Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for E2f7
Z-value: 0.85
Transcription factors associated with E2f7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2f7
|
ENSMUSG00000020185.17 | E2f7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2f7 | mm39_v1_chr10_+_110581293_110581433 | 0.74 | 1.2e-13 | Click! |
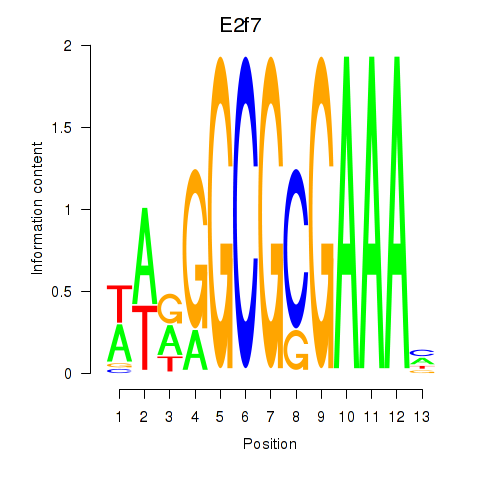
Activity profile of E2f7 motif
Sorted Z-values of E2f7 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of E2f7
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_69188716 | 6.95 |
ENSMUST00000119827.8
ENSMUST00000020099.13 |
Cdk1
|
cyclin-dependent kinase 1 |
| chr13_-_100912308 | 6.05 |
ENSMUST00000075550.4
|
Cenph
|
centromere protein H |
| chr4_+_108436639 | 5.40 |
ENSMUST00000102744.4
|
Orc1
|
origin recognition complex, subunit 1 |
| chr12_+_24758240 | 5.00 |
ENSMUST00000020980.12
|
Rrm2
|
ribonucleotide reductase M2 |
| chr11_+_98441923 | 4.76 |
ENSMUST00000081033.13
ENSMUST00000107511.8 ENSMUST00000107509.8 ENSMUST00000017339.12 |
Zpbp2
|
zona pellucida binding protein 2 |
| chr12_+_24758968 | 4.74 |
ENSMUST00000154588.2
|
Rrm2
|
ribonucleotide reductase M2 |
| chr2_+_72306503 | 4.59 |
ENSMUST00000102691.11
ENSMUST00000157019.2 |
Cdca7
|
cell division cycle associated 7 |
| chr13_-_55477535 | 4.41 |
ENSMUST00000021941.8
|
Mxd3
|
Max dimerization protein 3 |
| chr11_+_102139671 | 4.26 |
ENSMUST00000100392.5
|
Hrob
|
homologous recombination factor with OB-fold |
| chr2_+_11176891 | 4.25 |
ENSMUST00000028118.9
|
Prkcq
|
protein kinase C, theta |
| chr1_-_74544946 | 4.13 |
ENSMUST00000044260.11
ENSMUST00000186282.7 |
Usp37
|
ubiquitin specific peptidase 37 |
| chr8_+_75836187 | 3.98 |
ENSMUST00000164309.3
ENSMUST00000212426.2 ENSMUST00000212811.2 |
Mcm5
|
minichromosome maintenance complex component 5 |
| chr14_-_20438890 | 3.51 |
ENSMUST00000022345.7
|
Dnajc9
|
DnaJ heat shock protein family (Hsp40) member C9 |
| chr11_+_85202058 | 3.51 |
ENSMUST00000020835.16
|
Ppm1d
|
protein phosphatase 1D magnesium-dependent, delta isoform |
| chr6_-_88875646 | 3.49 |
ENSMUST00000058011.8
|
Mcm2
|
minichromosome maintenance complex component 2 |
| chr15_-_27681584 | 3.38 |
ENSMUST00000226145.2
ENSMUST00000226170.2 |
Otulinl
|
OTU deubiquitinase with linear linkage specificity like |
| chr12_-_76756772 | 3.38 |
ENSMUST00000166101.2
|
Sptb
|
spectrin beta, erythrocytic |
| chr2_-_157046386 | 3.09 |
ENSMUST00000029170.8
|
Rbl1
|
RB transcriptional corepressor like 1 |
| chr4_+_98812082 | 3.06 |
ENSMUST00000091358.11
|
Usp1
|
ubiquitin specific peptidase 1 |
| chr4_+_98812047 | 3.05 |
ENSMUST00000030289.9
|
Usp1
|
ubiquitin specific peptidase 1 |
| chr11_-_87295292 | 2.97 |
ENSMUST00000067692.13
|
Rad51c
|
RAD51 paralog C |
| chr17_-_35827676 | 2.87 |
ENSMUST00000160885.2
ENSMUST00000159009.2 ENSMUST00000161012.8 |
Tcf19
|
transcription factor 19 |
| chr15_-_27681643 | 2.86 |
ENSMUST00000100739.5
|
Otulinl
|
OTU deubiquitinase with linear linkage specificity like |
| chr6_+_30509826 | 2.80 |
ENSMUST00000031797.11
|
Ssmem1
|
serine-rich single-pass membrane protein 1 |
| chr4_+_126450728 | 2.72 |
ENSMUST00000048391.15
|
Clspn
|
claspin |
| chr13_-_22227114 | 2.60 |
ENSMUST00000091741.6
|
H2ac11
|
H2A clustered histone 11 |
| chr12_+_24758724 | 2.56 |
ENSMUST00000153058.8
|
Rrm2
|
ribonucleotide reductase M2 |
| chr10_+_11157326 | 2.56 |
ENSMUST00000070300.5
|
Fbxo30
|
F-box protein 30 |
| chr13_-_21937997 | 2.52 |
ENSMUST00000074752.4
|
H2ac15
|
H2A clustered histone 15 |
| chr11_-_6394352 | 2.51 |
ENSMUST00000093346.6
|
H2az2
|
H2A.Z histone variant 2 |
| chr10_+_115653152 | 2.50 |
ENSMUST00000080630.11
ENSMUST00000179196.3 ENSMUST00000035563.15 |
Tspan8
|
tetraspanin 8 |
| chr1_+_74545203 | 2.50 |
ENSMUST00000087215.7
|
Cnot9
|
CCR4-NOT transcription complex, subunit 9 |
| chr10_+_11157047 | 2.46 |
ENSMUST00000129456.8
|
Fbxo30
|
F-box protein 30 |
| chr14_-_52257113 | 2.45 |
ENSMUST00000166169.4
ENSMUST00000226605.2 |
Zfp219
|
zinc finger protein 219 |
| chr14_+_79689230 | 2.20 |
ENSMUST00000100359.3
ENSMUST00000226192.2 |
Kbtbd6
|
kelch repeat and BTB (POZ) domain containing 6 |
| chr3_+_108291145 | 2.20 |
ENSMUST00000090561.10
ENSMUST00000102629.8 ENSMUST00000128089.2 |
Psrc1
|
proline/serine-rich coiled-coil 1 |
| chr12_-_11315755 | 2.13 |
ENSMUST00000166117.4
ENSMUST00000219600.2 ENSMUST00000218487.2 |
Gen1
|
GEN1, Holliday junction 5' flap endonuclease |
| chr17_+_35827997 | 2.07 |
ENSMUST00000164242.9
ENSMUST00000045956.14 |
Cchcr1
|
coiled-coil alpha-helical rod protein 1 |
| chr13_-_22017677 | 2.05 |
ENSMUST00000081342.7
|
H2ac24
|
H2A clustered histone 24 |
| chr9_+_64188857 | 1.98 |
ENSMUST00000215031.2
ENSMUST00000213165.2 ENSMUST00000213289.2 ENSMUST00000216594.2 ENSMUST00000034964.7 |
Tipin
|
timeless interacting protein |
| chr11_-_77404160 | 1.88 |
ENSMUST00000060417.11
|
Trp53i13
|
transformation related protein 53 inducible protein 13 |
| chr3_+_88439616 | 1.87 |
ENSMUST00000172699.2
|
Mex3a
|
mex3 RNA binding family member A |
| chr13_+_21900554 | 1.82 |
ENSMUST00000070124.5
|
H2ac13
|
H2A clustered histone 13 |
| chr4_+_98812144 | 1.81 |
ENSMUST00000169053.2
|
Usp1
|
ubiquitin specific peptidase 1 |
| chr13_-_21967540 | 1.73 |
ENSMUST00000189457.2
|
H3c11
|
H3 clustered histone 11 |
| chr3_+_60408678 | 1.72 |
ENSMUST00000191747.6
ENSMUST00000194069.6 |
Mbnl1
|
muscleblind like splicing factor 1 |
| chr12_+_116369017 | 1.71 |
ENSMUST00000084828.5
ENSMUST00000222469.2 ENSMUST00000221114.2 ENSMUST00000221970.2 |
Ncapg2
|
non-SMC condensin II complex, subunit G2 |
| chr1_+_153300874 | 1.67 |
ENSMUST00000042373.12
|
Shcbp1l
|
Shc SH2-domain binding protein 1-like |
| chr1_-_88629843 | 1.64 |
ENSMUST00000159814.2
|
Arl4c
|
ADP-ribosylation factor-like 4C |
| chr17_-_35069136 | 1.59 |
ENSMUST00000046022.16
|
Skiv2l
|
superkiller viralicidic activity 2-like (S. cerevisiae) |
| chr17_+_35069347 | 1.58 |
ENSMUST00000097343.11
ENSMUST00000173357.8 ENSMUST00000173065.8 ENSMUST00000165953.3 |
Nelfe
|
negative elongation factor complex member E, Rdbp |
| chr15_+_82183143 | 1.58 |
ENSMUST00000023089.5
|
Wbp2nl
|
WBP2 N-terminal like |
| chr8_-_106074585 | 1.47 |
ENSMUST00000014922.5
|
Fhod1
|
formin homology 2 domain containing 1 |
| chr3_-_105594865 | 1.46 |
ENSMUST00000090680.11
|
Ddx20
|
DEAD box helicase 20 |
| chr11_+_95733109 | 1.42 |
ENSMUST00000107714.9
ENSMUST00000107711.8 |
Gngt2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr11_-_95733235 | 1.42 |
ENSMUST00000059026.10
|
Abi3
|
ABI family member 3 |
| chr3_-_143908111 | 1.42 |
ENSMUST00000121796.8
|
Lmo4
|
LIM domain only 4 |
| chr5_-_92496730 | 1.40 |
ENSMUST00000038816.13
ENSMUST00000118006.3 |
Cxcl10
|
chemokine (C-X-C motif) ligand 10 |
| chr18_+_44237474 | 1.39 |
ENSMUST00000081271.7
|
Spink12
|
serine peptidase inhibitor, Kazal type 12 |
| chr18_+_44237577 | 1.37 |
ENSMUST00000239465.2
|
Spink12
|
serine peptidase inhibitor, Kazal type 12 |
| chr1_-_128287347 | 1.29 |
ENSMUST00000190495.2
ENSMUST00000027601.11 |
Mcm6
|
minichromosome maintenance complex component 6 |
| chr17_-_35265702 | 1.25 |
ENSMUST00000097338.11
|
Msh5
|
mutS homolog 5 |
| chr13_-_47259652 | 1.25 |
ENSMUST00000021807.13
ENSMUST00000135278.8 |
Dek
|
DEK proto-oncogene (DNA binding) |
| chr3_+_14598848 | 1.24 |
ENSMUST00000108370.9
|
Lrrcc1
|
leucine rich repeat and coiled-coil domain containing 1 |
| chr9_+_54672032 | 1.20 |
ENSMUST00000034830.9
|
Crabp1
|
cellular retinoic acid binding protein I |
| chr3_-_143908060 | 1.20 |
ENSMUST00000121112.6
|
Lmo4
|
LIM domain only 4 |
| chr6_-_67014383 | 1.19 |
ENSMUST00000043098.9
|
Gadd45a
|
growth arrest and DNA-damage-inducible 45 alpha |
| chr11_-_6394385 | 1.18 |
ENSMUST00000109737.9
|
H2az2
|
H2A.Z histone variant 2 |
| chr14_-_66071337 | 1.18 |
ENSMUST00000225853.2
|
Esco2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr4_+_132495636 | 1.18 |
ENSMUST00000102561.11
|
Rpa2
|
replication protein A2 |
| chr16_+_76810588 | 1.16 |
ENSMUST00000239066.2
ENSMUST00000023580.8 |
Usp25
|
ubiquitin specific peptidase 25 |
| chr11_+_95733489 | 1.14 |
ENSMUST00000100532.10
ENSMUST00000036088.11 |
Gngt2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr7_+_27869115 | 1.14 |
ENSMUST00000042405.8
|
Fbl
|
fibrillarin |
| chr10_+_67374366 | 1.12 |
ENSMUST00000127820.2
|
Egr2
|
early growth response 2 |
| chr13_+_21994588 | 1.11 |
ENSMUST00000091745.6
|
H2ac23
|
H2A clustered histone 23 |
| chr11_-_106192627 | 1.09 |
ENSMUST00000103071.4
|
Gh
|
growth hormone |
| chr7_+_44465806 | 1.02 |
ENSMUST00000207103.2
ENSMUST00000118125.9 |
Nup62
Il4i1
|
nucleoporin 62 interleukin 4 induced 1 |
| chr4_+_108477117 | 1.02 |
ENSMUST00000030320.13
|
Cc2d1b
|
coiled-coil and C2 domain containing 1B |
| chr3_+_103187162 | 1.01 |
ENSMUST00000106860.6
|
Trim33
|
tripartite motif-containing 33 |
| chr11_-_69214593 | 0.97 |
ENSMUST00000092973.6
|
Cntrob
|
centrobin, centrosomal BRCA2 interacting protein |
| chr4_+_124608569 | 0.96 |
ENSMUST00000030734.5
|
Sf3a3
|
splicing factor 3a, subunit 3 |
| chr3_+_14598927 | 0.95 |
ENSMUST00000163660.8
|
Lrrcc1
|
leucine rich repeat and coiled-coil domain containing 1 |
| chr6_-_67014348 | 0.94 |
ENSMUST00000204369.2
|
Gadd45a
|
growth arrest and DNA-damage-inducible 45 alpha |
| chr1_+_180396478 | 0.93 |
ENSMUST00000027777.12
|
Parp1
|
poly (ADP-ribose) polymerase family, member 1 |
| chr9_+_21279802 | 0.91 |
ENSMUST00000214474.2
|
Ilf3
|
interleukin enhancer binding factor 3 |
| chr11_-_101357046 | 0.88 |
ENSMUST00000040430.8
|
Vat1
|
vesicle amine transport 1 |
| chr12_+_73333641 | 0.85 |
ENSMUST00000153941.8
ENSMUST00000122920.8 ENSMUST00000101313.4 |
Slc38a6
|
solute carrier family 38, member 6 |
| chr12_+_11315868 | 0.80 |
ENSMUST00000020931.6
|
Smc6
|
structural maintenance of chromosomes 6 |
| chr2_+_112092271 | 0.75 |
ENSMUST00000028553.4
|
Nop10
|
NOP10 ribonucleoprotein |
| chr11_+_87018079 | 0.72 |
ENSMUST00000139532.2
|
Trim37
|
tripartite motif-containing 37 |
| chr11_-_70300836 | 0.71 |
ENSMUST00000019065.10
ENSMUST00000135148.2 |
Pelp1
|
proline, glutamic acid and leucine rich protein 1 |
| chr13_-_21934675 | 0.57 |
ENSMUST00000102983.2
|
H4c12
|
H4 clustered histone 12 |
| chr6_-_134874778 | 0.57 |
ENSMUST00000165392.8
ENSMUST00000204880.3 ENSMUST00000203409.3 ENSMUST00000046255.14 ENSMUST00000111932.8 ENSMUST00000149375.8 ENSMUST00000116515.9 |
Gpr19
|
G protein-coupled receptor 19 |
| chr14_-_18897750 | 0.55 |
ENSMUST00000178728.2
|
Gm3005
|
predicted gene 3005 |
| chr9_+_81745723 | 0.49 |
ENSMUST00000057067.10
ENSMUST00000189832.7 ENSMUST00000189391.2 |
Mei4
|
meiotic double-stranded break formation protein 4 |
| chr9_+_46194293 | 0.49 |
ENSMUST00000074957.5
|
Bud13
|
BUD13 homolog |
| chr3_+_88049633 | 0.48 |
ENSMUST00000001455.13
ENSMUST00000119251.8 |
Mef2d
|
myocyte enhancer factor 2D |
| chr13_+_22227359 | 0.46 |
ENSMUST00000110452.2
|
H2bc11
|
H2B clustered histone 11 |
| chr14_-_18359247 | 0.42 |
ENSMUST00000170207.8
|
Gm8108
|
predicted gene 8108 |
| chr7_-_98305737 | 0.42 |
ENSMUST00000205911.2
ENSMUST00000038359.6 ENSMUST00000206611.2 ENSMUST00000206619.2 |
Emsy
|
EMSY, BRCA2-interacting transcriptional repressor |
| chr15_-_97991114 | 0.39 |
ENSMUST00000180657.2
|
Senp1
|
SUMO1/sentrin specific peptidase 1 |
| chr14_-_18659699 | 0.38 |
ENSMUST00000170480.8
|
Gm3002
|
predicted gene 3002 |
| chr3_+_152101698 | 0.32 |
ENSMUST00000200570.5
|
Zzz3
|
zinc finger, ZZ domain containing 3 |
| chr7_-_4687916 | 0.31 |
ENSMUST00000206306.2
ENSMUST00000205952.2 ENSMUST00000079970.6 |
Hspbp1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr9_-_87137515 | 0.28 |
ENSMUST00000093802.6
|
Cep162
|
centrosomal protein 162 |
| chr14_-_17398733 | 0.26 |
ENSMUST00000163719.8
|
Gm8281
|
predicted gene, 8281 |
| chr6_-_113717689 | 0.25 |
ENSMUST00000032440.6
|
Sec13
|
SEC13 homolog, nuclear pore and COPII coat complex component |
| chr4_+_119280002 | 0.21 |
ENSMUST00000094819.5
|
Zmynd12
|
zinc finger, MYND domain containing 12 |
| chr12_+_11316101 | 0.21 |
ENSMUST00000218866.2
|
Smc6
|
structural maintenance of chromosomes 6 |
| chr4_+_126450762 | 0.20 |
ENSMUST00000147675.2
|
Clspn
|
claspin |
| chr9_-_111100859 | 0.19 |
ENSMUST00000035079.10
|
Mlh1
|
mutL homolog 1 |
| chr11_+_26337194 | 0.17 |
ENSMUST00000136830.2
ENSMUST00000109509.8 |
Fancl
|
Fanconi anemia, complementation group L |
| chr15_-_81756076 | 0.17 |
ENSMUST00000023117.10
|
Phf5a
|
PHD finger protein 5A |
| chr4_-_108436514 | 0.16 |
ENSMUST00000079213.6
|
Prpf38a
|
PRP38 pre-mRNA processing factor 38 (yeast) domain containing A |
| chr7_-_98305986 | 0.12 |
ENSMUST00000205276.2
|
Emsy
|
EMSY, BRCA2-interacting transcriptional repressor |
| chr1_-_190915441 | 0.10 |
ENSMUST00000027941.14
|
Atf3
|
activating transcription factor 3 |
| chr13_+_21938258 | 0.06 |
ENSMUST00000091709.3
|
H2bc15
|
H2B clustered histone 15 |
| chr13_+_92491234 | 0.06 |
ENSMUST00000022218.6
|
Dhfr
|
dihydrofolate reductase |
| chrX_+_151789457 | 0.03 |
ENSMUST00000095755.4
|
Usp51
|
ubiquitin specific protease 51 |
| chr19_-_11806388 | 0.03 |
ENSMUST00000061235.3
|
Olfr1417
|
olfactory receptor 1417 |
| chr2_+_130509530 | 0.02 |
ENSMUST00000103193.5
|
Itpa
|
inosine triphosphatase (nucleoside triphosphate pyrophosphatase) |
| chr1_+_172348611 | 0.02 |
ENSMUST00000085894.12
ENSMUST00000161140.8 ENSMUST00000162988.8 |
Cfap45
|
cilia and flagella associated protein 45 |
| chr6_+_113054592 | 0.02 |
ENSMUST00000113157.8
|
Setd5
|
SET domain containing 5 |
| chr14_-_17742998 | 0.01 |
ENSMUST00000165619.8
|
Gm3252
|
predicted gene 3252 |
| chr4_+_33062999 | 0.01 |
ENSMUST00000108162.8
ENSMUST00000024035.9 |
Gabrr2
|
gamma-aminobutyric acid (GABA) C receptor, subunit rho 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.4 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.7 | 7.9 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.6 | 3.0 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.6 | 6.4 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.5 | 12.3 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.5 | 4.2 | GO:2000318 | positive regulation of T-helper 17 type immune response(GO:2000318) |
| 0.5 | 1.6 | GO:0035038 | female pronucleus assembly(GO:0035038) |
| 0.4 | 2.5 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.4 | 4.8 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.4 | 2.0 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.4 | 1.1 | GO:0034963 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.4 | 1.1 | GO:0021594 | rhombomere formation(GO:0021594) rhombomere 3 formation(GO:0021660) rhombomere 5 morphogenesis(GO:0021664) rhombomere 5 formation(GO:0021666) |
| 0.4 | 2.9 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.4 | 1.4 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.3 | 0.9 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.3 | 4.1 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.2 | 1.5 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.2 | 4.9 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.2 | 1.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.2 | 2.1 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.2 | 4.8 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.2 | 3.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 | 1.2 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.2 | 1.7 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.2 | 1.6 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.2 | 1.2 | GO:0044838 | cell quiescence(GO:0044838) |
| 0.2 | 2.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.7 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.1 | 0.9 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.1 | 1.6 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 2.6 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.1 | 0.5 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 | 4.0 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 1.0 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.1 | 0.7 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 0.2 | GO:0043060 | meiotic metaphase I plate congression(GO:0043060) meiotic metaphase plate congression(GO:0051311) |
| 0.1 | 2.9 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.1 | 1.0 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.1 | 1.7 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 3.4 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.1 | 1.2 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 1.2 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 2.2 | GO:0090224 | mitotic metaphase plate congression(GO:0007080) regulation of mitotic spindle organization(GO:0060236) regulation of spindle organization(GO:0090224) |
| 0.0 | 3.5 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 1.1 | GO:0090031 | positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 0.0 | 2.5 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 1.5 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 1.6 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.4 | GO:0016926 | regulation of definitive erythrocyte differentiation(GO:0010724) protein desumoylation(GO:0016926) |
| 0.0 | 0.1 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 2.1 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 1.0 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.7 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.9 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.1 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 12.3 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.8 | 8.9 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.6 | 3.0 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.5 | 1.6 | GO:0055087 | Ski complex(GO:0055087) |
| 0.4 | 3.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.4 | 1.6 | GO:0032021 | NELF complex(GO:0032021) |
| 0.3 | 4.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.3 | 2.0 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.3 | 1.0 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.2 | 0.7 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.2 | 1.8 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 1.7 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 1.1 | GO:0001651 | dense fibrillar component(GO:0001651) granular component(GO:0001652) |
| 0.1 | 1.7 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 9.1 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 1.5 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 2.4 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.1 | 5.0 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 1.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 0.3 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.1 | 1.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 0.2 | GO:0005715 | chiasma(GO:0005712) late recombination nodule(GO:0005715) |
| 0.1 | 0.7 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.7 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 2.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 5.1 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 3.0 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 4.4 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 1.5 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 1.2 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 12.3 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.9 | 6.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.7 | 3.5 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.6 | 5.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.5 | 2.9 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.4 | 7.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.4 | 1.1 | GO:0036009 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.3 | 1.0 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.3 | 6.9 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.2 | 1.4 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.2 | 4.2 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 1.7 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 1.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.1 | 12.5 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 1.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 1.2 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.1 | 0.7 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 0.4 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 0.9 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 3.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 1.2 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 3.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.7 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 2.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 4.2 | GO:0070035 | ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.0 | 1.0 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 1.6 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 1.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 1.7 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.8 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 7.8 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 2.6 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 2.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 5.9 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 2.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 2.1 | GO:0001047 | core promoter binding(GO:0001047) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 6.9 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.3 | 8.1 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.2 | 9.6 | PID ATR PATHWAY | ATR signaling pathway |
| 0.2 | 20.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 2.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 4.2 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 2.1 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 2.6 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 2.5 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.7 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.9 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 1.4 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 1.1 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 2.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 24.7 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.6 | 8.8 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.6 | 7.9 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.3 | 6.0 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.2 | 3.1 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 5.4 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 4.2 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 3.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 2.6 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 1.9 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 2.1 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 1.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |