Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for E2f3
Z-value: 1.00
Transcription factors associated with E2f3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2f3
|
ENSMUSG00000016477.19 | E2f3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2f3 | mm39_v1_chr13_-_30170031_30170048 | 0.36 | 1.7e-03 | Click! |
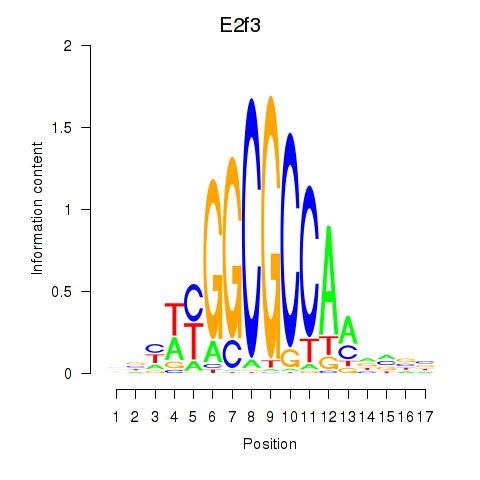
Activity profile of E2f3 motif
Sorted Z-values of E2f3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of E2f3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_113957362 | 15.52 |
ENSMUST00000202555.2
|
Selplg
|
selectin, platelet (p-selectin) ligand |
| chr5_-_113957318 | 13.83 |
ENSMUST00000201194.4
|
Selplg
|
selectin, platelet (p-selectin) ligand |
| chr11_+_11635908 | 9.15 |
ENSMUST00000065433.12
|
Ikzf1
|
IKAROS family zinc finger 1 |
| chr2_-_126975804 | 5.67 |
ENSMUST00000110387.4
|
Ncaph
|
non-SMC condensin I complex, subunit H |
| chr12_+_24758968 | 5.05 |
ENSMUST00000154588.2
|
Rrm2
|
ribonucleotide reductase M2 |
| chr12_-_69274936 | 4.55 |
ENSMUST00000221411.2
ENSMUST00000021359.7 |
Pole2
|
polymerase (DNA directed), epsilon 2 (p59 subunit) |
| chr4_+_43957401 | 4.32 |
ENSMUST00000030202.14
|
Glipr2
|
GLI pathogenesis-related 2 |
| chr6_+_120440159 | 4.13 |
ENSMUST00000002976.5
|
Il17ra
|
interleukin 17 receptor A |
| chr8_+_123294740 | 4.11 |
ENSMUST00000006760.3
|
Cdt1
|
chromatin licensing and DNA replication factor 1 |
| chr2_+_109111083 | 4.09 |
ENSMUST00000028527.8
|
Kif18a
|
kinesin family member 18A |
| chr5_-_65855511 | 4.06 |
ENSMUST00000201948.4
|
Pds5a
|
PDS5 cohesin associated factor A |
| chr12_+_24758724 | 4.04 |
ENSMUST00000153058.8
|
Rrm2
|
ribonucleotide reductase M2 |
| chr4_+_43957677 | 4.01 |
ENSMUST00000107855.2
|
Glipr2
|
GLI pathogenesis-related 2 |
| chrX_-_50294652 | 3.96 |
ENSMUST00000114875.8
|
Mbnl3
|
muscleblind like splicing factor 3 |
| chr17_+_34372046 | 3.95 |
ENSMUST00000114232.4
|
H2-DMb1
|
histocompatibility 2, class II, locus Mb1 |
| chr11_-_101442663 | 3.88 |
ENSMUST00000017290.11
|
Brca1
|
breast cancer 1, early onset |
| chr12_+_24758240 | 3.81 |
ENSMUST00000020980.12
|
Rrm2
|
ribonucleotide reductase M2 |
| chr7_+_13012735 | 3.78 |
ENSMUST00000098814.13
ENSMUST00000146998.9 |
Lig1
|
ligase I, DNA, ATP-dependent |
| chr2_+_117942357 | 3.66 |
ENSMUST00000039559.9
|
Thbs1
|
thrombospondin 1 |
| chr1_-_95595280 | 3.63 |
ENSMUST00000043336.11
|
St8sia4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 |
| chr13_-_55477535 | 3.39 |
ENSMUST00000021941.8
|
Mxd3
|
Max dimerization protein 3 |
| chr18_+_11790409 | 3.38 |
ENSMUST00000047322.8
|
Rbbp8
|
retinoblastoma binding protein 8, endonuclease |
| chrX_-_101232978 | 3.36 |
ENSMUST00000033683.8
|
Rps4x
|
ribosomal protein S4, X-linked |
| chr2_+_172821620 | 3.32 |
ENSMUST00000109125.8
ENSMUST00000109126.5 ENSMUST00000050442.15 |
Spo11
|
SPO11 initiator of meiotic double stranded breaks |
| chr15_-_79967543 | 3.31 |
ENSMUST00000081650.15
|
Rpl3
|
ribosomal protein L3 |
| chr10_+_20223516 | 3.27 |
ENSMUST00000169712.3
ENSMUST00000217608.2 |
Mtfr2
|
mitochondrial fission regulator 2 |
| chr9_-_22043083 | 3.25 |
ENSMUST00000069330.14
ENSMUST00000217643.2 |
Acp5
|
acid phosphatase 5, tartrate resistant |
| chr11_+_60066681 | 3.20 |
ENSMUST00000102688.2
|
Rai1
|
retinoic acid induced 1 |
| chr3_+_68912302 | 3.20 |
ENSMUST00000136502.8
ENSMUST00000107803.7 |
Smc4
|
structural maintenance of chromosomes 4 |
| chr17_-_71782296 | 3.14 |
ENSMUST00000127430.2
|
Smchd1
|
SMC hinge domain containing 1 |
| chr16_-_4536992 | 3.08 |
ENSMUST00000115851.10
|
Nmral1
|
NmrA-like family domain containing 1 |
| chr10_-_34003933 | 3.02 |
ENSMUST00000062784.8
|
Calhm6
|
calcium homeostasis modulator family member 6 |
| chr16_-_4536943 | 3.02 |
ENSMUST00000120056.8
ENSMUST00000074970.8 |
Nmral1
|
NmrA-like family domain containing 1 |
| chr10_-_93425553 | 2.86 |
ENSMUST00000020203.7
|
Snrpf
|
small nuclear ribonucleoprotein polypeptide F |
| chr9_-_22042930 | 2.86 |
ENSMUST00000213815.2
|
Acp5
|
acid phosphatase 5, tartrate resistant |
| chr18_+_11790888 | 2.82 |
ENSMUST00000234499.2
|
Rbbp8
|
retinoblastoma binding protein 8, endonuclease |
| chr2_+_150751475 | 2.78 |
ENSMUST00000028948.5
|
Gins1
|
GINS complex subunit 1 (Psf1 homolog) |
| chr9_+_64188857 | 2.72 |
ENSMUST00000215031.2
ENSMUST00000213165.2 ENSMUST00000213289.2 ENSMUST00000216594.2 ENSMUST00000034964.7 |
Tipin
|
timeless interacting protein |
| chr9_+_44151962 | 2.72 |
ENSMUST00000092426.5
ENSMUST00000217221.2 ENSMUST00000213891.2 |
Ccdc153
|
coiled-coil domain containing 153 |
| chr15_-_83479312 | 2.69 |
ENSMUST00000016901.5
|
Ttll12
|
tubulin tyrosine ligase-like family, member 12 |
| chr5_-_65855199 | 2.63 |
ENSMUST00000031104.7
|
Pds5a
|
PDS5 cohesin associated factor A |
| chr13_-_51947685 | 2.58 |
ENSMUST00000110040.9
ENSMUST00000021900.14 |
Sema4d
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
| chr9_+_56979307 | 2.56 |
ENSMUST00000169879.8
|
Sin3a
|
transcriptional regulator, SIN3A (yeast) |
| chr6_-_34887743 | 2.55 |
ENSMUST00000081214.12
|
Wdr91
|
WD repeat domain 91 |
| chr5_-_100867520 | 2.44 |
ENSMUST00000112908.2
ENSMUST00000045617.15 |
Hpse
|
heparanase |
| chr9_+_44152029 | 2.40 |
ENSMUST00000217510.2
ENSMUST00000216632.2 |
Ccdc153
|
coiled-coil domain containing 153 |
| chr18_+_67933169 | 2.31 |
ENSMUST00000025425.7
|
Cep192
|
centrosomal protein 192 |
| chr3_+_97920819 | 2.27 |
ENSMUST00000079812.8
|
Notch2
|
notch 2 |
| chr10_-_60983404 | 2.25 |
ENSMUST00000122259.8
|
Sgpl1
|
sphingosine phosphate lyase 1 |
| chr19_+_40883127 | 2.23 |
ENSMUST00000050092.7
|
Zfp518a
|
zinc finger protein 518A |
| chr12_-_21423607 | 2.22 |
ENSMUST00000064536.13
|
Adam17
|
a disintegrin and metallopeptidase domain 17 |
| chr10_-_60983438 | 2.22 |
ENSMUST00000092498.12
ENSMUST00000137833.2 ENSMUST00000155919.8 |
Sgpl1
|
sphingosine phosphate lyase 1 |
| chr11_+_115455260 | 2.20 |
ENSMUST00000021085.11
|
Nup85
|
nucleoporin 85 |
| chr12_-_21423524 | 2.17 |
ENSMUST00000232107.2
|
Adam17
|
a disintegrin and metallopeptidase domain 17 |
| chr15_-_67048595 | 2.17 |
ENSMUST00000229213.2
|
St3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr5_-_68004743 | 2.15 |
ENSMUST00000072971.13
ENSMUST00000113652.8 ENSMUST00000113651.8 ENSMUST00000037380.15 |
Atp8a1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1 |
| chr15_-_67048673 | 2.11 |
ENSMUST00000229028.2
|
St3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chrX_-_104972938 | 2.01 |
ENSMUST00000198448.5
ENSMUST00000199233.5 ENSMUST00000134507.8 ENSMUST00000154866.8 ENSMUST00000128968.8 ENSMUST00000134381.8 ENSMUST00000150914.8 |
Atrx
|
ATRX, chromatin remodeler |
| chr3_-_95125190 | 1.95 |
ENSMUST00000136139.8
|
Gabpb2
|
GA repeat binding protein, beta 2 |
| chrX_-_104973003 | 1.95 |
ENSMUST00000130980.2
ENSMUST00000113573.8 |
Atrx
|
ATRX, chromatin remodeler |
| chr6_-_88818832 | 1.92 |
ENSMUST00000032169.8
|
Abtb1
|
ankyrin repeat and BTB (POZ) domain containing 1 |
| chr3_-_84387700 | 1.92 |
ENSMUST00000194027.2
ENSMUST00000107689.7 |
Fhdc1
|
FH2 domain containing 1 |
| chr3_-_51248032 | 1.91 |
ENSMUST00000062009.14
ENSMUST00000194641.6 |
Elf2
|
E74-like factor 2 |
| chr2_+_31560725 | 1.91 |
ENSMUST00000038474.14
ENSMUST00000137156.2 |
Exosc2
|
exosome component 2 |
| chr16_+_91282121 | 1.89 |
ENSMUST00000023689.11
ENSMUST00000117748.8 |
Ifnar1
|
interferon (alpha and beta) receptor 1 |
| chr12_-_21423551 | 1.86 |
ENSMUST00000101551.10
|
Adam17
|
a disintegrin and metallopeptidase domain 17 |
| chr17_-_33007238 | 1.85 |
ENSMUST00000159086.10
|
Zfp871
|
zinc finger protein 871 |
| chr10_+_75152705 | 1.82 |
ENSMUST00000105420.3
|
Adora2a
|
adenosine A2a receptor |
| chr9_-_57673128 | 1.82 |
ENSMUST00000065330.8
|
Clk3
|
CDC-like kinase 3 |
| chr4_+_11486002 | 1.82 |
ENSMUST00000108307.3
|
Virma
|
vir like m6A methyltransferase associated |
| chr12_-_4924341 | 1.79 |
ENSMUST00000137337.8
ENSMUST00000045921.14 |
Mfsd2b
|
major facilitator superfamily domain containing 2B |
| chr3_-_95125002 | 1.77 |
ENSMUST00000107209.8
|
Gabpb2
|
GA repeat binding protein, beta 2 |
| chr4_+_11485945 | 1.74 |
ENSMUST00000055372.14
ENSMUST00000059914.13 |
Virma
|
vir like m6A methyltransferase associated |
| chr17_+_27136065 | 1.74 |
ENSMUST00000078961.6
|
Kifc5b
|
kinesin family member C5B |
| chr17_+_24971952 | 1.69 |
ENSMUST00000044922.8
ENSMUST00000234202.2 |
Hs3st6
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 6 |
| chr2_+_34999497 | 1.69 |
ENSMUST00000028235.11
ENSMUST00000156933.8 ENSMUST00000028237.15 ENSMUST00000113032.8 |
Cntrl
|
centriolin |
| chr4_-_123558516 | 1.67 |
ENSMUST00000147030.2
|
Macf1
|
microtubule-actin crosslinking factor 1 |
| chr15_-_89033761 | 1.66 |
ENSMUST00000088823.5
|
Mapk11
|
mitogen-activated protein kinase 11 |
| chr1_-_91340884 | 1.65 |
ENSMUST00000086851.2
|
Hes6
|
hairy and enhancer of split 6 |
| chr15_+_73384407 | 1.62 |
ENSMUST00000043414.12
|
Dennd3
|
DENN/MADD domain containing 3 |
| chr12_-_100865783 | 1.58 |
ENSMUST00000053668.10
|
Gpr68
|
G protein-coupled receptor 68 |
| chr17_+_29251602 | 1.57 |
ENSMUST00000130216.3
|
Srsf3
|
serine and arginine-rich splicing factor 3 |
| chr18_-_52662728 | 1.56 |
ENSMUST00000025409.9
|
Lox
|
lysyl oxidase |
| chr16_+_91444730 | 1.55 |
ENSMUST00000119368.8
ENSMUST00000114037.9 ENSMUST00000114036.9 ENSMUST00000122302.8 |
Son
|
Son DNA binding protein |
| chr14_-_30348153 | 1.49 |
ENSMUST00000112211.9
ENSMUST00000112210.11 |
Prkcd
|
protein kinase C, delta |
| chr10_-_80512117 | 1.49 |
ENSMUST00000200082.5
|
Mknk2
|
MAP kinase-interacting serine/threonine kinase 2 |
| chr13_-_21586858 | 1.49 |
ENSMUST00000117721.8
ENSMUST00000070785.16 ENSMUST00000116433.2 ENSMUST00000223831.2 ENSMUST00000116434.11 ENSMUST00000224820.2 |
Zkscan3
|
zinc finger with KRAB and SCAN domains 3 |
| chr1_-_191307648 | 1.48 |
ENSMUST00000027933.11
|
Dtl
|
denticleless E3 ubiquitin protein ligase |
| chr3_-_68911807 | 1.47 |
ENSMUST00000154741.8
ENSMUST00000148031.2 |
Ift80
|
intraflagellar transport 80 |
| chr4_-_140805613 | 1.46 |
ENSMUST00000030760.15
|
Necap2
|
NECAP endocytosis associated 2 |
| chr19_+_53588808 | 1.46 |
ENSMUST00000025930.10
|
Smc3
|
structural maintenance of chromosomes 3 |
| chr18_+_31922173 | 1.46 |
ENSMUST00000025106.5
ENSMUST00000234146.2 |
Polr2d
|
polymerase (RNA) II (DNA directed) polypeptide D |
| chr3_-_68911835 | 1.42 |
ENSMUST00000107812.8
|
Ift80
|
intraflagellar transport 80 |
| chr2_+_52747855 | 1.40 |
ENSMUST00000155586.9
ENSMUST00000090952.11 ENSMUST00000127122.9 ENSMUST00000049483.14 ENSMUST00000050719.13 |
Fmnl2
|
formin-like 2 |
| chrX_-_104972844 | 1.38 |
ENSMUST00000198441.5
ENSMUST00000137453.8 |
Atrx
|
ATRX, chromatin remodeler |
| chr1_-_128287347 | 1.36 |
ENSMUST00000190495.2
ENSMUST00000027601.11 |
Mcm6
|
minichromosome maintenance complex component 6 |
| chr17_+_34258411 | 1.35 |
ENSMUST00000087497.11
ENSMUST00000131134.9 ENSMUST00000235819.2 ENSMUST00000114255.9 ENSMUST00000114252.9 ENSMUST00000237989.2 |
Col11a2
|
collagen, type XI, alpha 2 |
| chr3_-_84063067 | 1.34 |
ENSMUST00000047368.8
|
Mnd1
|
meiotic nuclear divisions 1 |
| chr3_-_68911886 | 1.34 |
ENSMUST00000169064.8
|
Ift80
|
intraflagellar transport 80 |
| chr5_-_68004702 | 1.32 |
ENSMUST00000135930.8
|
Atp8a1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1 |
| chr17_-_56891576 | 1.28 |
ENSMUST00000075510.12
|
Safb2
|
scaffold attachment factor B2 |
| chr8_-_70449018 | 1.28 |
ENSMUST00000065169.12
ENSMUST00000212478.2 |
Gatad2a
|
GATA zinc finger domain containing 2A |
| chr4_-_34614883 | 1.24 |
ENSMUST00000140334.2
ENSMUST00000108142.8 ENSMUST00000048706.10 |
Orc3
|
origin recognition complex, subunit 3 |
| chr3_+_66892979 | 1.23 |
ENSMUST00000162362.8
ENSMUST00000065074.14 ENSMUST00000065047.13 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr7_-_5016237 | 1.22 |
ENSMUST00000208944.2
|
Fiz1
|
Flt3 interacting zinc finger protein 1 |
| chr4_+_57821050 | 1.20 |
ENSMUST00000238994.2
|
Pakap
|
paralemmin A kinase anchor protein |
| chr6_-_72322116 | 1.19 |
ENSMUST00000070345.5
|
Usp39
|
ubiquitin specific peptidase 39 |
| chr16_+_91282183 | 1.18 |
ENSMUST00000129878.2
|
Ifnar1
|
interferon (alpha and beta) receptor 1 |
| chr9_-_62888156 | 1.17 |
ENSMUST00000098651.6
ENSMUST00000214830.2 |
Pias1
|
protein inhibitor of activated STAT 1 |
| chr9_-_20888054 | 1.10 |
ENSMUST00000054197.7
|
S1pr2
|
sphingosine-1-phosphate receptor 2 |
| chr1_+_156386414 | 1.09 |
ENSMUST00000166172.9
ENSMUST00000027888.13 |
Abl2
|
v-abl Abelson murine leukemia viral oncogene 2 (arg, Abelson-related gene) |
| chr10_+_80062468 | 1.09 |
ENSMUST00000130260.2
|
Pwwp3a
|
PWWP domain containing 3A, DNA repair factor |
| chr10_-_80754016 | 1.08 |
ENSMUST00000057623.14
ENSMUST00000179022.8 |
Lmnb2
|
lamin B2 |
| chr15_+_99122742 | 1.06 |
ENSMUST00000041415.5
|
Kcnh3
|
potassium voltage-gated channel, subfamily H (eag-related), member 3 |
| chr13_-_21715202 | 1.06 |
ENSMUST00000156674.3
ENSMUST00000110481.3 ENSMUST00000045228.12 |
Zkscan8
|
zinc finger with KRAB and SCAN domains 8 |
| chr19_-_5416626 | 1.06 |
ENSMUST00000237167.2
|
Banf1
|
BAF nuclear assembly factor 1 |
| chr11_+_65698001 | 1.06 |
ENSMUST00000071465.9
ENSMUST00000018491.8 |
Zkscan6
|
zinc finger with KRAB and SCAN domains 6 |
| chr6_-_71417607 | 1.05 |
ENSMUST00000002292.15
|
Rmnd5a
|
required for meiotic nuclear division 5 homolog A |
| chr11_-_86435579 | 1.05 |
ENSMUST00000138810.3
ENSMUST00000058286.9 ENSMUST00000154617.8 |
Rps6kb1
|
ribosomal protein S6 kinase, polypeptide 1 |
| chrX_-_101114906 | 1.03 |
ENSMUST00000188731.2
|
Rtl5
|
retrotransposon Gag like 5 |
| chr11_+_21189277 | 1.02 |
ENSMUST00000109578.8
ENSMUST00000006221.14 |
Vps54
|
VPS54 GARP complex subunit |
| chr2_-_168432111 | 1.01 |
ENSMUST00000029057.13
ENSMUST00000074618.10 |
Nfatc2
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 2 |
| chr9_+_21914083 | 1.00 |
ENSMUST00000216344.2
|
Prkcsh
|
protein kinase C substrate 80K-H |
| chr1_-_91326490 | 1.00 |
ENSMUST00000190519.2
ENSMUST00000027534.13 ENSMUST00000187306.7 |
Ilkap
|
integrin-linked kinase-associated serine/threonine phosphatase 2C |
| chr1_-_95595245 | 0.99 |
ENSMUST00000189556.2
|
St8sia4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 |
| chr7_-_80338600 | 0.98 |
ENSMUST00000122255.8
|
Crtc3
|
CREB regulated transcription coactivator 3 |
| chr18_+_75500600 | 0.97 |
ENSMUST00000026999.10
|
Smad7
|
SMAD family member 7 |
| chrX_+_93768175 | 0.96 |
ENSMUST00000101388.4
|
Zxdb
|
zinc finger, X-linked, duplicated B |
| chr10_-_30476658 | 0.96 |
ENSMUST00000019927.7
|
Trmt11
|
tRNA methyltransferase 11 |
| chr18_+_44513547 | 0.92 |
ENSMUST00000202306.2
ENSMUST00000025350.10 |
Dcp2
|
decapping mRNA 2 |
| chrX_-_101114794 | 0.91 |
ENSMUST00000113631.2
|
Rtl5
|
retrotransposon Gag like 5 |
| chr17_+_64907697 | 0.91 |
ENSMUST00000086723.10
|
Man2a1
|
mannosidase 2, alpha 1 |
| chr4_+_109137561 | 0.88 |
ENSMUST00000177089.8
ENSMUST00000175776.8 ENSMUST00000132165.9 |
Eps15
|
epidermal growth factor receptor pathway substrate 15 |
| chr9_-_44876817 | 0.88 |
ENSMUST00000214761.2
ENSMUST00000213666.2 ENSMUST00000213890.2 ENSMUST00000125642.8 ENSMUST00000213193.2 ENSMUST00000117506.9 ENSMUST00000138559.9 ENSMUST00000117549.8 |
Ube4a
|
ubiquitination factor E4A |
| chr17_-_80369626 | 0.88 |
ENSMUST00000184635.8
|
Hnrnpll
|
heterogeneous nuclear ribonucleoprotein L-like |
| chr8_-_57940834 | 0.88 |
ENSMUST00000034022.4
|
Sap30
|
sin3 associated polypeptide |
| chr9_+_63509925 | 0.88 |
ENSMUST00000041551.9
|
Aagab
|
alpha- and gamma-adaptin binding protein |
| chr8_-_70448872 | 0.87 |
ENSMUST00000177851.9
|
Gatad2a
|
GATA zinc finger domain containing 2A |
| chrX_-_8042129 | 0.87 |
ENSMUST00000143984.2
|
Tbc1d25
|
TBC1 domain family, member 25 |
| chr15_+_84116231 | 0.87 |
ENSMUST00000023072.7
|
Parvb
|
parvin, beta |
| chr5_-_148336711 | 0.86 |
ENSMUST00000048116.15
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr5_-_92653377 | 0.84 |
ENSMUST00000031377.9
|
Scarb2
|
scavenger receptor class B, member 2 |
| chr1_-_4563821 | 0.84 |
ENSMUST00000191939.2
|
Sox17
|
SRY (sex determining region Y)-box 17 |
| chr4_+_109137462 | 0.83 |
ENSMUST00000102729.10
|
Eps15
|
epidermal growth factor receptor pathway substrate 15 |
| chr2_+_151753119 | 0.82 |
ENSMUST00000028955.6
|
Angpt4
|
angiopoietin 4 |
| chr18_-_84607615 | 0.79 |
ENSMUST00000125763.3
|
Zfp407
|
zinc finger protein 407 |
| chr3_-_41696906 | 0.79 |
ENSMUST00000026866.15
|
Sclt1
|
sodium channel and clathrin linker 1 |
| chrX_+_20570145 | 0.77 |
ENSMUST00000033383.3
|
Usp11
|
ubiquitin specific peptidase 11 |
| chrX_+_73326303 | 0.77 |
ENSMUST00000069722.13
ENSMUST00000124200.8 ENSMUST00000114180.11 ENSMUST00000132437.8 |
Taz
|
tafazzin |
| chr14_+_73381003 | 0.76 |
ENSMUST00000110952.10
|
Rcbtb2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr19_+_3373285 | 0.75 |
ENSMUST00000025835.6
|
Cpt1a
|
carnitine palmitoyltransferase 1a, liver |
| chr11_-_62348599 | 0.75 |
ENSMUST00000127471.9
|
Ncor1
|
nuclear receptor co-repressor 1 |
| chr9_+_22099386 | 0.75 |
ENSMUST00000217301.2
ENSMUST00000178901.8 |
Zfp872
|
zinc finger protein 872 |
| chr8_-_120331936 | 0.74 |
ENSMUST00000093099.13
|
Taf1c
|
TATA-box binding protein associated factor, RNA polymerase I, C |
| chr17_+_36152383 | 0.71 |
ENSMUST00000082337.13
|
Mdc1
|
mediator of DNA damage checkpoint 1 |
| chr3_+_66893031 | 0.70 |
ENSMUST00000046542.13
ENSMUST00000162693.8 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr11_-_84020382 | 0.69 |
ENSMUST00000018795.13
|
Tada2a
|
transcriptional adaptor 2A |
| chr16_-_78173645 | 0.69 |
ENSMUST00000023570.14
|
Btg3
|
BTG anti-proliferation factor 3 |
| chr4_+_116565706 | 0.68 |
ENSMUST00000030452.13
ENSMUST00000106462.9 |
Ccdc163
|
coiled-coil domain containing 163 |
| chr10_+_33780993 | 0.68 |
ENSMUST00000169670.8
|
Rsph4a
|
radial spoke head 4 homolog A (Chlamydomonas) |
| chr9_+_113760376 | 0.68 |
ENSMUST00000214095.2
ENSMUST00000116492.3 ENSMUST00000216558.2 |
Ubp1
|
upstream binding protein 1 |
| chr5_-_34093678 | 0.66 |
ENSMUST00000030993.8
|
Nelfa
|
negative elongation factor complex member A, Whsc2 |
| chr4_+_116565784 | 0.65 |
ENSMUST00000138305.8
ENSMUST00000125671.8 ENSMUST00000130828.8 |
Ccdc163
|
coiled-coil domain containing 163 |
| chr5_-_39801940 | 0.65 |
ENSMUST00000152057.2
ENSMUST00000053116.7 |
Hs3st1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
| chr7_+_34818709 | 0.64 |
ENSMUST00000205391.2
ENSMUST00000042985.11 |
Cebpa
|
CCAAT/enhancer binding protein (C/EBP), alpha |
| chr19_-_4319170 | 0.64 |
ENSMUST00000037992.16
ENSMUST00000113852.6 ENSMUST00000236794.2 |
Ssh3
|
slingshot protein phosphatase 3 |
| chr10_+_43354807 | 0.64 |
ENSMUST00000167488.9
|
Bend3
|
BEN domain containing 3 |
| chr17_-_80369762 | 0.64 |
ENSMUST00000061331.14
|
Hnrnpll
|
heterogeneous nuclear ribonucleoprotein L-like |
| chr2_-_168432235 | 0.63 |
ENSMUST00000109184.8
|
Nfatc2
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 2 |
| chr17_-_24877431 | 0.63 |
ENSMUST00000095544.6
|
Npw
|
neuropeptide W |
| chr19_+_5416769 | 0.62 |
ENSMUST00000025759.9
|
Eif1ad
|
eukaryotic translation initiation factor 1A domain containing |
| chr3_-_95125051 | 0.60 |
ENSMUST00000107204.8
|
Gabpb2
|
GA repeat binding protein, beta 2 |
| chr17_-_48145466 | 0.60 |
ENSMUST00000066368.13
|
Mdfi
|
MyoD family inhibitor |
| chr3_+_33853941 | 0.60 |
ENSMUST00000099153.10
|
Ttc14
|
tetratricopeptide repeat domain 14 |
| chr10_+_43355113 | 0.58 |
ENSMUST00000040147.8
|
Bend3
|
BEN domain containing 3 |
| chr8_+_71995554 | 0.55 |
ENSMUST00000034272.9
|
Mvb12a
|
multivesicular body subunit 12A |
| chr3_+_33854305 | 0.55 |
ENSMUST00000196139.5
ENSMUST00000200271.5 ENSMUST00000198529.5 ENSMUST00000117915.8 ENSMUST00000108210.9 ENSMUST00000196975.5 |
Ttc14
|
tetratricopeptide repeat domain 14 |
| chr15_+_99870661 | 0.54 |
ENSMUST00000100206.4
|
Larp4
|
La ribonucleoprotein domain family, member 4 |
| chr12_+_111505216 | 0.53 |
ENSMUST00000050993.11
|
Eif5
|
eukaryotic translation initiation factor 5 |
| chr2_-_180929828 | 0.52 |
ENSMUST00000049032.13
|
Gmeb2
|
glucocorticoid modulatory element binding protein 2 |
| chr8_+_106587212 | 0.52 |
ENSMUST00000008594.9
|
Nutf2
|
nuclear transport factor 2 |
| chr7_+_100021425 | 0.52 |
ENSMUST00000098259.11
ENSMUST00000051777.15 |
C2cd3
|
C2 calcium-dependent domain containing 3 |
| chr11_-_72097821 | 0.52 |
ENSMUST00000204457.3
|
Gm43951
|
predicted gene, 43951 |
| chr16_-_44153288 | 0.52 |
ENSMUST00000136381.8
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr8_+_84626715 | 0.52 |
ENSMUST00000141158.8
|
Adgrl1
|
adhesion G protein-coupled receptor L1 |
| chr9_+_72892850 | 0.51 |
ENSMUST00000150826.9
ENSMUST00000085350.11 ENSMUST00000140675.8 |
Ccpg1
|
cell cycle progression 1 |
| chr8_-_124586159 | 0.50 |
ENSMUST00000034452.12
|
Ccsap
|
centriole, cilia and spindle associated protein |
| chr10_+_75152908 | 0.50 |
ENSMUST00000219044.2
|
Adora2a
|
adenosine A2a receptor |
| chr16_-_43709968 | 0.48 |
ENSMUST00000023387.14
|
Qtrt2
|
queuine tRNA-ribosyltransferase accessory subunit 2 |
| chr6_-_97594498 | 0.48 |
ENSMUST00000113355.9
|
Frmd4b
|
FERM domain containing 4B |
| chr2_+_115412148 | 0.48 |
ENSMUST00000166472.8
ENSMUST00000110918.3 |
Cdin1
|
CDAN1 interacting nuclease 1 |
| chr18_+_47245204 | 0.47 |
ENSMUST00000234633.2
|
Hspe1-rs1
|
heat shock protein 1 (chaperonin 10), related sequence 1 |
| chr5_-_148336574 | 0.47 |
ENSMUST00000202457.4
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr11_-_20199359 | 0.47 |
ENSMUST00000050611.14
ENSMUST00000109596.8 ENSMUST00000162811.2 |
Cep68
|
centrosomal protein 68 |
| chr1_-_16689527 | 0.46 |
ENSMUST00000182554.9
|
Ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr12_+_80992294 | 0.46 |
ENSMUST00000110354.8
ENSMUST00000110352.10 ENSMUST00000110351.8 ENSMUST00000110356.3 |
Srsf5
|
serine and arginine-rich splicing factor 5 |
| chr9_-_105837842 | 0.46 |
ENSMUST00000190193.8
ENSMUST00000165165.3 |
Col6a5
|
collagen, type VI, alpha 5 |
| chrX_-_73129195 | 0.43 |
ENSMUST00000140399.3
ENSMUST00000123362.8 ENSMUST00000100750.10 ENSMUST00000033770.13 |
Mecp2
|
methyl CpG binding protein 2 |
| chr13_+_54722833 | 0.40 |
ENSMUST00000156024.2
|
Arl10
|
ADP-ribosylation factor-like 10 |
| chr18_+_9957906 | 0.37 |
ENSMUST00000025137.9
|
Thoc1
|
THO complex 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.9 | 29.3 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 3.1 | 9.2 | GO:0045660 | positive regulation of neutrophil differentiation(GO:0045660) |
| 1.4 | 5.7 | GO:2000371 | regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 1.4 | 4.1 | GO:0072708 | mediator complex assembly(GO:0036034) DNA replication preinitiation complex assembly(GO:0071163) response to sorbitol(GO:0072708) regulation of mediator complex assembly(GO:2001176) positive regulation of mediator complex assembly(GO:2001178) |
| 1.2 | 1.2 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 1.2 | 3.7 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) regulation of cGMP-mediated signaling(GO:0010752) |
| 1.1 | 3.3 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 1.0 | 3.1 | GO:0060821 | inactivation of X chromosome by DNA methylation(GO:0060821) |
| 1.0 | 6.3 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.9 | 4.5 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.9 | 5.3 | GO:0035127 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.8 | 2.4 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.8 | 3.9 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.8 | 6.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.8 | 2.3 | GO:0035622 | intrahepatic bile duct development(GO:0035622) cholangiocyte proliferation(GO:1990705) |
| 0.7 | 6.6 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.7 | 6.2 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.7 | 4.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.6 | 2.6 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.6 | 2.6 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.6 | 12.9 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.5 | 2.7 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.5 | 0.5 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.5 | 1.5 | GO:0031990 | mRNA export from nucleus in response to heat stress(GO:0031990) |
| 0.5 | 1.9 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.5 | 2.3 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.4 | 4.2 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.4 | 1.5 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.4 | 3.2 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.4 | 4.6 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.4 | 3.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.4 | 1.1 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 0.3 | 1.6 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.3 | 8.0 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.3 | 3.1 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.3 | 0.8 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.3 | 3.6 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.2 | 3.9 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.2 | 0.9 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.2 | 2.2 | GO:0021995 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.2 | 4.5 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.2 | 0.8 | GO:0060807 | cardiogenic plate morphogenesis(GO:0003142) regulation of transcription from RNA polymerase II promoter involved in definitive endodermal cell fate specification(GO:0060807) |
| 0.2 | 1.7 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.2 | 1.4 | GO:0060023 | soft palate development(GO:0060023) |
| 0.2 | 0.2 | GO:1990009 | retinal cell apoptotic process(GO:1990009) |
| 0.2 | 0.7 | GO:0072362 | regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.2 | 1.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.2 | 1.0 | GO:0033262 | regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.2 | 0.8 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.2 | 1.7 | GO:0075509 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.2 | 1.2 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.4 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.1 | 1.5 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.1 | 1.0 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.1 | 1.6 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 4.1 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.1 | 1.0 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.1 | 0.5 | GO:0006168 | adenine salvage(GO:0006168) adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.1 | 1.4 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 8.3 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.1 | 1.0 | GO:2000320 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.1 | 0.4 | GO:1904046 | negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 0.1 | 4.0 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 1.4 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.1 | 0.4 | GO:0045829 | negative regulation of isotype switching(GO:0045829) |
| 0.1 | 2.3 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.1 | 3.2 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 3.3 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 0.7 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.1 | 0.9 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.6 | GO:1903772 | regulation of viral budding via host ESCRT complex(GO:1903772) |
| 0.1 | 0.8 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 4.3 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 1.3 | GO:0015809 | arginine transport(GO:0015809) |
| 0.1 | 1.7 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 1.5 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.1 | 0.4 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 1.2 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 | 0.9 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.1 | 1.0 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 0.7 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 2.9 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 0.5 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.1 | 0.6 | GO:0000050 | urea cycle(GO:0000050) |
| 0.1 | 0.9 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.1 | 1.6 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 0.3 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.1 | 0.7 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.1 | 0.8 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.8 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.1 | 0.5 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.1 | 1.0 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 3.3 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 1.3 | GO:0060008 | Sertoli cell differentiation(GO:0060008) |
| 0.0 | 2.2 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 0.2 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 0.1 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.2 | GO:0071681 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 2.3 | GO:0007041 | lysosomal transport(GO:0007041) |
| 0.0 | 0.5 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 1.9 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 1.9 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 1.5 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 1.4 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 1.6 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.6 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.3 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.1 | GO:0021592 | fourth ventricle development(GO:0021592) initiation of neural tube closure(GO:0021993) |
| 0.0 | 1.7 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.0 | 0.5 | GO:0035584 | calcium-mediated signaling using intracellular calcium source(GO:0035584) |
| 0.0 | 3.0 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.2 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.5 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.2 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.6 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 0.9 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.7 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 1.4 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.5 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 12.9 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 1.7 | 29.3 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 1.2 | 1.2 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 1.1 | 5.7 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.9 | 2.8 | GO:0000811 | GINS complex(GO:0000811) |
| 0.9 | 4.5 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.9 | 5.3 | GO:1990707 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 0.8 | 3.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.8 | 9.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.5 | 2.7 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.5 | 4.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.5 | 3.6 | GO:0036396 | MIS complex(GO:0036396) |
| 0.5 | 2.9 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.5 | 3.2 | GO:0000796 | condensin complex(GO:0000796) |
| 0.4 | 3.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.4 | 3.9 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.3 | 3.9 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 1.5 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.2 | 0.6 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.2 | 1.5 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.2 | 1.1 | GO:0034657 | GID complex(GO:0034657) |
| 0.2 | 4.2 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.2 | 1.0 | GO:0000938 | GARP complex(GO:0000938) |
| 0.2 | 1.0 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.2 | 0.8 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.2 | 0.7 | GO:0032021 | NELF complex(GO:0032021) |
| 0.2 | 1.9 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.2 | 2.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.2 | 1.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.7 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 0.7 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 4.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 2.3 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 3.4 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.1 | 0.5 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.1 | 2.2 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 1.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.8 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 2.3 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 4.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 1.5 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 1.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 6.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 0.9 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 1.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.4 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 3.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.4 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 4.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 12.5 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.4 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 3.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.3 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 4.1 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 3.6 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 0.8 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 4.6 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 1.2 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 3.6 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.5 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.7 | GO:0005871 | kinesin complex(GO:0005871) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 12.9 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 1.1 | 3.4 | GO:0004637 | phosphoribosylamine-glycine ligase activity(GO:0004637) |
| 0.9 | 3.7 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.8 | 3.3 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.8 | 5.7 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.8 | 4.6 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.8 | 5.3 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.6 | 6.3 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.5 | 2.2 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.5 | 4.3 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.5 | 4.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.5 | 6.3 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.4 | 2.3 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.4 | 3.8 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.4 | 1.5 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) TIR domain binding(GO:0070976) |
| 0.4 | 1.5 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.3 | 5.6 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.3 | 0.9 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.3 | 2.0 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.3 | 1.0 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.3 | 1.6 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.3 | 0.8 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.2 | 2.4 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.2 | 9.2 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.2 | 1.9 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.2 | 3.3 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.2 | 4.5 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.2 | 0.8 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.2 | 2.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.2 | 1.0 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.2 | 0.9 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.2 | 1.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.2 | 4.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 1.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.2 | 2.8 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.2 | 0.5 | GO:0003999 | adenine binding(GO:0002055) adenine phosphoribosyltransferase activity(GO:0003999) |
| 0.1 | 1.3 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 4.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 2.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 3.0 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 0.8 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 1.0 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.1 | 1.0 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 1.2 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.1 | 1.5 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 3.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 0.5 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 0.7 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 1.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 1.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 1.4 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) |
| 0.1 | 0.4 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 1.7 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 1.4 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 3.4 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 2.8 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 0.5 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 2.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 2.3 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.2 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.9 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 4.8 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.7 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.3 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.7 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 1.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 1.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.5 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.2 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 3.2 | GO:0035326 | enhancer binding(GO:0035326) |
| 0.0 | 1.5 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 6.6 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 1.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 1.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 1.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 1.2 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.4 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 29.3 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.3 | 12.4 | PID ATM PATHWAY | ATM pathway |
| 0.3 | 3.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.2 | 8.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 5.2 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 13.8 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 5.2 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 10.0 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 1.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 6.3 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 2.8 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 1.7 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 3.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 3.8 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 2.0 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 3.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 2.8 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 4.0 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.0 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 1.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 1.9 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 2.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 17.3 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.6 | 8.5 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.5 | 8.6 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.4 | 11.0 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.3 | 4.5 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.3 | 4.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.2 | 2.9 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.2 | 4.5 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.2 | 4.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 1.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 1.2 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 3.1 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 4.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 2.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 4.2 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 2.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 6.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 4.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 2.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 2.2 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.1 | 6.7 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 1.0 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 1.8 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 2.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.7 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.7 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 1.0 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.8 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 5.2 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 1.5 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.9 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 2.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 2.3 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 0.7 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.9 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 1.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.8 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |