Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for E2f6
Z-value: 0.81
Transcription factors associated with E2f6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2f6
|
ENSMUSG00000057469.9 | E2f6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2f6 | mm39_v1_chr12_+_16860931_16861002 | -0.37 | 1.5e-03 | Click! |
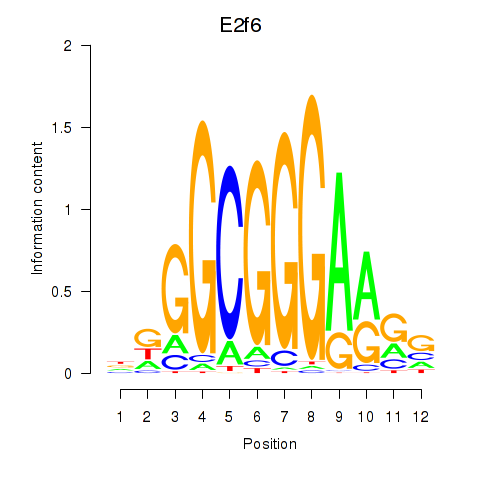
Activity profile of E2f6 motif
Sorted Z-values of E2f6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of E2f6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_74498551 | 6.85 |
ENSMUST00000001872.5
|
Hoxd13
|
homeobox D13 |
| chr12_-_57592907 | 5.29 |
ENSMUST00000044380.8
|
Foxa1
|
forkhead box A1 |
| chr10_+_43455157 | 5.07 |
ENSMUST00000058714.10
|
Cd24a
|
CD24a antigen |
| chr4_-_11386756 | 4.79 |
ENSMUST00000108313.8
ENSMUST00000108311.9 |
Esrp1
|
epithelial splicing regulatory protein 1 |
| chr3_-_30563611 | 4.59 |
ENSMUST00000173899.8
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr1_+_75336965 | 4.45 |
ENSMUST00000027409.10
|
Des
|
desmin |
| chr4_-_11386679 | 4.42 |
ENSMUST00000043781.14
ENSMUST00000108310.8 |
Esrp1
|
epithelial splicing regulatory protein 1 |
| chr18_+_56840813 | 4.04 |
ENSMUST00000025486.9
|
Lmnb1
|
lamin B1 |
| chr4_-_55532453 | 3.44 |
ENSMUST00000132746.2
ENSMUST00000107619.3 |
Klf4
|
Kruppel-like factor 4 (gut) |
| chr7_-_83533497 | 3.35 |
ENSMUST00000094216.5
|
Tlnrd1
|
talin rod domain containing 1 |
| chr4_-_11386394 | 2.92 |
ENSMUST00000155519.2
|
Esrp1
|
epithelial splicing regulatory protein 1 |
| chr13_-_55979191 | 2.89 |
ENSMUST00000021968.7
|
Pitx1
|
paired-like homeodomain transcription factor 1 |
| chr9_+_65797519 | 2.81 |
ENSMUST00000045802.7
|
Pclaf
|
PCNA clamp associated factor |
| chr7_-_132725041 | 2.63 |
ENSMUST00000171022.8
|
Ctbp2
|
C-terminal binding protein 2 |
| chr2_+_31204314 | 2.59 |
ENSMUST00000113532.9
|
Hmcn2
|
hemicentin 2 |
| chr5_-_144294854 | 2.58 |
ENSMUST00000055190.8
|
Baiap2l1
|
BAI1-associated protein 2-like 1 |
| chr7_-_132725075 | 2.54 |
ENSMUST00000163601.8
ENSMUST00000033269.15 ENSMUST00000124096.8 |
Ctbp2
Fgfr2
|
C-terminal binding protein 2 fibroblast growth factor receptor 2 |
| chr17_+_64907697 | 2.50 |
ENSMUST00000086723.10
|
Man2a1
|
mannosidase 2, alpha 1 |
| chr7_-_132725137 | 2.48 |
ENSMUST00000170459.8
|
Ctbp2
|
C-terminal binding protein 2 |
| chr11_+_85723377 | 2.42 |
ENSMUST00000000095.7
|
Tbx2
|
T-box 2 |
| chr13_+_74269554 | 2.37 |
ENSMUST00000036208.7
ENSMUST00000225423.2 ENSMUST00000221703.2 |
Slc9a3
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 |
| chr11_-_75918551 | 2.36 |
ENSMUST00000021207.7
|
Rflnb
|
refilin B |
| chr18_+_75500600 | 2.34 |
ENSMUST00000026999.10
|
Smad7
|
SMAD family member 7 |
| chr2_+_78699360 | 2.32 |
ENSMUST00000028398.14
|
Ube2e3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr9_+_65537834 | 2.29 |
ENSMUST00000055844.10
|
Rbpms2
|
RNA binding protein with multiple splicing 2 |
| chr1_+_74430575 | 2.27 |
ENSMUST00000027367.14
|
Ctdsp1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 |
| chr1_-_180641430 | 2.22 |
ENSMUST00000162814.8
|
H3f3a
|
H3.3 histone A |
| chr7_-_99002430 | 2.21 |
ENSMUST00000094154.6
|
Serpinh1
|
serine (or cysteine) peptidase inhibitor, clade H, member 1 |
| chr17_-_35827676 | 2.20 |
ENSMUST00000160885.2
ENSMUST00000159009.2 ENSMUST00000161012.8 |
Tcf19
|
transcription factor 19 |
| chr19_-_45731290 | 2.14 |
ENSMUST00000111927.8
|
Fgf8
|
fibroblast growth factor 8 |
| chr4_-_127224591 | 2.11 |
ENSMUST00000046532.4
|
Gjb3
|
gap junction protein, beta 3 |
| chr13_-_111945499 | 2.10 |
ENSMUST00000109267.9
|
Map3k1
|
mitogen-activated protein kinase kinase kinase 1 |
| chr6_-_125471666 | 2.10 |
ENSMUST00000032492.9
|
Cd9
|
CD9 antigen |
| chr8_+_106245368 | 2.08 |
ENSMUST00000034363.7
|
Hsd11b2
|
hydroxysteroid 11-beta dehydrogenase 2 |
| chr14_+_99536111 | 2.08 |
ENSMUST00000005279.8
|
Klf5
|
Kruppel-like factor 5 |
| chr4_-_43523745 | 2.07 |
ENSMUST00000150592.2
|
Tpm2
|
tropomyosin 2, beta |
| chrX_+_141464722 | 2.07 |
ENSMUST00000112896.9
|
Tmem164
|
transmembrane protein 164 |
| chr7_+_24069680 | 2.05 |
ENSMUST00000205428.2
ENSMUST00000171904.3 ENSMUST00000205626.2 |
Kcnn4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr9_-_85209340 | 1.97 |
ENSMUST00000187711.2
|
Tent5a
|
terminal nucleotidyltransferase 5A |
| chr11_+_21189277 | 1.96 |
ENSMUST00000109578.8
ENSMUST00000006221.14 |
Vps54
|
VPS54 GARP complex subunit |
| chr7_-_99002204 | 1.96 |
ENSMUST00000208292.2
ENSMUST00000207989.2 ENSMUST00000208749.2 ENSMUST00000169437.9 ENSMUST00000208119.2 ENSMUST00000207849.2 |
Serpinh1
|
serine (or cysteine) peptidase inhibitor, clade H, member 1 |
| chr9_+_118755521 | 1.94 |
ENSMUST00000073109.12
|
Ctdspl
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr8_+_121811091 | 1.93 |
ENSMUST00000181504.2
|
Foxf1
|
forkhead box F1 |
| chr6_-_47571901 | 1.93 |
ENSMUST00000081721.13
ENSMUST00000114618.8 ENSMUST00000114616.8 |
Ezh2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr14_+_56062422 | 1.90 |
ENSMUST00000172271.9
|
Nfatc4
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 4 |
| chr6_+_135042649 | 1.89 |
ENSMUST00000050104.8
|
Gprc5a
|
G protein-coupled receptor, family C, group 5, member A |
| chr1_-_64776890 | 1.89 |
ENSMUST00000116133.4
ENSMUST00000063982.7 |
Fzd5
|
frizzled class receptor 5 |
| chr9_+_45230370 | 1.86 |
ENSMUST00000034597.8
|
Tmprss13
|
transmembrane protease, serine 13 |
| chr15_-_85918378 | 1.85 |
ENSMUST00000016172.10
|
Celsr1
|
cadherin, EGF LAG seven-pass G-type receptor 1 |
| chr15_-_77726333 | 1.84 |
ENSMUST00000016771.13
|
Myh9
|
myosin, heavy polypeptide 9, non-muscle |
| chr6_+_42326934 | 1.82 |
ENSMUST00000203401.3
ENSMUST00000164375.4 |
Zyx
|
zyxin |
| chr8_-_58106057 | 1.82 |
ENSMUST00000034021.12
|
Galnt7
|
polypeptide N-acetylgalactosaminyltransferase 7 |
| chr7_+_142660971 | 1.82 |
ENSMUST00000009689.11
|
Kcnq1
|
potassium voltage-gated channel, subfamily Q, member 1 |
| chr4_-_114766070 | 1.81 |
ENSMUST00000068654.5
|
Foxd2
|
forkhead box D2 |
| chrX_+_72527208 | 1.80 |
ENSMUST00000033741.15
ENSMUST00000169489.2 |
Bgn
|
biglycan |
| chr3_-_88366159 | 1.79 |
ENSMUST00000147200.8
ENSMUST00000169222.8 |
Sema4a
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr14_+_56062252 | 1.78 |
ENSMUST00000024179.6
|
Nfatc4
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 4 |
| chr1_-_180641159 | 1.77 |
ENSMUST00000162118.8
ENSMUST00000159685.2 ENSMUST00000161308.8 |
H3f3a
|
H3.3 histone A |
| chr2_-_65955338 | 1.73 |
ENSMUST00000028378.4
|
Galnt3
|
polypeptide N-acetylgalactosaminyltransferase 3 |
| chr2_+_103800553 | 1.72 |
ENSMUST00000111140.3
ENSMUST00000111139.3 |
Lmo2
|
LIM domain only 2 |
| chr6_-_125290809 | 1.71 |
ENSMUST00000032489.8
|
Ltbr
|
lymphotoxin B receptor |
| chr6_+_43242516 | 1.69 |
ENSMUST00000031750.14
|
Arhgef5
|
Rho guanine nucleotide exchange factor (GEF) 5 |
| chr7_-_28661751 | 1.69 |
ENSMUST00000068045.14
ENSMUST00000217157.2 |
Actn4
|
actinin alpha 4 |
| chr5_+_137348423 | 1.67 |
ENSMUST00000111055.9
|
Ephb4
|
Eph receptor B4 |
| chr5_-_147244074 | 1.67 |
ENSMUST00000031650.4
|
Cdx2
|
caudal type homeobox 2 |
| chr7_-_126625657 | 1.66 |
ENSMUST00000205568.2
|
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr4_-_129142208 | 1.65 |
ENSMUST00000052602.6
|
C77080
|
expressed sequence C77080 |
| chr3_+_122523219 | 1.64 |
ENSMUST00000200389.2
|
Pde5a
|
phosphodiesterase 5A, cGMP-specific |
| chr7_-_132725575 | 1.64 |
ENSMUST00000171968.8
|
Ctbp2
|
C-terminal binding protein 2 |
| chr2_+_103800459 | 1.64 |
ENSMUST00000111143.8
ENSMUST00000138815.2 |
Lmo2
|
LIM domain only 2 |
| chr17_+_47747657 | 1.63 |
ENSMUST00000150819.3
|
AI661453
|
expressed sequence AI661453 |
| chr10_+_126899468 | 1.63 |
ENSMUST00000120226.8
ENSMUST00000133115.8 |
Cdk4
|
cyclin-dependent kinase 4 |
| chr19_-_45731312 | 1.61 |
ENSMUST00000026241.12
ENSMUST00000026240.14 ENSMUST00000111928.8 |
Fgf8
|
fibroblast growth factor 8 |
| chr6_+_42326760 | 1.61 |
ENSMUST00000203652.3
ENSMUST00000070635.13 |
Zyx
|
zyxin |
| chr6_+_17306334 | 1.60 |
ENSMUST00000007799.13
ENSMUST00000115456.6 |
Cav1
|
caveolin 1, caveolae protein |
| chr4_-_59549314 | 1.59 |
ENSMUST00000148331.9
ENSMUST00000030076.12 |
Ptbp3
|
polypyrimidine tract binding protein 3 |
| chr1_+_40720731 | 1.59 |
ENSMUST00000192345.2
|
Slc9a2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 2 |
| chr4_-_131776368 | 1.58 |
ENSMUST00000105981.9
ENSMUST00000084253.10 ENSMUST00000141291.3 |
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr11_-_102208615 | 1.57 |
ENSMUST00000107117.9
|
Ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr17_-_25179635 | 1.57 |
ENSMUST00000024981.9
|
Jpt2
|
Jupiter microtubule associated homolog 2 |
| chrX_+_72830668 | 1.56 |
ENSMUST00000002090.3
|
Ssr4
|
signal sequence receptor, delta |
| chr5_+_137348363 | 1.55 |
ENSMUST00000061244.15
|
Ephb4
|
Eph receptor B4 |
| chrX_+_141464393 | 1.54 |
ENSMUST00000112889.8
ENSMUST00000101198.9 ENSMUST00000112891.8 ENSMUST00000087333.9 |
Tmem164
|
transmembrane protein 164 |
| chr13_-_100912308 | 1.54 |
ENSMUST00000075550.4
|
Cenph
|
centromere protein H |
| chrX_-_140508177 | 1.54 |
ENSMUST00000067841.8
|
Irs4
|
insulin receptor substrate 4 |
| chr2_-_155676765 | 1.54 |
ENSMUST00000029143.7
ENSMUST00000239423.2 |
Fam83c
|
family with sequence similarity 83, member C |
| chr17_+_47747540 | 1.53 |
ENSMUST00000037701.13
|
AI661453
|
expressed sequence AI661453 |
| chr5_+_137348638 | 1.52 |
ENSMUST00000166239.8
ENSMUST00000111054.2 |
Ephb4
|
Eph receptor B4 |
| chr1_+_172309337 | 1.52 |
ENSMUST00000127052.8
|
Igsf9
|
immunoglobulin superfamily, member 9 |
| chr11_+_72851989 | 1.52 |
ENSMUST00000163326.8
ENSMUST00000108485.9 ENSMUST00000021142.8 ENSMUST00000108486.8 ENSMUST00000108484.8 |
Atp2a3
|
ATPase, Ca++ transporting, ubiquitous |
| chr14_+_55777723 | 1.51 |
ENSMUST00000048781.4
ENSMUST00000226519.2 |
Pck2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr17_+_57369231 | 1.51 |
ENSMUST00000097299.10
ENSMUST00000169543.8 ENSMUST00000163763.2 |
Crb3
|
crumbs family member 3 |
| chr6_+_17306379 | 1.50 |
ENSMUST00000115455.3
|
Cav1
|
caveolin 1, caveolae protein |
| chr1_+_172309766 | 1.49 |
ENSMUST00000135267.8
ENSMUST00000052629.13 ENSMUST00000111235.8 |
Igsf9
|
immunoglobulin superfamily, member 9 |
| chr1_+_86454431 | 1.49 |
ENSMUST00000045897.15
ENSMUST00000186255.7 ENSMUST00000188699.7 |
Ptma
|
prothymosin alpha |
| chr7_-_132724889 | 1.48 |
ENSMUST00000166439.8
|
Ctbp2
|
C-terminal binding protein 2 |
| chr1_+_151447124 | 1.47 |
ENSMUST00000148810.8
|
Niban1
|
niban apoptosis regulator 1 |
| chr8_-_58106027 | 1.47 |
ENSMUST00000110316.3
|
Galnt7
|
polypeptide N-acetylgalactosaminyltransferase 7 |
| chr4_+_53440516 | 1.47 |
ENSMUST00000107651.9
ENSMUST00000107647.8 |
Slc44a1
|
solute carrier family 44, member 1 |
| chr3_-_89325594 | 1.46 |
ENSMUST00000029679.4
|
Cks1b
|
CDC28 protein kinase 1b |
| chr1_-_71692320 | 1.45 |
ENSMUST00000186940.7
ENSMUST00000188894.7 ENSMUST00000188674.7 ENSMUST00000189821.7 ENSMUST00000187938.7 ENSMUST00000190780.7 ENSMUST00000186736.2 ENSMUST00000055226.13 ENSMUST00000186129.7 |
Fn1
|
fibronectin 1 |
| chr9_-_110571645 | 1.44 |
ENSMUST00000006005.12
|
Pth1r
|
parathyroid hormone 1 receptor |
| chr11_+_62539657 | 1.44 |
ENSMUST00000127589.2
ENSMUST00000155759.3 |
Mmgt2
|
membrane magnesium transporter 2 |
| chr10_+_126899396 | 1.44 |
ENSMUST00000006911.12
|
Cdk4
|
cyclin-dependent kinase 4 |
| chr3_-_30563831 | 1.43 |
ENSMUST00000173495.8
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr6_-_70769135 | 1.42 |
ENSMUST00000066134.6
|
Rpia
|
ribose 5-phosphate isomerase A |
| chr9_-_21709796 | 1.42 |
ENSMUST00000213691.2
|
Kank2
|
KN motif and ankyrin repeat domains 2 |
| chr13_+_30320446 | 1.41 |
ENSMUST00000047311.16
|
Mboat1
|
membrane bound O-acyltransferase domain containing 1 |
| chr6_+_113448388 | 1.41 |
ENSMUST00000058300.14
|
Il17rc
|
interleukin 17 receptor C |
| chr4_-_59549243 | 1.40 |
ENSMUST00000173699.8
ENSMUST00000173884.8 ENSMUST00000102883.11 ENSMUST00000174586.8 |
Ptbp3
|
polypyrimidine tract binding protein 3 |
| chrX_-_47543029 | 1.40 |
ENSMUST00000114958.8
|
Elf4
|
E74-like factor 4 (ets domain transcription factor) |
| chr13_-_55983946 | 1.40 |
ENSMUST00000173618.2
|
Pitx1
|
paired-like homeodomain transcription factor 1 |
| chr10_-_4337435 | 1.40 |
ENSMUST00000100077.5
|
Zbtb2
|
zinc finger and BTB domain containing 2 |
| chr5_-_137529465 | 1.40 |
ENSMUST00000150063.9
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr17_-_28569721 | 1.40 |
ENSMUST00000156862.3
|
Tead3
|
TEA domain family member 3 |
| chr7_-_28981335 | 1.39 |
ENSMUST00000108236.5
ENSMUST00000098604.12 |
Spint2
|
serine protease inhibitor, Kunitz type 2 |
| chr3_-_30563919 | 1.38 |
ENSMUST00000172697.8
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr4_-_133694607 | 1.37 |
ENSMUST00000105893.8
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr3_-_89300936 | 1.37 |
ENSMUST00000124783.8
ENSMUST00000126027.8 |
Zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr19_-_3625698 | 1.36 |
ENSMUST00000172362.3
ENSMUST00000025846.16 ENSMUST00000226109.2 ENSMUST00000113997.9 |
Ppp6r3
|
protein phosphatase 6, regulatory subunit 3 |
| chr7_+_144391786 | 1.36 |
ENSMUST00000155320.8
|
Fgf3
|
fibroblast growth factor 3 |
| chrX_+_72830607 | 1.36 |
ENSMUST00000166518.8
|
Ssr4
|
signal sequence receptor, delta |
| chr8_+_57964956 | 1.35 |
ENSMUST00000210871.2
|
Hmgb2
|
high mobility group box 2 |
| chr15_+_82225380 | 1.35 |
ENSMUST00000050349.3
|
Pheta2
|
PH domain containing endocytic trafficking adaptor 2 |
| chr1_+_86454511 | 1.34 |
ENSMUST00000188533.2
|
Ptma
|
prothymosin alpha |
| chr9_-_35028100 | 1.34 |
ENSMUST00000034537.8
|
St3gal4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr2_+_139520098 | 1.34 |
ENSMUST00000184404.8
ENSMUST00000099307.4 |
Ism1
|
isthmin 1, angiogenesis inhibitor |
| chr4_-_133694543 | 1.33 |
ENSMUST00000123234.8
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr17_-_34694911 | 1.33 |
ENSMUST00000065841.5
|
Btnl4
|
butyrophilin-like 4 |
| chr5_-_137529251 | 1.32 |
ENSMUST00000132525.8
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chrX_+_99773523 | 1.31 |
ENSMUST00000019503.14
|
Gdpd2
|
glycerophosphodiester phosphodiesterase domain containing 2 |
| chr8_-_106074585 | 1.31 |
ENSMUST00000014922.5
|
Fhod1
|
formin homology 2 domain containing 1 |
| chr4_-_91260182 | 1.30 |
ENSMUST00000176362.2
|
Elavl2
|
ELAV like RNA binding protein 1 |
| chr2_-_34803988 | 1.30 |
ENSMUST00000028232.7
ENSMUST00000202907.2 |
Phf19
|
PHD finger protein 19 |
| chr3_-_88366351 | 1.29 |
ENSMUST00000165898.8
ENSMUST00000127436.8 |
Sema4a
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr16_+_57369595 | 1.29 |
ENSMUST00000159414.2
|
Filip1l
|
filamin A interacting protein 1-like |
| chr7_-_28078671 | 1.29 |
ENSMUST00000209061.2
|
Zfp36
|
zinc finger protein 36 |
| chr6_+_42326980 | 1.28 |
ENSMUST00000203849.2
|
Zyx
|
zyxin |
| chrX_+_99773784 | 1.28 |
ENSMUST00000113744.2
|
Gdpd2
|
glycerophosphodiester phosphodiesterase domain containing 2 |
| chr9_-_57552844 | 1.28 |
ENSMUST00000216979.2
ENSMUST00000034863.8 |
Csk
|
c-src tyrosine kinase |
| chr8_+_84724130 | 1.27 |
ENSMUST00000095228.5
|
Samd1
|
sterile alpha motif domain containing 1 |
| chr15_-_102165884 | 1.27 |
ENSMUST00000043172.15
|
Rarg
|
retinoic acid receptor, gamma |
| chr18_+_64473091 | 1.27 |
ENSMUST00000175965.10
|
Onecut2
|
one cut domain, family member 2 |
| chr15_+_61857226 | 1.27 |
ENSMUST00000161976.8
ENSMUST00000022971.8 |
Myc
|
myelocytomatosis oncogene |
| chr6_-_51989456 | 1.26 |
ENSMUST00000078214.8
ENSMUST00000204778.3 |
Skap2
|
src family associated phosphoprotein 2 |
| chr19_+_6356486 | 1.26 |
ENSMUST00000025681.8
|
Cdc42bpg
|
CDC42 binding protein kinase gamma (DMPK-like) |
| chr11_-_115427007 | 1.25 |
ENSMUST00000118155.8
ENSMUST00000153892.2 |
Sumo2
|
small ubiquitin-like modifier 2 |
| chr15_+_61857390 | 1.24 |
ENSMUST00000159327.2
ENSMUST00000167731.8 |
Myc
|
myelocytomatosis oncogene |
| chr9_-_57552392 | 1.24 |
ENSMUST00000216934.2
|
Csk
|
c-src tyrosine kinase |
| chr8_-_106553822 | 1.23 |
ENSMUST00000239468.2
ENSMUST00000041400.6 |
Ranbp10
|
RAN binding protein 10 |
| chr5_-_134776101 | 1.23 |
ENSMUST00000015138.13
|
Eln
|
elastin |
| chr8_+_54530530 | 1.23 |
ENSMUST00000033919.6
|
Vegfc
|
vascular endothelial growth factor C |
| chr2_-_34262012 | 1.22 |
ENSMUST00000113132.9
ENSMUST00000040638.15 |
Pbx3
|
pre B cell leukemia homeobox 3 |
| chr9_-_66951025 | 1.22 |
ENSMUST00000113695.8
|
Tpm1
|
tropomyosin 1, alpha |
| chr11_+_96242422 | 1.22 |
ENSMUST00000100523.7
|
Hoxb2
|
homeobox B2 |
| chr6_+_17306414 | 1.21 |
ENSMUST00000150901.2
|
Cav1
|
caveolin 1, caveolae protein |
| chr2_+_13579092 | 1.21 |
ENSMUST00000193675.2
|
Vim
|
vimentin |
| chr7_-_132724344 | 1.21 |
ENSMUST00000167218.8
|
Ctbp2
|
C-terminal binding protein 2 |
| chr12_+_8821404 | 1.21 |
ENSMUST00000171158.8
|
Sdc1
|
syndecan 1 |
| chr6_+_54303837 | 1.21 |
ENSMUST00000059138.6
|
Prr15
|
proline rich 15 |
| chr11_+_118913788 | 1.20 |
ENSMUST00000026662.8
|
Cbx2
|
chromobox 2 |
| chr6_+_42326714 | 1.20 |
ENSMUST00000203846.3
|
Zyx
|
zyxin |
| chr11_+_98798627 | 1.20 |
ENSMUST00000092706.13
|
Cdc6
|
cell division cycle 6 |
| chr15_+_59520199 | 1.20 |
ENSMUST00000067543.8
|
Trib1
|
tribbles pseudokinase 1 |
| chr11_+_102495189 | 1.20 |
ENSMUST00000057893.7
|
Fzd2
|
frizzled class receptor 2 |
| chr12_+_8821317 | 1.19 |
ENSMUST00000020911.14
|
Sdc1
|
syndecan 1 |
| chrX_-_11946785 | 1.18 |
ENSMUST00000115513.9
ENSMUST00000115512.9 ENSMUST00000065143.14 ENSMUST00000124033.8 |
Bcor
|
BCL6 interacting corepressor |
| chr4_-_11007635 | 1.18 |
ENSMUST00000054776.4
|
Plekhf2
|
pleckstrin homology domain containing, family F (with FYVE domain) member 2 |
| chr5_+_32293145 | 1.16 |
ENSMUST00000031017.11
|
Fosl2
|
fos-like antigen 2 |
| chr18_-_64794338 | 1.16 |
ENSMUST00000025482.10
|
Atp8b1
|
ATPase, class I, type 8B, member 1 |
| chr18_-_14105591 | 1.16 |
ENSMUST00000234025.2
|
Zfp521
|
zinc finger protein 521 |
| chr9_-_66951151 | 1.16 |
ENSMUST00000113696.8
|
Tpm1
|
tropomyosin 1, alpha |
| chr13_-_104057016 | 1.16 |
ENSMUST00000022222.12
|
Erbin
|
Erbb2 interacting protein |
| chr11_+_94900677 | 1.16 |
ENSMUST00000055947.10
|
Samd14
|
sterile alpha motif domain containing 14 |
| chr17_-_28569574 | 1.15 |
ENSMUST00000114799.8
ENSMUST00000219703.3 |
Tead3
|
TEA domain family member 3 |
| chr9_+_100525501 | 1.15 |
ENSMUST00000146312.8
ENSMUST00000129269.8 |
Stag1
|
stromal antigen 1 |
| chr13_+_48816466 | 1.15 |
ENSMUST00000021813.5
|
Barx1
|
BarH-like homeobox 1 |
| chr12_+_24758240 | 1.15 |
ENSMUST00000020980.12
|
Rrm2
|
ribonucleotide reductase M2 |
| chr8_-_11362731 | 1.15 |
ENSMUST00000033898.10
|
Col4a1
|
collagen, type IV, alpha 1 |
| chr5_+_117457126 | 1.15 |
ENSMUST00000111967.8
|
Vsig10
|
V-set and immunoglobulin domain containing 10 |
| chr10_-_128237087 | 1.13 |
ENSMUST00000042666.13
|
Slc39a5
|
solute carrier family 39 (metal ion transporter), member 5 |
| chr7_-_28661648 | 1.12 |
ENSMUST00000127210.8
|
Actn4
|
actinin alpha 4 |
| chr19_-_5713728 | 1.12 |
ENSMUST00000169854.2
|
Sipa1
|
signal-induced proliferation associated gene 1 |
| chr7_+_121464254 | 1.12 |
ENSMUST00000033161.7
|
Scnn1b
|
sodium channel, nonvoltage-gated 1 beta |
| chr15_-_102165740 | 1.11 |
ENSMUST00000135466.2
|
Rarg
|
retinoic acid receptor, gamma |
| chr7_+_36397426 | 1.11 |
ENSMUST00000021641.8
|
Tshz3
|
teashirt zinc finger family member 3 |
| chr10_-_128361731 | 1.10 |
ENSMUST00000026427.8
|
Esyt1
|
extended synaptotagmin-like protein 1 |
| chr17_+_35295909 | 1.10 |
ENSMUST00000013910.5
|
Ly6g6e
|
lymphocyte antigen 6 complex, locus G6E |
| chr1_-_75119277 | 1.09 |
ENSMUST00000168720.8
ENSMUST00000041213.12 ENSMUST00000189809.2 |
Cnppd1
|
cyclin Pas1/PHO80 domain containing 1 |
| chr2_+_143757193 | 1.09 |
ENSMUST00000103172.4
|
Dstn
|
destrin |
| chr3_+_89325750 | 1.08 |
ENSMUST00000039110.12
ENSMUST00000125036.8 ENSMUST00000191485.7 ENSMUST00000154791.8 |
Shc1
|
src homology 2 domain-containing transforming protein C1 |
| chr17_+_29020064 | 1.08 |
ENSMUST00000004985.11
|
Brpf3
|
bromodomain and PHD finger containing, 3 |
| chr13_-_40887244 | 1.08 |
ENSMUST00000110193.9
|
Tfap2a
|
transcription factor AP-2, alpha |
| chr9_+_100525637 | 1.08 |
ENSMUST00000041418.13
|
Stag1
|
stromal antigen 1 |
| chr11_-_70128678 | 1.08 |
ENSMUST00000108575.9
|
0610010K14Rik
|
RIKEN cDNA 0610010K14 gene |
| chr7_+_28050077 | 1.07 |
ENSMUST00000082134.6
|
Rps16
|
ribosomal protein S16 |
| chr9_+_108216233 | 1.07 |
ENSMUST00000082429.8
|
Gpx1
|
glutathione peroxidase 1 |
| chr3_+_89986831 | 1.06 |
ENSMUST00000029549.16
|
Tpm3
|
tropomyosin 3, gamma |
| chr14_+_54669054 | 1.06 |
ENSMUST00000089688.6
ENSMUST00000225641.2 |
Mmp14
|
matrix metallopeptidase 14 (membrane-inserted) |
| chr3_-_86049988 | 1.06 |
ENSMUST00000029722.7
|
Rps3a1
|
ribosomal protein S3A1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.2 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 1.7 | 5.1 | GO:0034118 | erythrocyte aggregation(GO:0034117) regulation of erythrocyte aggregation(GO:0034118) |
| 1.1 | 4.4 | GO:0045415 | negative regulation of interleukin-8 biosynthetic process(GO:0045415) |
| 1.1 | 4.3 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 1.0 | 4.0 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 1.0 | 4.9 | GO:0031509 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 1.0 | 12.5 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.9 | 3.7 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.9 | 5.1 | GO:1903609 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.8 | 2.5 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.8 | 2.4 | GO:0048627 | myoblast development(GO:0048627) |
| 0.8 | 4.7 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.8 | 3.8 | GO:0060128 | corticotropin hormone secreting cell differentiation(GO:0060128) |
| 0.7 | 2.9 | GO:0009609 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) response to selenium ion(GO:0010269) |
| 0.7 | 1.4 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.7 | 2.8 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.7 | 2.1 | GO:0002017 | regulation of blood volume by renal aldosterone(GO:0002017) |
| 0.7 | 0.7 | GO:0072554 | blood vessel lumenization(GO:0072554) |
| 0.7 | 2.0 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.7 | 7.4 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.7 | 2.7 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.6 | 1.9 | GO:0007439 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.6 | 3.8 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.6 | 1.9 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.6 | 2.5 | GO:0090095 | lactic acid secretion(GO:0046722) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.6 | 1.8 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.6 | 3.0 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.6 | 1.8 | GO:0072347 | response to anesthetic(GO:0072347) |
| 0.6 | 2.4 | GO:1900158 | negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.6 | 6.8 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.6 | 1.7 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.5 | 2.2 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.5 | 1.6 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.5 | 3.2 | GO:0060853 | arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) Notch signaling pathway involved in arterial endothelial cell fate commitment(GO:0060853) blood vessel endothelial cell fate specification(GO:0097101) |
| 0.5 | 0.9 | GO:0061314 | Notch signaling involved in heart development(GO:0061314) |
| 0.5 | 1.4 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.5 | 1.4 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.5 | 0.5 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.5 | 1.8 | GO:1900756 | protein processing in phagocytic vesicle(GO:1900756) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 0.5 | 0.5 | GO:0060994 | regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) |
| 0.5 | 7.3 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.5 | 1.8 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.5 | 1.4 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.4 | 2.2 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.4 | 3.1 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.4 | 1.3 | GO:1904582 | positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.4 | 3.4 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.4 | 1.2 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.4 | 1.6 | GO:0003017 | lymph circulation(GO:0003017) |
| 0.4 | 1.9 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.4 | 1.9 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.4 | 2.3 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.4 | 1.1 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.4 | 2.2 | GO:0061209 | cell proliferation involved in mesonephros development(GO:0061209) |
| 0.4 | 2.5 | GO:0042997 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) negative regulation of Golgi to plasma membrane protein transport(GO:0042997) negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.4 | 1.4 | GO:0015904 | tetracycline transport(GO:0015904) |
| 0.4 | 1.4 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.4 | 0.7 | GO:0070368 | positive regulation of hepatocyte differentiation(GO:0070368) |
| 0.4 | 3.9 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.3 | 1.0 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.3 | 2.4 | GO:0003256 | regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003256) |
| 0.3 | 1.0 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.3 | 0.7 | GO:0070094 | positive regulation of glucagon secretion(GO:0070094) |
| 0.3 | 2.0 | GO:1990839 | response to endothelin(GO:1990839) |
| 0.3 | 2.0 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.3 | 1.6 | GO:0072081 | proximal/distal pattern formation involved in nephron development(GO:0072047) specification of nephron tubule identity(GO:0072081) specification of loop of Henle identity(GO:0072086) |
| 0.3 | 1.9 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.3 | 0.3 | GO:1904783 | positive regulation of NMDA glutamate receptor activity(GO:1904783) |
| 0.3 | 0.9 | GO:1904828 | regulation of hydrogen sulfide biosynthetic process(GO:1904826) positive regulation of hydrogen sulfide biosynthetic process(GO:1904828) |
| 0.3 | 1.5 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.3 | 2.4 | GO:0015871 | choline transport(GO:0015871) |
| 0.3 | 0.9 | GO:2000077 | transepithelial water transport(GO:0035377) positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) positive regulation of establishment of Sertoli cell barrier(GO:1904446) negative regulation of type B pancreatic cell development(GO:2000077) |
| 0.3 | 3.8 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.3 | 1.2 | GO:0017126 | nucleologenesis(GO:0017126) |
| 0.3 | 1.7 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.3 | 12.4 | GO:0060441 | epithelial tube branching involved in lung morphogenesis(GO:0060441) |
| 0.3 | 3.4 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.3 | 1.6 | GO:0002678 | positive regulation of chronic inflammatory response(GO:0002678) positive regulation of oocyte development(GO:0060282) |
| 0.3 | 1.4 | GO:0021763 | subthalamic nucleus development(GO:0021763) superior vena cava morphogenesis(GO:0060578) |
| 0.3 | 2.7 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.3 | 2.2 | GO:0015675 | nickel cation transport(GO:0015675) |
| 0.3 | 1.9 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.3 | 2.1 | GO:0044838 | cell quiescence(GO:0044838) |
| 0.3 | 2.9 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.3 | 2.1 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.3 | 2.3 | GO:2000320 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.3 | 3.9 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.3 | 0.5 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.2 | 1.2 | GO:1903677 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.2 | 3.0 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.2 | 1.2 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.2 | 1.1 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.2 | 3.4 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.2 | 1.6 | GO:0003383 | apical constriction(GO:0003383) |
| 0.2 | 0.9 | GO:0097298 | regulation of nucleus size(GO:0097298) |
| 0.2 | 0.7 | GO:1904093 | regulation of autophagic cell death(GO:1904092) negative regulation of autophagic cell death(GO:1904093) |
| 0.2 | 0.9 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.2 | 0.9 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.2 | 1.1 | GO:0043974 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.2 | 1.9 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.2 | 0.6 | GO:1904632 | negative regulation of interleukin-12 biosynthetic process(GO:0045083) response to diterpene(GO:1904629) cellular response to diterpene(GO:1904630) response to glucoside(GO:1904631) cellular response to glucoside(GO:1904632) |
| 0.2 | 1.9 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.2 | 1.0 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.2 | 1.0 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.2 | 3.7 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.2 | 0.6 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.2 | 3.2 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.2 | 0.6 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.2 | 0.8 | GO:0060709 | glycogen cell differentiation involved in embryonic placenta development(GO:0060709) |
| 0.2 | 0.8 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.2 | 0.6 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.2 | 3.4 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.2 | 0.6 | GO:1990428 | miRNA transport(GO:1990428) |
| 0.2 | 1.8 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.2 | 0.2 | GO:0002085 | inhibition of neuroepithelial cell differentiation(GO:0002085) |
| 0.2 | 0.6 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.2 | 1.2 | GO:0070425 | negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 0.2 | 0.4 | GO:1903433 | regulation of constitutive secretory pathway(GO:1903433) |
| 0.2 | 4.2 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.2 | 1.5 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.2 | 0.6 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 | 2.8 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.2 | 0.6 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.2 | 1.7 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.2 | 0.4 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.2 | 0.6 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 0.2 | 0.5 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) rRNA export from nucleus(GO:0006407) |
| 0.2 | 0.9 | GO:2000327 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.2 | 0.9 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.2 | 1.1 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.2 | 0.7 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.2 | 2.0 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.2 | 0.7 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.2 | 0.7 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.2 | 0.5 | GO:0010767 | regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) |
| 0.2 | 0.5 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.2 | 0.9 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.2 | 2.3 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.2 | 4.0 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.2 | 0.5 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.2 | 0.7 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.2 | 0.5 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.2 | 0.5 | GO:1990117 | release of matrix enzymes from mitochondria(GO:0032976) B cell receptor apoptotic signaling pathway(GO:1990117) |
| 0.2 | 0.7 | GO:0040030 | regulation of molecular function, epigenetic(GO:0040030) |
| 0.2 | 0.5 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 0.2 | 1.0 | GO:0014826 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) |
| 0.2 | 0.3 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 0.2 | 2.8 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.2 | 0.2 | GO:1903699 | tarsal gland development(GO:1903699) |
| 0.2 | 0.7 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.2 | 0.7 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.2 | 0.7 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.2 | 0.6 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.2 | 0.3 | GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 0.2 | 0.5 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.2 | 0.3 | GO:0097369 | sodium ion import(GO:0097369) |
| 0.2 | 3.0 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.2 | 0.5 | GO:0045660 | positive regulation of neutrophil differentiation(GO:0045660) |
| 0.2 | 0.2 | GO:1905073 | occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 0.2 | 0.9 | GO:0032954 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.2 | 4.5 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.2 | 1.5 | GO:0070813 | hydrogen sulfide metabolic process(GO:0070813) |
| 0.2 | 0.8 | GO:0043091 | regulation of amino acid import(GO:0010958) L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.2 | 0.6 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.2 | 1.1 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) |
| 0.2 | 2.6 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.1 | 0.9 | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) |
| 0.1 | 0.6 | GO:0034552 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.1 | 0.6 | GO:0090289 | regulation of osteoclast proliferation(GO:0090289) positive regulation of osteoclast proliferation(GO:0090290) |
| 0.1 | 1.7 | GO:0035878 | nail development(GO:0035878) |
| 0.1 | 0.6 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.1 | 0.4 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.1 | 1.4 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.1 | 3.1 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.1 | 1.3 | GO:0032075 | positive regulation of nuclease activity(GO:0032075) |
| 0.1 | 0.6 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.1 | 1.0 | GO:0033578 | protein glycosylation in Golgi(GO:0033578) |
| 0.1 | 1.4 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.1 | 0.7 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.1 | 0.3 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.1 | 1.2 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 | 0.9 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 1.0 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.1 | 0.4 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 0.2 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.1 | 1.9 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 0.6 | GO:1902732 | positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.1 | 2.2 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.1 | 0.7 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 1.0 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 | 0.7 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.1 | 0.5 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.1 | 0.3 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.1 | 0.5 | GO:0060022 | hard palate development(GO:0060022) |
| 0.1 | 0.5 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.1 | 0.7 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.6 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.1 | 0.5 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 | 0.6 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.1 | 0.9 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 | 0.8 | GO:0060430 | lung saccule development(GO:0060430) |
| 0.1 | 0.3 | GO:0046832 | negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.1 | 1.1 | GO:0015791 | polyol transport(GO:0015791) |
| 0.1 | 0.7 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.1 | 0.8 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.1 | 0.7 | GO:2001045 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.4 | GO:0097296 | mineralocorticoid receptor signaling pathway(GO:0031959) activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.1 | 1.7 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 0.7 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.1 | 0.9 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 0.9 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 | 0.2 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 0.1 | 2.0 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.1 | 1.5 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 0.7 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.1 | 0.5 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.1 | 3.2 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 0.5 | GO:0030202 | heparin metabolic process(GO:0030202) |
| 0.1 | 3.0 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.1 | 1.1 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 0.9 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.1 | 0.7 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.1 | 0.6 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 | 2.7 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 2.6 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 0.5 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.1 | 1.6 | GO:2001046 | positive regulation of integrin-mediated signaling pathway(GO:2001046) |
| 0.1 | 2.2 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.1 | 0.7 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.1 | 0.3 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.1 | 0.5 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.1 | 0.5 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.6 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.1 | 0.3 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 0.2 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.1 | 2.1 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.1 | 1.1 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.1 | 0.4 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.1 | 3.0 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.1 | 0.4 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 0.4 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.1 | 1.7 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 1.6 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.1 | 0.8 | GO:0042538 | hyperosmotic salinity response(GO:0042538) |
| 0.1 | 1.0 | GO:2000489 | hepatic stellate cell activation(GO:0035733) regulation of hepatic stellate cell activation(GO:2000489) |
| 0.1 | 0.3 | GO:0010645 | atrial ventricular junction remodeling(GO:0003294) regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) regulation of renin secretion into blood stream(GO:1900133) |
| 0.1 | 0.7 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.1 | 0.4 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 | 0.9 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.4 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.1 | 0.2 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.1 | 0.1 | GO:0072131 | kidney mesenchyme morphogenesis(GO:0072131) metanephric mesenchyme morphogenesis(GO:0072133) |
| 0.1 | 0.9 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.1 | 0.8 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 0.2 | GO:0060722 | spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) |
| 0.1 | 1.2 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 1.5 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.1 | 1.6 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.1 | 0.8 | GO:0070932 | histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.1 | 0.5 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.1 | 0.5 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.1 | 0.3 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.1 | 0.8 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 1.4 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.1 | 1.3 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 0.2 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 | 1.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 1.5 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.1 | 1.7 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.1 | 0.9 | GO:0007320 | insemination(GO:0007320) |
| 0.1 | 0.5 | GO:2000561 | CD4-positive, alpha-beta T cell proliferation(GO:0035739) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.1 | 0.5 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.1 | 4.9 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.1 | 0.7 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.1 | 0.1 | GO:0051385 | response to mineralocorticoid(GO:0051385) |
| 0.1 | 0.5 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 0.7 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.1 | 1.3 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 0.4 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.1 | 0.6 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 0.7 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.1 | 0.7 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 0.4 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 1.5 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 0.4 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 0.5 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.1 | 0.7 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.1 | 0.4 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 0.1 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) renal outer medulla development(GO:0072054) |
| 0.1 | 0.3 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.1 | 0.6 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 0.3 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.1 | 0.3 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.1 | 0.4 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.1 | 0.4 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.1 | 0.7 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) |
| 0.1 | 0.2 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 0.7 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 0.2 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.6 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 1.1 | GO:0050891 | multicellular organismal water homeostasis(GO:0050891) |
| 0.1 | 2.1 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.5 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 2.3 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 2.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 0.3 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.1 | 0.8 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.1 | 0.3 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.1 | 0.4 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 0.7 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.1 | 0.3 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 0.4 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.1 | 5.6 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.1 | 0.5 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.1 | 0.6 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.1 | 0.1 | GO:1901536 | negative regulation of DNA demethylation(GO:1901536) |
| 0.1 | 0.6 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.1 | 0.6 | GO:0048485 | sympathetic nervous system development(GO:0048485) |
| 0.1 | 0.1 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.1 | 1.0 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.3 | GO:0007549 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.1 | 0.5 | GO:0021562 | vestibulocochlear nerve development(GO:0021562) |
| 0.1 | 0.4 | GO:1900025 | negative regulation of substrate adhesion-dependent cell spreading(GO:1900025) |
| 0.0 | 0.2 | GO:0070431 | nucleotide-binding domain, leucine rich repeat containing receptor signaling pathway(GO:0035872) nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070431) |
| 0.0 | 0.6 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.4 | GO:0061430 | bone trabecula formation(GO:0060346) bone trabecula morphogenesis(GO:0061430) |
| 0.0 | 1.0 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 0.1 | GO:0002842 | positive regulation of T cell mediated immune response to tumor cell(GO:0002842) |
| 0.0 | 0.5 | GO:0045060 | negative thymic T cell selection(GO:0045060) |
| 0.0 | 0.6 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.3 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.3 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.9 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.0 | 0.3 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.4 | GO:1902510 | regulation of apoptotic DNA fragmentation(GO:1902510) |
| 0.0 | 0.2 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.0 | 0.6 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.4 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 | 0.4 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.8 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.0 | 0.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0021769 | orbitofrontal cortex development(GO:0021769) fibroblast growth factor receptor signaling pathway involved in orbitofrontal cortex development(GO:0035607) |
| 0.0 | 0.3 | GO:2000173 | negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.0 | 0.7 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 1.0 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.1 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.4 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.0 | 0.2 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.0 | 0.3 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.2 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.6 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 1.1 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 1.2 | GO:0032330 | regulation of chondrocyte differentiation(GO:0032330) |
| 0.0 | 0.0 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.8 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.4 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.3 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 | 0.4 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.4 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.0 | 0.8 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 0.2 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.2 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.1 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.0 | 0.1 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.0 | 0.5 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.0 | 0.1 | GO:0060527 | prostate glandular acinus morphogenesis(GO:0060526) prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis(GO:0060527) |
| 0.0 | 0.8 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.6 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.3 | GO:0006569 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) |
| 0.0 | 0.1 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.5 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0071317 | cellular response to isoquinoline alkaloid(GO:0071317) |
| 0.0 | 0.2 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.7 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.2 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.9 | GO:0002029 | desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) negative adaptation of signaling pathway(GO:0022401) |
| 0.0 | 1.1 | GO:0048146 | positive regulation of fibroblast proliferation(GO:0048146) |
| 0.0 | 0.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.9 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 1.1 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.4 | GO:0021930 | cell proliferation in external granule layer(GO:0021924) cerebellar granule cell precursor proliferation(GO:0021930) |
| 0.0 | 0.1 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.1 | GO:2001245 | negative regulation of phospholipid biosynthetic process(GO:0071072) negative regulation of phospholipid metabolic process(GO:1903726) regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 | 1.0 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.5 | GO:1904293 | negative regulation of ERAD pathway(GO:1904293) |
| 0.0 | 0.1 | GO:1903898 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.0 | 0.3 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.0 | 0.4 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.2 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 1.1 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 1.3 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 1.0 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 2.1 | GO:0006939 | smooth muscle contraction(GO:0006939) |
| 0.0 | 0.1 | GO:0033599 | regulation of mammary gland epithelial cell proliferation(GO:0033599) |
| 0.0 | 0.8 | GO:0045682 | regulation of epidermis development(GO:0045682) |
| 0.0 | 0.3 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.0 | 0.2 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.0 | 0.8 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.8 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.5 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.0 | 0.1 | GO:0031440 | regulation of mRNA 3'-end processing(GO:0031440) |
| 0.0 | 0.2 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.8 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.1 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.0 | 0.3 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.4 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 1.0 | GO:0007492 | endoderm development(GO:0007492) |
| 0.0 | 0.9 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 1.3 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 2.3 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.8 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.2 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.1 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.0 | 0.3 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.2 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.9 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.7 | GO:0048536 | spleen development(GO:0048536) |
| 0.0 | 0.7 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.4 | GO:0039529 | RIG-I signaling pathway(GO:0039529) |
| 0.0 | 0.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.2 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.1 | GO:1990441 | mRNA splicing via endonucleolytic cleavage and ligation involved in unfolded protein response(GO:0030969) regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) mRNA splicing, via endonucleolytic cleavage and ligation(GO:0070054) mRNA endonucleolytic cleavage involved in unfolded protein response(GO:0070055) response to L-glutamate(GO:1902065) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) negative regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990441) |
| 0.0 | 0.5 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 | 0.1 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.1 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.0 | 0.3 | GO:0032682 | negative regulation of chemokine production(GO:0032682) |
| 0.0 | 0.2 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.2 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.4 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.0 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.1 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.0 | 0.7 | GO:0048661 | positive regulation of smooth muscle cell proliferation(GO:0048661) |
| 0.0 | 0.1 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.1 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 0.9 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 0.0 | 0.9 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.4 | GO:0006084 | acetyl-CoA metabolic process(GO:0006084) |
| 0.0 | 0.6 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 0.2 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 0.3 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.0 | 0.1 | GO:0001675 | acrosome assembly(GO:0001675) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.9 | GO:0001740 | Barr body(GO:0001740) |
| 0.7 | 2.0 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.6 | 3.8 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.6 | 6.5 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.6 | 4.0 | GO:0005638 | lamin filament(GO:0005638) |
| 0.5 | 7.6 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.4 | 2.9 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.4 | 2.8 | GO:0098645 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.4 | 2.0 | GO:0000938 | GARP complex(GO:0000938) |
| 0.4 | 3.1 | GO:0097413 | Lewy body(GO:0097413) |
| 0.4 | 1.8 | GO:0031251 | PAN complex(GO:0031251) |
| 0.3 | 7.9 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.3 | 5.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.3 | 0.9 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.3 | 2.4 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.3 | 0.8 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.3 | 1.8 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.3 | 1.8 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.2 | 1.2 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.2 | 0.7 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.2 | 1.2 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.2 | 2.7 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.2 | 0.9 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.2 | 0.8 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.2 | 1.0 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.2 | 1.6 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.2 | 0.8 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.2 | 2.5 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.2 | 0.6 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 1.9 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.2 | 3.9 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.2 | 1.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 5.6 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.2 | 0.9 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.2 | 0.5 | GO:0097144 | BAX complex(GO:0097144) |
| 0.2 | 0.5 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.2 | 0.5 | GO:0097132 | cyclin D2-CDK6 complex(GO:0097132) |
| 0.2 | 2.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.2 | 1.4 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.2 | 1.4 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 0.7 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.1 | 9.5 | GO:0000791 | euchromatin(GO:0000791) |
| 0.1 | 3.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 1.0 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 0.7 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.1 | 1.1 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 0.4 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 0.7 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.5 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 3.6 | GO:0045120 | pronucleus(GO:0045120) |
| 0.1 | 1.5 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 0.4 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.1 | 1.3 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 2.5 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 2.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.7 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 0.6 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 0.7 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 0.9 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 0.7 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 1.8 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 2.5 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 12.3 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.1 | 0.6 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 4.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 1.9 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 0.7 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 6.5 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 2.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 2.4 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 1.6 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 7.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 0.4 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 0.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.3 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 0.7 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 39.8 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 2.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.1 | GO:0044317 | rod spherule(GO:0044317) |
| 0.1 | 0.8 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.2 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 1.0 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 0.9 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 10.4 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 0.6 | GO:0030663 | COPI vesicle coat(GO:0030126) COPI-coated vesicle membrane(GO:0030663) |
| 0.1 | 0.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 1.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 0.4 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 2.1 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.1 | 0.4 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 0.1 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.1 | 0.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.2 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.9 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.3 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.2 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.3 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 1.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 2.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 3.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.7 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.3 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 1.5 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.1 | GO:0002095 | caveolar macromolecular signaling complex(GO:0002095) |
| 0.0 | 1.2 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 1.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.3 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.7 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 3.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.4 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.1 | GO:0060473 | cortical granule(GO:0060473) |
| 0.0 | 1.0 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.3 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.3 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.6 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 1.2 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.3 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 1.2 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 1.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.3 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 2.9 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.4 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 2.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.8 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.5 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.5 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.6 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 1.1 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 2.0 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.6 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 2.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.5 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.3 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.8 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 2.9 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 0.0 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.7 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 0.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.9 | GO:0000502 | proteasome complex(GO:0000502) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 12.5 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.9 | 5.1 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.8 | 3.8 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.7 | 2.2 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.7 | 2.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.7 | 3.4 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.6 | 3.8 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.6 | 1.9 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.5 | 1.4 | GO:0031753 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.5 | 2.7 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.4 | 3.0 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.4 | 2.5 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.4 | 2.5 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.4 | 1.4 | GO:0008493 | tetracycline transporter activity(GO:0008493) |
| 0.3 | 2.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.3 | 1.0 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.3 | 3.4 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.3 | 3.1 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.3 | 2.4 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.3 | 1.9 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.3 | 1.0 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.3 | 1.5 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.3 | 0.9 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.3 | 2.9 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.3 | 2.8 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.3 | 2.2 | GO:0015099 | nickel cation transmembrane transporter activity(GO:0015099) |
| 0.3 | 0.5 | GO:0005333 | norepinephrine transmembrane transporter activity(GO:0005333) |
| 0.3 | 1.8 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.3 | 2.6 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.2 | 1.0 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.2 | 1.5 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.2 | 0.7 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.2 | 2.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.2 | 1.5 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 0.7 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.2 | 0.7 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.2 | 1.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.2 | 1.0 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.2 | 1.4 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.2 | 0.8 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.2 | 4.4 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.2 | 0.6 | GO:0015292 | uniporter activity(GO:0015292) |
| 0.2 | 1.3 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.2 | 4.9 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.2 | 0.9 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.2 | 1.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.2 | 1.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.2 | 1.9 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.2 | 1.5 | GO:0016672 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) |
| 0.2 | 0.5 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.2 | 3.6 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.2 | 0.7 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.2 | 2.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.2 | 0.5 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.2 | 3.4 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.2 | 1.1 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.2 | 1.4 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.2 | 12.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.2 | 1.4 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 1.8 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 3.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 2.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 1.4 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.1 | 1.4 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 5.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 2.8 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.1 | 1.4 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 0.5 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.1 | 0.4 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 0.9 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 0.5 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.1 | 0.4 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.1 | 1.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 0.8 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 3.0 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.9 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 3.8 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 2.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.5 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 0.5 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.1 | 0.4 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.1 | 2.5 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.1 | 8.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.6 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 0.9 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.1 | 1.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 1.7 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 3.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 1.4 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 0.8 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 2.0 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 0.3 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.1 | 0.1 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.1 | 0.3 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.1 | 5.1 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 0.7 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 1.2 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 0.9 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.7 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 0.3 | GO:0070540 | stearic acid binding(GO:0070540) |
| 0.1 | 0.3 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.5 | GO:0030977 | taurine binding(GO:0030977) |
| 0.1 | 0.8 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 0.2 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.1 | 1.4 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 0.3 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 0.8 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 1.5 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 0.6 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 3.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 1.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.4 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 0.6 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.1 | 6.3 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.1 | 0.4 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.1 | 0.4 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.1 | 0.3 | GO:0086077 | gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.1 | 1.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 1.6 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 12.3 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.1 | 0.7 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 0.6 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.1 | 3.0 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.1 | 0.2 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.1 | 2.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 0.8 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.3 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.1 | 0.7 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 1.4 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.1 | 0.3 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.1 | 1.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 5.0 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 0.6 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.1 | 0.8 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 1.8 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 1.8 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 1.4 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 0.3 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.1 | 0.6 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.3 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.1 | 0.4 | GO:0038049 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.1 | 0.4 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 0.3 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 2.1 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 0.2 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 2.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.1 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 0.2 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.1 | 0.2 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.1 | 1.8 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 0.6 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.2 | GO:0008127 | quercetin 2,3-dioxygenase activity(GO:0008127) |
| 0.1 | 0.8 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 3.1 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 0.3 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.1 | 0.6 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 1.0 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.4 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 30.2 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 1.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.3 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.6 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 1.2 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.1 | 0.2 | GO:0052895 | norspermine:oxygen oxidoreductase activity(GO:0052894) N1-acetylspermine:oxygen oxidoreductase (N1-acetylspermidine-forming) activity(GO:0052895) |
| 0.1 | 0.5 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.5 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.2 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.0 | 1.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 1.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.8 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 1.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.3 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.2 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.7 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.9 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.2 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.9 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 1.0 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 11.1 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.5 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 1.3 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 2.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.7 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 8.6 | GO:0000987 | core promoter proximal region sequence-specific DNA binding(GO:0000987) |
| 0.0 | 0.2 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.9 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.1 | GO:0070002 | glutamic-type peptidase activity(GO:0070002) |
| 0.0 | 0.2 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 1.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.4 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 1.1 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 1.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 5.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 1.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.3 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.1 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.0 | 0.3 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.6 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.4 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.2 | GO:0004448 | isocitrate dehydrogenase activity(GO:0004448) |
| 0.0 | 0.3 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.8 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 1.2 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.1 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 1.5 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.9 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 3.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.7 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.8 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.6 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.4 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.2 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.4 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.0 | 4.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 3.4 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.7 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.3 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 0.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0035614 | U1 snRNA binding(GO:0030619) snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.2 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.2 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.5 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 0.3 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.4 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 3.0 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) |
| 0.0 | 1.7 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 2.9 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 2.4 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.3 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.5 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.1 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 6.5 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.2 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.0 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.5 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.3 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.5 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.4 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.8 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.5 | GO:0003785 | actin monomer binding(GO:0003785) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 7.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.2 | 6.7 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.2 | 2.9 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 10.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 7.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 5.2 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.1 | 5.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 12.0 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 6.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 3.6 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 12.3 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 1.8 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 5.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 2.0 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 4.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 2.6 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 1.4 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.1 | 3.4 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 3.4 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 1.9 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 2.8 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 5.2 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 0.4 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.1 | 2.7 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.1 | 4.0 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 2.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 0.8 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 0.4 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 1.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 4.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 2.8 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 2.1 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 5.4 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 1.7 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 0.8 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 0.9 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 1.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 2.8 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 4.1 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 4.2 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 0.3 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 2.0 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 1.8 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 1.5 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.3 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 4.7 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.8 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.4 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.5 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 6.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 8.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.9 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.3 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.2 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.3 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.9 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.5 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.9 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.6 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.2 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.6 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.2 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.4 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 7.6 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.3 | 0.9 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.3 | 4.3 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.3 | 8.1 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.2 | 5.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.2 | 14.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 4.8 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.2 | 5.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 7.6 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.2 | 3.8 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.2 | 2.5 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.2 | 2.0 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 2.4 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 2.7 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 2.9 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 1.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 1.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 2.1 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 4.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 2.6 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 3.5 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.1 | 2.0 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 2.7 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 5.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 3.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 3.5 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.1 | 1.1 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.1 | 6.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 2.9 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 2.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.9 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 3.2 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 1.8 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 1.4 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 2.0 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.1 | 0.5 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 4.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 0.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 1.0 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 0.5 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 3.0 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 0.7 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 2.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 0.7 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 0.9 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 1.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 1.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 0.6 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 1.3 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.1 | 8.0 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.1 | 1.0 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 0.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.0 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 5.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.8 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.5 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 1.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.7 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.0 | 0.1 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 1.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.8 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.6 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.7 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.7 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.3 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 2.2 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 4.6 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 2.4 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 1.0 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 1.4 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.0 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 2.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.4 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 1.1 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 1.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.9 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.4 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 2.6 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.1 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.1 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.2 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.2 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.6 | REACTOME TRANSPORT OF GLUCOSE AND OTHER SUGARS BILE SALTS AND ORGANIC ACIDS METAL IONS AND AMINE COMPOUNDS | Genes involved in Transport of glucose and other sugars, bile salts and organic acids, metal ions and amine compounds |