110709.7 |
Pms2
|
PMS1 homolog2, mismatch repair system component |
| chr2_+_132658055 | 4.57 |
ENSMUST00000028831.15
ENSMUST00000066559.6 |
Mcm8
|
minichromosome maintenance 8 homologous recombination repair factor |
| chr18_+_66591604 | 4.52 |
ENSMUST00000025399.9
ENSMUST00000237161.2 ENSMUST00000236933.2 |
Pmaip1
|
phorbol-12-myristate-13-acetate-induced protein 1 |
| chr1_+_74545203 | 4.52 |
ENSMUST00000087215.7
|
Cnot9
|
CCR4-NOT transcription complex, subunit 9 |
| chr7_-_99770280 | 4.50 |
ENSMUST00000208184.2
|
Pold3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chr1_+_131838220 | 4.49 |
ENSMUST00000189946.7
|
Nucks1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr8_+_106363141 | 4.47 |
ENSMUST00000005841.16
|
Ctcf
|
CCCTC-binding factor |
| chr2_-_91067312 | 4.46 |
ENSMUST00000028696.5
|
Ddb2
|
damage specific DNA binding protein 2 |
| chr2_+_84867783 | 4.44 |
ENSMUST00000168266.8
ENSMUST00000130729.3 |
Ssrp1
|
structure specific recognition protein 1 |
| chr13_-_47259266 | 4.44 |
ENSMUST00000129352.3
|
Dek
|
DEK proto-oncogene (DNA binding) |
| chr1_-_38168697 | 4.39 |
ENSMUST00000027251.12
|
Rev1
|
REV1, DNA directed polymerase |
| chr2_+_53082079 | 4.36 |
ENSMUST00000028336.7
|
Arl6ip6
|
ADP-ribosylation factor-like 6 interacting protein 6 |
| chr5_+_137744228 | 4.35 |
ENSMUST00000100539.10
|
Tsc22d4
|
TSC22 domain family, member 4 |
| chr6_-_148847633 | 4.32 |
ENSMUST00000132696.8
|
Sinhcaf
|
SIN3-HDAC complex associated factor |
| chr15_-_81756076 | 4.30 |
ENSMUST00000023117.10
|
Phf5a
|
PHD finger protein 5A |
| chr15_+_58004753 | 4.27 |
ENSMUST00000067563.9
|
Wdyhv1
|
WDYHV motif containing 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 20.2 | 60.7 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 13.3 | 39.8 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 13.1 | 157.4 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 12.3 | 49.0 | GO:0006272 | leading strand elongation(GO:0006272) |
| 11.5 | 34.6 | GO:0051439 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051439) |
| 10.8 | 64.8 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 8.4 | 25.3 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 7.3 | 130.9 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 7.0 | 7.0 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 6.5 | 26.1 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 6.4 | 146.4 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 5.7 | 28.4 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 5.5 | 49.8 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 4.7 | 51.9 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 4.6 | 69.5 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 4.5 | 13.6 | GO:0035622 | intrahepatic bile duct development(GO:0035622) cholangiocyte proliferation(GO:1990705) |
| 4.3 | 17.2 | GO:0046722 | lactic acid secretion(GO:0046722) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 4.3 | 17.0 | GO:0048478 | replication fork protection(GO:0048478) |
| 4.1 | 12.2 | GO:0010767 | regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) |
| 3.7 | 18.4 | GO:0046125 | pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 3.7 | 14.7 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 3.6 | 35.5 | GO:0019985 | translesion synthesis(GO:0019985) |
| 3.3 | 108.4 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 3.1 | 18.8 | GO:0044838 | cell quiescence(GO:0044838) |
| 3.1 | 9.3 | GO:0060821 | inactivation of X chromosome by DNA methylation(GO:0060821) |
| 3.0 | 3.0 | GO:0010159 | specification of organ position(GO:0010159) |
| 2.8 | 11.3 | GO:0019046 | release from viral latency(GO:0019046) regulation of DNA strand elongation(GO:0060382) |
| 2.7 | 18.8 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 2.6 | 31.7 | GO:0033504 | floor plate development(GO:0033504) |
| 2.6 | 21.1 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 2.5 | 7.6 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 2.4 | 7.3 | GO:1904093 | regulation of autophagic cell death(GO:1904092) negative regulation of autophagic cell death(GO:1904093) |
| 2.4 | 7.1 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 2.3 | 23.0 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 2.3 | 6.8 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 2.3 | 15.8 | GO:0061010 | gall bladder development(GO:0061010) |
| 2.2 | 8.9 | GO:0031662 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) response to monosodium glutamate(GO:1904008) cellular response to monosodium glutamate(GO:1904009) |
| 2.2 | 2.2 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 2.1 | 10.7 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 2.1 | 19.1 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 2.0 | 19.9 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 1.9 | 23.3 | GO:0048102 | autophagic cell death(GO:0048102) |
| 1.9 | 1.9 | GO:0031990 | mRNA export from nucleus in response to heat stress(GO:0031990) |
| 1.8 | 29.4 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 1.7 | 10.3 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 1.7 | 11.6 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 1.6 | 8.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 1.6 | 3.2 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 1.5 | 10.8 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 1.5 | 9.3 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 1.5 | 13.3 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 1.4 | 7.1 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 1.4 | 25.7 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 1.4 | 5.6 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 1.4 | 23.8 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 1.4 | 8.3 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) DNA replication, removal of RNA primer(GO:0043137) |
| 1.4 | 9.6 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 1.3 | 5.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 1.3 | 12.6 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 1.3 | 52.7 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 1.3 | 12.5 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 1.2 | 7.4 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 1.1 | 25.1 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 1.1 | 4.5 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 1.1 | 7.8 | GO:0042148 | strand invasion(GO:0042148) |
| 1.1 | 3.3 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 1.1 | 5.4 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 1.1 | 8.6 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 1.0 | 5.1 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 1.0 | 5.9 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 1.0 | 7.7 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 1.0 | 7.6 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.9 | 10.2 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.9 | 6.8 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.8 | 9.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.8 | 16.2 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.8 | 8.1 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.8 | 7.8 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.8 | 9.2 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 0.8 | 20.4 | GO:0043486 | histone exchange(GO:0043486) |
| 0.8 | 4.5 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.7 | 4.2 | GO:1903436 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.7 | 42.3 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.7 | 38.1 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.7 | 8.2 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.7 | 15.0 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.7 | 8.7 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.6 | 2.4 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.6 | 1.8 | GO:0019389 | glucuronoside metabolic process(GO:0019389) |
| 0.5 | 6.5 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.5 | 16.2 | GO:1902751 | positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.5 | 7.5 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.5 | 2.1 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.5 | 2.5 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.5 | 9.2 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.4 | 11.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.4 | 1.2 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.4 | 1.2 | GO:0060939 | transforming growth factor beta receptor complex assembly(GO:0007181) cardiac fibroblast cell differentiation(GO:0060935) cardiac fibroblast cell development(GO:0060936) epicardium-derived cardiac fibroblast cell differentiation(GO:0060938) epicardium-derived cardiac fibroblast cell development(GO:0060939) |
| 0.4 | 2.0 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.4 | 3.0 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.4 | 4.8 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.4 | 4.8 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.4 | 4.7 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.4 | 2.5 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.4 | 1.4 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.4 | 3.2 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.4 | 1.1 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.3 | 3.1 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.3 | 1.4 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.3 | 15.9 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.3 | 5.1 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.3 | 2.8 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.3 | 0.3 | GO:0072554 | blood vessel lumenization(GO:0072554) |
| 0.3 | 0.9 | GO:1990258 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.3 | 9.8 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.3 | 3.5 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.3 | 10.8 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.3 | 5.6 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.3 | 1.4 | GO:1904349 | positive regulation of small intestine smooth muscle contraction(GO:1904349) |
| 0.3 | 5.0 | GO:0072189 | ureter development(GO:0072189) |
| 0.3 | 1.9 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.3 | 0.5 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.3 | 18.7 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.3 | 1.6 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.3 | 5.1 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.3 | 16.5 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.2 | 6.9 | GO:0007099 | centriole replication(GO:0007099) |
| 0.2 | 5.1 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.2 | 7.1 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.2 | 1.4 | GO:1903912 | negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.2 | 1.6 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.2 | 0.6 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.2 | 3.8 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.2 | 5.6 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.2 | 5.2 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.2 | 24.2 | GO:0035308 | negative regulation of protein dephosphorylation(GO:0035308) |
| 0.2 | 11.0 | GO:0042491 | auditory receptor cell differentiation(GO:0042491) |
| 0.2 | 2.1 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.2 | 2.5 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.1 | 2.7 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 1.6 | GO:1904251 | regulation of bile acid metabolic process(GO:1904251) |
| 0.1 | 0.4 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.1 | 11.5 | GO:0007569 | cell aging(GO:0007569) |
| 0.1 | 2.3 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 5.8 | GO:0038066 | p38MAPK cascade(GO:0038066) |
| 0.1 | 2.8 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 0.4 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.1 | 2.6 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.1 | 4.7 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 7.3 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.1 | 5.4 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 33.9 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.1 | 2.8 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.1 | 2.4 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.1 | 1.0 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 | 2.0 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.1 | 0.3 | GO:0048880 | proprioception(GO:0019230) sensory system development(GO:0048880) |
| 0.1 | 2.8 | GO:0048255 | mRNA stabilization(GO:0048255) |
| 0.1 | 1.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 5.8 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.1 | 0.7 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.1 | 0.5 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 1.1 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 3.7 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.1 | 2.1 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 17.8 | GO:0048511 | rhythmic process(GO:0048511) |
| 0.0 | 2.3 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.5 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) |
| 0.0 | 0.5 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.0 | 1.7 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 1.7 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.0 | 0.4 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 11.9 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 0.3 | GO:0048148 | behavioral response to cocaine(GO:0048148) |
| 0.0 | 5.0 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.7 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 23.2 | 139.2 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 13.0 | 208.6 | GO:0042555 | MCM complex(GO:0042555) |
| 9.9 | 49.7 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 9.1 | 27.4 | GO:0000811 | GINS complex(GO:0000811) |
| 8.0 | 39.8 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 4.8 | 24.0 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 4.1 | 32.9 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 4.0 | 27.8 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 3.7 | 25.8 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 3.2 | 12.8 | GO:0035061 | interchromatin granule(GO:0035061) |
| 2.9 | 23.3 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 2.3 | 9.3 | GO:0001740 | Barr body(GO:0001740) |
| 2.2 | 13.3 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 2.2 | 144.1 | GO:0005657 | replication fork(GO:0005657) |
| 2.1 | 25.4 | GO:0000796 | condensin complex(GO:0000796) |
| 2.0 | 11.7 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 1.9 | 11.4 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 1.8 | 16.5 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 1.4 | 8.2 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 1.4 | 38.0 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 1.3 | 8.9 | GO:0005638 | lamin filament(GO:0005638) |
| 1.1 | 7.5 | GO:0036396 | MIS complex(GO:0036396) |
| 1.0 | 5.1 | GO:0031251 | PAN complex(GO:0031251) |
| 1.0 | 6.9 | GO:0098536 | deuterosome(GO:0098536) |
| 0.9 | 4.6 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.9 | 3.5 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.8 | 2.5 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.8 | 24.2 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.8 | 2.5 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.8 | 5.6 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.8 | 7.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.8 | 24.5 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.7 | 50.2 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.7 | 7.1 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.7 | 1.4 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.7 | 4.7 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.6 | 6.1 | GO:0000801 | central element(GO:0000801) |
| 0.6 | 3.7 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.6 | 9.4 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.6 | 10.9 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.5 | 112.1 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.5 | 38.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.4 | 16.7 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.4 | 1.2 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.4 | 4.5 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.4 | 2.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.4 | 4.0 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.4 | 4.2 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.4 | 19.2 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.3 | 3.8 | GO:0000243 | commitment complex(GO:0000243) |
| 0.3 | 28.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.3 | 6.2 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.3 | 122.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.3 | 2.2 | GO:0000803 | sex chromosome(GO:0000803) |
| 0.3 | 5.1 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.3 | 1.6 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.2 | 3.9 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.2 | 1.4 | GO:0034709 | methylosome(GO:0034709) |
| 0.2 | 9.1 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.2 | 15.5 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.2 | 1.9 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.2 | 26.8 | GO:0005814 | centriole(GO:0005814) |
| 0.2 | 4.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.2 | 42.5 | GO:0005819 | spindle(GO:0005819) |
| 0.2 | 5.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.2 | 2.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.2 | 11.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 12.6 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.1 | 2.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 9.6 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.1 | 1.4 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 2.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 1.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 9.8 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.1 | 38.3 | GO:0000785 | chromatin(GO:0000785) |
| 0.1 | 6.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 1.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 7.6 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 41.7 | GO:0005813 | centrosome(GO:0005813) |
| 0.1 | 40.9 | GO:0005694 | chromosome(GO:0005694) |
| 0.1 | 6.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 3.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 7.2 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 0.4 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 5.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 134.7 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.1 | 0.5 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 0.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 3.3 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 2.8 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 12.1 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 178.4 | GO:0005634 | nucleus(GO:0005634) |
| 0.0 | 6.3 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.9 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.3 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 23.2 | 139.2 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 12.4 | 87.0 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 11.9 | 59.4 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 6.6 | 39.5 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 5.9 | 11.8 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 5.5 | 22.0 | GO:0036033 | mediator complex binding(GO:0036033) |
| 4.6 | 18.4 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 4.6 | 41.0 | GO:0000150 | recombinase activity(GO:0000150) |
| 4.2 | 76.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 4.1 | 16.3 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 3.8 | 34.6 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 3.2 | 19.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 3.1 | 31.2 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 3.0 | 12.2 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 2.7 | 27.5 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 2.7 | 13.3 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 2.6 | 133.7 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 2.5 | 12.6 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 2.5 | 52.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 2.4 | 7.2 | GO:0004798 | thymidylate kinase activity(GO:0004798) |
| 2.0 | 6.0 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 1.9 | 5.8 | GO:0070773 | protein-N-terminal glutamine amidohydrolase activity(GO:0070773) |
| 1.8 | 57.0 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 1.7 | 6.8 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 1.6 | 7.8 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 1.5 | 12.0 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 1.3 | 58.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 1.3 | 23.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 1.2 | 4.8 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 1.1 | 3.2 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 1.0 | 18.3 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 1.0 | 16.1 | GO:0031386 | protein tag(GO:0031386) |
| 1.0 | 7.6 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.9 | 8.2 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.9 | 2.7 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.9 | 10.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.8 | 26.1 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.7 | 9.6 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.7 | 5.7 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.6 | 8.4 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.6 | 3.9 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.6 | 19.8 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.6 | 9.8 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.6 | 4.6 | GO:0032407 | MutSalpha complex binding(GO:0032407) |
| 0.6 | 60.4 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.5 | 154.8 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.5 | 34.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.5 | 1.4 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.5 | 64.4 | GO:0042393 | histone binding(GO:0042393) |
| 0.5 | 7.8 | GO:0005522 | profilin binding(GO:0005522) |
| 0.5 | 59.2 | GO:0004386 | helicase activity(GO:0004386) |
| 0.4 | 2.2 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.4 | 5.1 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.4 | 18.5 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.4 | 1.2 | GO:0004512 | inositol-3-phosphate synthase activity(GO:0004512) intramolecular lyase activity(GO:0016872) |
| 0.4 | 14.8 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.4 | 2.8 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.3 | 2.4 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.3 | 13.6 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.3 | 38.1 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.3 | 5.0 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.3 | 0.9 | GO:0036009 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.3 | 1.5 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.3 | 15.2 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.3 | 7.3 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.3 | 6.9 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.3 | 1.6 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.3 | 2.3 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.3 | 2.5 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.2 | 6.6 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.2 | 2.9 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.2 | 3.3 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.2 | 65.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.2 | 5.1 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.2 | 7.6 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.2 | 5.4 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.2 | 1.2 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.2 | 13.6 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.2 | 8.6 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.2 | 4.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.2 | 0.7 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.2 | 3.5 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.2 | 2.0 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.2 | 1.0 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.2 | 71.1 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.2 | 5.3 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.2 | 1.4 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.1 | 2.5 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 4.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 3.0 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 0.7 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 2.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 1.4 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 2.6 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 1.2 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 1.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 5.8 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 1.8 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 43.6 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.1 | 1.7 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.1 | 1.6 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 7.1 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 3.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 2.0 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 41.5 | GO:0044212 | transcription regulatory region DNA binding(GO:0044212) |
| 0.1 | 3.4 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 4.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 7.8 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 7.7 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.1 | 2.4 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 7.3 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 18.0 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.1 | 20.7 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 1.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 13.8 | GO:0016887 | ATPase activity(GO:0016887) |
| 0.0 | 0.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 1.0 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.4 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 1.1 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 3.7 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 1.4 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 2.2 | GO:0017124 | SH3 domain binding(GO:0017124) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.9 | 79.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 5.1 | 92.5 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 4.1 | 202.9 | PID ATR PATHWAY | ATR signaling pathway |
| 2.8 | 286.4 | PID E2F PATHWAY | E2F transcription factor network |
| 1.2 | 18.8 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 1.1 | 31.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.9 | 58.9 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.9 | 25.0 | PID MYC PATHWAY | C-MYC pathway |
| 0.9 | 50.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.7 | 18.3 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.5 | 7.6 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.5 | 10.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.4 | 21.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.4 | 27.1 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.3 | 6.2 | PID ATM PATHWAY | ATM pathway |
| 0.3 | 34.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.3 | 10.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.3 | 6.0 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.3 | 13.9 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.3 | 1.1 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.3 | 5.1 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.2 | 9.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 7.8 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.2 | 13.9 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.2 | 16.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.2 | 2.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 8.1 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 7.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 8.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 7.2 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.1 | 4.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 4.8 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 10.0 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 1.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.8 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 2.2 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 2.4 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 1.2 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.8 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.5 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.1 | 241.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 14.1 | 309.7 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 7.3 | 101.7 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 4.4 | 74.5 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 4.2 | 8.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 2.4 | 12.2 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 2.4 | 12.0 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 1.9 | 37.0 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 1.7 | 12.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 1.5 | 24.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 1.2 | 12.5 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 1.2 | 7.3 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 1.2 | 13.9 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 1.2 | 38.1 | REACTOME KINESINS | Genes involved in Kinesins |
| 1.1 | 49.0 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.9 | 18.3 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.9 | 10.9 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.9 | 8.9 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.7 | 32.9 | REACTOME RNA POL I TRANSCRIPTION | Genes involved in RNA Polymerase I Transcription |
| 0.7 | 18.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.7 | 35.0 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.6 | 12.6 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.5 | 62.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.5 | 35.4 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.5 | 11.9 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.5 | 13.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.4 | 7.6 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.4 | 16.3 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.3 | 12.1 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.2 | 4.3 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.2 | 4.8 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.2 | 50.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.2 | 5.1 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.2 | 5.8 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.2 | 5.2 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.2 | 2.6 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.2 | 2.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.2 | 6.3 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.2 | 2.5 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 4.6 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.1 | 2.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 14.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 1.6 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 11.3 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 1.4 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 5.3 | REACTOME MITOTIC G2 G2 M PHASES | Genes involved in Mitotic G2-G2/M phases |
| 0.1 | 6.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 0.9 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 1.4 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 1.6 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 1.4 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.5 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |