Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
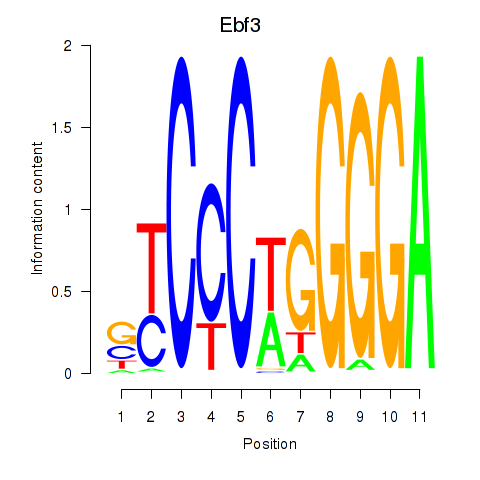
Results for Ebf3
Z-value: 1.72
Transcription factors associated with Ebf3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ebf3
|
ENSMUSG00000010476.15 | Ebf3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ebf3 | mm39_v1_chr7_-_136915602_136915715 | 0.27 | 2.2e-02 | Click! |
Activity profile of Ebf3 motif
Sorted Z-values of Ebf3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Ebf3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_102255999 | 9.76 |
ENSMUST00000006749.10
|
Slc4a1
|
solute carrier family 4 (anion exchanger), member 1 |
| chr11_-_97944239 | 8.24 |
ENSMUST00000017544.9
|
Stac2
|
SH3 and cysteine rich domain 2 |
| chr11_+_104122216 | 7.89 |
ENSMUST00000106992.10
|
Mapt
|
microtubule-associated protein tau |
| chrX_+_10351360 | 7.71 |
ENSMUST00000076354.13
ENSMUST00000115526.2 |
Tspan7
|
tetraspanin 7 |
| chr11_-_53371050 | 7.29 |
ENSMUST00000104955.4
|
Sowaha
|
sosondowah ankyrin repeat domain family member A |
| chr7_+_24596806 | 7.07 |
ENSMUST00000003469.8
|
Cd79a
|
CD79A antigen (immunoglobulin-associated alpha) |
| chr11_+_104122399 | 6.85 |
ENSMUST00000132977.8
ENSMUST00000132245.8 ENSMUST00000100347.11 |
Mapt
|
microtubule-associated protein tau |
| chr11_+_104122341 | 6.47 |
ENSMUST00000106993.10
|
Mapt
|
microtubule-associated protein tau |
| chr4_-_129015027 | 6.34 |
ENSMUST00000030572.10
|
Hpca
|
hippocalcin |
| chr14_-_70864666 | 6.08 |
ENSMUST00000022694.17
|
Dmtn
|
dematin actin binding protein |
| chr14_-_70866385 | 5.99 |
ENSMUST00000228824.2
|
Dmtn
|
dematin actin binding protein |
| chr17_+_44499451 | 5.96 |
ENSMUST00000024755.7
|
Clic5
|
chloride intracellular channel 5 |
| chr1_+_87192067 | 5.93 |
ENSMUST00000027472.7
|
Efhd1
|
EF hand domain containing 1 |
| chr11_+_104122291 | 5.79 |
ENSMUST00000145227.8
|
Mapt
|
microtubule-associated protein tau |
| chr15_+_89383799 | 5.62 |
ENSMUST00000109309.9
|
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr12_-_31763859 | 5.49 |
ENSMUST00000057783.6
ENSMUST00000236002.2 ENSMUST00000174480.3 ENSMUST00000176710.2 |
Gpr22
|
G protein-coupled receptor 22 |
| chrX_+_7504913 | 5.29 |
ENSMUST00000128890.2
|
Syp
|
synaptophysin |
| chr4_-_129015493 | 5.25 |
ENSMUST00000135763.2
ENSMUST00000149763.3 ENSMUST00000164649.8 |
Hpca
|
hippocalcin |
| chr17_-_24863907 | 4.78 |
ENSMUST00000234505.2
|
Slc9a3r2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 |
| chr1_-_173195236 | 4.74 |
ENSMUST00000005470.5
ENSMUST00000111220.8 |
Cadm3
|
cell adhesion molecule 3 |
| chr10_+_112107026 | 4.61 |
ENSMUST00000219301.2
ENSMUST00000092175.4 |
Kcnc2
|
potassium voltage gated channel, Shaw-related subfamily, member 2 |
| chr13_+_55547498 | 4.59 |
ENSMUST00000057167.9
|
Slc34a1
|
solute carrier family 34 (sodium phosphate), member 1 |
| chr18_-_35781422 | 4.59 |
ENSMUST00000237462.2
|
Mzb1
|
marginal zone B and B1 cell-specific protein 1 |
| chr12_-_113860566 | 4.58 |
ENSMUST00000103474.5
|
Ighv7-1
|
immunoglobulin heavy variable 7-1 |
| chr6_+_68279392 | 4.56 |
ENSMUST00000103322.3
|
Igkv2-109
|
immunoglobulin kappa variable 2-109 |
| chr12_-_41536430 | 4.52 |
ENSMUST00000043884.6
|
Lrrn3
|
leucine rich repeat protein 3, neuronal |
| chr17_-_24863956 | 4.49 |
ENSMUST00000019684.13
|
Slc9a3r2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 |
| chr14_-_34310602 | 4.46 |
ENSMUST00000064098.14
ENSMUST00000090040.12 ENSMUST00000022330.9 ENSMUST00000022327.13 |
Ldb3
|
LIM domain binding 3 |
| chr6_-_68840015 | 4.36 |
ENSMUST00000103336.2
|
Igkv1-88
|
immunoglobulin kappa chain variable 1-88 |
| chr6_+_67586695 | 4.32 |
ENSMUST00000103303.3
|
Igkv1-135
|
immunoglobulin kappa variable 1-135 |
| chr17_-_48758538 | 4.31 |
ENSMUST00000024794.12
|
Tspo2
|
translocator protein 2 |
| chr7_-_25315299 | 4.30 |
ENSMUST00000098663.4
ENSMUST00000238895.2 |
Erich4
|
glutamate rich 4 |
| chr2_+_55327110 | 4.11 |
ENSMUST00000112633.3
ENSMUST00000112632.2 |
Kcnj3
|
potassium inwardly-rectifying channel, subfamily J, member 3 |
| chr2_-_52448552 | 4.10 |
ENSMUST00000102760.10
ENSMUST00000102761.9 |
Cacnb4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr12_-_113912416 | 4.09 |
ENSMUST00000103464.3
|
Ighv4-1
|
immunoglobulin heavy variable 4-1 |
| chr12_-_113823290 | 4.04 |
ENSMUST00000103459.5
|
Ighv5-17
|
immunoglobulin heavy variable 5-17 |
| chr13_+_55097200 | 4.02 |
ENSMUST00000026994.14
ENSMUST00000109994.9 |
Unc5a
|
unc-5 netrin receptor A |
| chr9_-_66950991 | 4.01 |
ENSMUST00000113689.8
ENSMUST00000113684.8 |
Tpm1
|
tropomyosin 1, alpha |
| chr14_-_34310637 | 3.96 |
ENSMUST00000227819.2
|
Ldb3
|
LIM domain binding 3 |
| chr8_+_95498822 | 3.93 |
ENSMUST00000211956.2
ENSMUST00000211947.2 |
Cx3cl1
|
chemokine (C-X3-C motif) ligand 1 |
| chr9_-_37344542 | 3.88 |
ENSMUST00000115038.3
|
Robo3
|
roundabout guidance receptor 3 |
| chr4_+_129030710 | 3.83 |
ENSMUST00000102600.4
|
Fndc5
|
fibronectin type III domain containing 5 |
| chr11_+_92989229 | 3.81 |
ENSMUST00000107859.8
ENSMUST00000107861.8 ENSMUST00000042943.13 ENSMUST00000107858.9 |
Car10
|
carbonic anhydrase 10 |
| chrX_+_165021919 | 3.73 |
ENSMUST00000060210.14
ENSMUST00000112233.8 |
Gpm6b
|
glycoprotein m6b |
| chr10_-_67748461 | 3.68 |
ENSMUST00000064656.8
|
Zfp365
|
zinc finger protein 365 |
| chr17_+_87270504 | 3.66 |
ENSMUST00000024956.15
|
Rhoq
|
ras homolog family member Q |
| chr4_+_137408975 | 3.63 |
ENSMUST00000047243.12
|
Rap1gap
|
Rap1 GTPase-activating protein |
| chr17_+_50816296 | 3.63 |
ENSMUST00000043938.8
|
Plcl2
|
phospholipase C-like 2 |
| chrX_+_74425990 | 3.63 |
ENSMUST00000033541.5
|
Fundc2
|
FUN14 domain containing 2 |
| chr17_-_74017410 | 3.62 |
ENSMUST00000112591.3
ENSMUST00000024858.12 |
Galnt14
|
polypeptide N-acetylgalactosaminyltransferase 14 |
| chr11_+_83300481 | 3.55 |
ENSMUST00000175848.8
ENSMUST00000108140.10 |
Rasl10b
|
RAS-like, family 10, member B |
| chr12_-_113700190 | 3.55 |
ENSMUST00000103452.3
ENSMUST00000192264.2 |
Ighv5-9-1
|
immunoglobulin heavy variable 5-9-1 |
| chr17_+_87270707 | 3.55 |
ENSMUST00000139344.2
|
Rhoq
|
ras homolog family member Q |
| chr5_+_37403098 | 3.54 |
ENSMUST00000031004.11
|
Crmp1
|
collapsin response mediator protein 1 |
| chrX_+_158242121 | 3.49 |
ENSMUST00000112470.3
ENSMUST00000043151.12 ENSMUST00000156172.3 |
Map7d2
|
MAP7 domain containing 2 |
| chr14_-_34310438 | 3.47 |
ENSMUST00000228044.2
ENSMUST00000022328.14 |
Ldb3
|
LIM domain binding 3 |
| chr1_-_193052533 | 3.46 |
ENSMUST00000169907.8
|
Camk1g
|
calcium/calmodulin-dependent protein kinase I gamma |
| chr7_-_30614249 | 3.46 |
ENSMUST00000190950.7
ENSMUST00000187137.7 ENSMUST00000190638.7 |
Mag
|
myelin-associated glycoprotein |
| chr6_-_126512375 | 3.40 |
ENSMUST00000060972.5
|
Kcna5
|
potassium voltage-gated channel, shaker-related subfamily, member 5 |
| chr9_+_95519654 | 3.34 |
ENSMUST00000015498.9
|
Pcolce2
|
procollagen C-endopeptidase enhancer 2 |
| chr4_-_141143313 | 3.34 |
ENSMUST00000006378.9
ENSMUST00000105788.2 |
Clcnkb
|
chloride channel, voltage-sensitive Kb |
| chr5_+_135835713 | 3.33 |
ENSMUST00000126232.8
|
Srrm3
|
serine/arginine repetitive matrix 3 |
| chr7_-_126014027 | 3.31 |
ENSMUST00000032968.7
ENSMUST00000206325.2 |
Cd19
|
CD19 antigen |
| chr12_-_113561594 | 3.25 |
ENSMUST00000103444.3
|
Ighv5-4
|
immunoglobulin heavy variable 5-4 |
| chr18_-_31580436 | 3.20 |
ENSMUST00000025110.5
|
Syt4
|
synaptotagmin IV |
| chr5_-_8417982 | 3.18 |
ENSMUST00000088761.11
ENSMUST00000115386.8 ENSMUST00000050166.14 ENSMUST00000046838.14 ENSMUST00000115388.9 ENSMUST00000088744.12 ENSMUST00000115385.2 |
Adam22
|
a disintegrin and metallopeptidase domain 22 |
| chr1_-_193052568 | 3.17 |
ENSMUST00000016323.11
|
Camk1g
|
calcium/calmodulin-dependent protein kinase I gamma |
| chr19_-_17316906 | 3.14 |
ENSMUST00000169897.2
|
Gcnt1
|
glucosaminyl (N-acetyl) transferase 1, core 2 |
| chr11_+_63019799 | 3.13 |
ENSMUST00000108702.8
|
Pmp22
|
peripheral myelin protein 22 |
| chr17_+_48666919 | 3.13 |
ENSMUST00000224001.2
ENSMUST00000024792.8 ENSMUST00000225849.2 |
Treml1
|
triggering receptor expressed on myeloid cells-like 1 |
| chr2_-_13496624 | 3.05 |
ENSMUST00000091436.7
|
Cubn
|
cubilin (intrinsic factor-cobalamin receptor) |
| chr2_+_55325931 | 3.03 |
ENSMUST00000067101.10
|
Kcnj3
|
potassium inwardly-rectifying channel, subfamily J, member 3 |
| chr11_+_101137786 | 3.01 |
ENSMUST00000107282.4
|
Ramp2
|
receptor (calcitonin) activity modifying protein 2 |
| chr7_+_19144950 | 3.00 |
ENSMUST00000208710.2
ENSMUST00000003643.3 |
Ckm
|
creatine kinase, muscle |
| chr3_-_107667499 | 3.00 |
ENSMUST00000153114.2
ENSMUST00000118593.8 ENSMUST00000120243.8 |
Csf1
|
colony stimulating factor 1 (macrophage) |
| chr1_+_75376714 | 2.95 |
ENSMUST00000113589.8
|
Speg
|
SPEG complex locus |
| chr1_+_167426019 | 2.93 |
ENSMUST00000111386.8
ENSMUST00000111384.8 |
Rxrg
|
retinoid X receptor gamma |
| chr7_-_119078472 | 2.92 |
ENSMUST00000209095.2
ENSMUST00000033263.6 ENSMUST00000207261.2 |
Umod
|
uromodulin |
| chr2_-_28511941 | 2.92 |
ENSMUST00000028156.8
ENSMUST00000164290.8 |
Gfi1b
|
growth factor independent 1B |
| chr5_-_144698443 | 2.91 |
ENSMUST00000061446.8
|
Tmem130
|
transmembrane protein 130 |
| chr6_+_68518603 | 2.91 |
ENSMUST00000168090.3
ENSMUST00000103326.3 |
Igkv1-99
|
immunoglobulin kappa variable 1-99 |
| chr5_-_24806960 | 2.86 |
ENSMUST00000030791.12
|
Smarcd3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr6_-_69282389 | 2.84 |
ENSMUST00000103350.3
|
Igkv4-68
|
immunoglobulin kappa variable 4-68 |
| chr7_+_127661835 | 2.83 |
ENSMUST00000106242.10
ENSMUST00000120355.8 ENSMUST00000106240.9 ENSMUST00000098015.10 |
Itgam
Gm49368
|
integrin alpha M predicted gene, 49368 |
| chr6_-_69415741 | 2.83 |
ENSMUST00000103354.3
|
Igkv4-59
|
immunoglobulin kappa variable 4-59 |
| chr11_-_98220466 | 2.82 |
ENSMUST00000041685.7
|
Neurod2
|
neurogenic differentiation 2 |
| chr19_-_45804446 | 2.81 |
ENSMUST00000079431.10
ENSMUST00000026247.13 ENSMUST00000162528.9 |
Kcnip2
|
Kv channel-interacting protein 2 |
| chr16_+_13721016 | 2.81 |
ENSMUST00000128757.8
|
Mpv17l
|
Mpv17 transgene, kidney disease mutant-like |
| chr5_-_18054781 | 2.79 |
ENSMUST00000170051.8
|
Cd36
|
CD36 molecule |
| chr19_+_6468761 | 2.78 |
ENSMUST00000113462.8
ENSMUST00000077182.13 ENSMUST00000236635.2 ENSMUST00000113461.8 |
Nrxn2
|
neurexin II |
| chr15_+_82140224 | 2.78 |
ENSMUST00000143238.2
|
1500009C09Rik
|
RIKEN cDNA 1500009C09 gene |
| chrX_+_165021897 | 2.77 |
ENSMUST00000112235.8
|
Gpm6b
|
glycoprotein m6b |
| chr11_-_102837514 | 2.75 |
ENSMUST00000057849.6
|
C1ql1
|
complement component 1, q subcomponent-like 1 |
| chr11_+_83299963 | 2.72 |
ENSMUST00000021022.10
|
Rasl10b
|
RAS-like, family 10, member B |
| chrX_+_73352694 | 2.69 |
ENSMUST00000130581.2
|
Gdi1
|
guanosine diphosphate (GDP) dissociation inhibitor 1 |
| chr5_-_18054702 | 2.68 |
ENSMUST00000165232.8
|
Cd36
|
CD36 molecule |
| chr17_-_26240827 | 2.68 |
ENSMUST00000118828.8
|
Rab11fip3
|
RAB11 family interacting protein 3 (class II) |
| chr17_+_27904155 | 2.66 |
ENSMUST00000231669.2
ENSMUST00000097360.3 ENSMUST00000231236.2 |
Pacsin1
|
protein kinase C and casein kinase substrate in neurons 1 |
| chr15_-_98575332 | 2.65 |
ENSMUST00000120997.2
ENSMUST00000109149.9 ENSMUST00000003451.11 |
Rnd1
|
Rho family GTPase 1 |
| chr16_+_18210495 | 2.63 |
ENSMUST00000239548.1
|
ARVCF
|
armadillo repeat deleted in velocardiofacial syndrome |
| chr4_+_101353742 | 2.62 |
ENSMUST00000154120.9
ENSMUST00000106930.8 |
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr15_+_78798116 | 2.61 |
ENSMUST00000089378.5
|
Pdxp
|
pyridoxal (pyridoxine, vitamin B6) phosphatase |
| chr19_+_45139098 | 2.59 |
ENSMUST00000026236.11
|
Tlx1
|
T cell leukemia, homeobox 1 |
| chr2_-_113659360 | 2.53 |
ENSMUST00000024005.8
|
Scg5
|
secretogranin V |
| chr12_-_114487525 | 2.53 |
ENSMUST00000103495.3
|
Ighv10-3
|
immunoglobulin heavy variable V10-3 |
| chrX_+_55833061 | 2.50 |
ENSMUST00000151033.2
|
Fhl1
|
four and a half LIM domains 1 |
| chr9_-_66951025 | 2.50 |
ENSMUST00000113695.8
|
Tpm1
|
tropomyosin 1, alpha |
| chr6_-_69204417 | 2.50 |
ENSMUST00000103346.3
|
Igkv4-72
|
immunoglobulin kappa chain variable 4-72 |
| chr18_+_37888770 | 2.48 |
ENSMUST00000061279.10
|
Pcdhga11
|
protocadherin gamma subfamily A, 11 |
| chr1_+_40844739 | 2.47 |
ENSMUST00000114765.4
|
Tmem182
|
transmembrane protein 182 |
| chr9_+_89791943 | 2.46 |
ENSMUST00000189545.2
ENSMUST00000034909.11 ENSMUST00000034912.6 |
Rasgrf1
|
RAS protein-specific guanine nucleotide-releasing factor 1 |
| chr15_+_102011352 | 2.46 |
ENSMUST00000169627.9
|
Tns2
|
tensin 2 |
| chr1_-_38875757 | 2.44 |
ENSMUST00000147695.9
|
Lonrf2
|
LON peptidase N-terminal domain and ring finger 2 |
| chr12_-_113625906 | 2.42 |
ENSMUST00000103448.3
|
Ighv5-9
|
immunoglobulin heavy variable 5-9 |
| chr1_-_36748985 | 2.41 |
ENSMUST00000043951.10
|
Actr1b
|
ARP1 actin-related protein 1B, centractin beta |
| chr9_-_44199428 | 2.40 |
ENSMUST00000160384.2
|
Abcg4
|
ATP binding cassette subfamily G member 4 |
| chr10_-_60055082 | 2.38 |
ENSMUST00000135158.9
|
Chst3
|
carbohydrate sulfotransferase 3 |
| chr6_-_48685108 | 2.37 |
ENSMUST00000126422.3
ENSMUST00000119315.2 ENSMUST00000053661.7 |
Gimap6
|
GTPase, IMAP family member 6 |
| chr1_+_167425953 | 2.36 |
ENSMUST00000015987.10
|
Rxrg
|
retinoid X receptor gamma |
| chr15_+_102011415 | 2.36 |
ENSMUST00000046144.10
|
Tns2
|
tensin 2 |
| chr12_-_113790741 | 2.35 |
ENSMUST00000103457.3
ENSMUST00000192877.2 |
Ighv5-15
|
immunoglobulin heavy variable 5-15 |
| chr12_-_114057841 | 2.34 |
ENSMUST00000103471.2
ENSMUST00000195884.2 |
Ighv9-1
|
immunoglobulin heavy variable 9-1 |
| chr7_+_45434755 | 2.34 |
ENSMUST00000233503.2
ENSMUST00000120005.10 ENSMUST00000211609.2 |
Lmtk3
|
lemur tyrosine kinase 3 |
| chr7_-_100581314 | 2.32 |
ENSMUST00000107032.3
|
Arhgef17
|
Rho guanine nucleotide exchange factor (GEF) 17 |
| chr3_-_87934772 | 2.31 |
ENSMUST00000005014.9
|
Hapln2
|
hyaluronan and proteoglycan link protein 2 |
| chr12_-_113589576 | 2.30 |
ENSMUST00000103446.2
|
Ighv5-6
|
immunoglobulin heavy variable 5-6 |
| chr1_-_57008986 | 2.27 |
ENSMUST00000176759.2
ENSMUST00000177424.2 |
Satb2
|
special AT-rich sequence binding protein 2 |
| chr11_+_101137231 | 2.27 |
ENSMUST00000122006.8
ENSMUST00000151830.2 |
Ramp2
|
receptor (calcitonin) activity modifying protein 2 |
| chr10_+_101517348 | 2.24 |
ENSMUST00000179929.8
ENSMUST00000219195.2 ENSMUST00000127504.9 |
Mgat4c
|
MGAT4 family, member C |
| chr5_+_134128543 | 2.24 |
ENSMUST00000016088.9
|
Castor2
|
cytosolic arginine sensor for mTORC1 subunit 2 |
| chr2_-_27974889 | 2.23 |
ENSMUST00000028179.15
ENSMUST00000117486.8 ENSMUST00000135472.2 |
Fcnb
|
ficolin B |
| chr7_+_127661807 | 2.22 |
ENSMUST00000064821.14
|
Itgam
|
integrin alpha M |
| chr16_+_20408886 | 2.22 |
ENSMUST00000232279.2
ENSMUST00000232474.2 |
Vwa5b2
|
von Willebrand factor A domain containing 5B2 |
| chr5_+_150042092 | 2.21 |
ENSMUST00000200960.4
ENSMUST00000202530.4 |
Fry
|
FRY microtubule binding protein |
| chr5_+_117501557 | 2.20 |
ENSMUST00000111959.2
|
Wsb2
|
WD repeat and SOCS box-containing 2 |
| chr7_-_44320244 | 2.20 |
ENSMUST00000048102.15
|
Myh14
|
myosin, heavy polypeptide 14 |
| chr7_-_119078330 | 2.19 |
ENSMUST00000207460.2
|
Umod
|
uromodulin |
| chr19_+_37538843 | 2.19 |
ENSMUST00000066439.8
ENSMUST00000238817.2 |
Exoc6
|
exocyst complex component 6 |
| chr1_+_89382491 | 2.18 |
ENSMUST00000027521.15
ENSMUST00000190096.7 |
Agap1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| chr11_-_69728560 | 2.17 |
ENSMUST00000108634.9
|
Nlgn2
|
neuroligin 2 |
| chr10_+_115979787 | 2.16 |
ENSMUST00000105271.9
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr4_+_9269285 | 2.14 |
ENSMUST00000038841.14
|
Clvs1
|
clavesin 1 |
| chr9_-_66956425 | 2.14 |
ENSMUST00000113687.8
ENSMUST00000113693.8 ENSMUST00000113701.8 ENSMUST00000034928.12 ENSMUST00000113685.10 ENSMUST00000030185.5 ENSMUST00000050905.16 ENSMUST00000113705.8 ENSMUST00000113697.8 ENSMUST00000113707.9 |
Tpm1
|
tropomyosin 1, alpha |
| chr10_-_8638215 | 2.13 |
ENSMUST00000212553.2
|
Sash1
|
SAM and SH3 domain containing 1 |
| chr17_+_44112679 | 2.12 |
ENSMUST00000229744.2
|
Rcan2
|
regulator of calcineurin 2 |
| chr6_+_30639217 | 2.05 |
ENSMUST00000031806.10
|
Cpa1
|
carboxypeptidase A1, pancreatic |
| chr11_-_97934368 | 2.05 |
ENSMUST00000131519.2
|
Stac2
|
SH3 and cysteine rich domain 2 |
| chr11_+_76900091 | 2.04 |
ENSMUST00000129572.3
|
Slc6a4
|
solute carrier family 6 (neurotransmitter transporter, serotonin), member 4 |
| chr6_-_42301574 | 2.03 |
ENSMUST00000031891.15
ENSMUST00000143278.8 |
Fam131b
|
family with sequence similarity 131, member B |
| chr13_-_49462694 | 2.03 |
ENSMUST00000110087.9
|
Fgd3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr6_-_69245427 | 2.02 |
ENSMUST00000103348.3
|
Igkv4-70
|
immunoglobulin kappa chain variable 4-70 |
| chr19_+_4281953 | 2.01 |
ENSMUST00000025773.5
|
Pold4
|
polymerase (DNA-directed), delta 4 |
| chr15_+_99599978 | 2.01 |
ENSMUST00000023759.6
|
Smarcd1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr10_-_90959853 | 2.01 |
ENSMUST00000170810.8
ENSMUST00000076694.13 |
Slc25a3
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 3 |
| chr1_+_135768595 | 1.99 |
ENSMUST00000112087.9
ENSMUST00000178854.8 ENSMUST00000027671.12 ENSMUST00000179863.8 ENSMUST00000112085.9 ENSMUST00000112086.3 |
Tnnt2
|
troponin T2, cardiac |
| chr15_+_4404965 | 1.97 |
ENSMUST00000061925.5
|
Plcxd3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr6_+_103488291 | 1.97 |
ENSMUST00000204321.2
|
Chl1
|
cell adhesion molecule L1-like |
| chr15_+_99600149 | 1.96 |
ENSMUST00000229236.2
|
Smarcd1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr10_-_127587576 | 1.94 |
ENSMUST00000079692.6
|
Gpr182
|
G protein-coupled receptor 182 |
| chr6_+_110622533 | 1.93 |
ENSMUST00000071076.13
ENSMUST00000172951.2 |
Grm7
|
glutamate receptor, metabotropic 7 |
| chr14_-_60324265 | 1.93 |
ENSMUST00000080368.13
|
Atp8a2
|
ATPase, aminophospholipid transporter-like, class I, type 8A, member 2 |
| chr19_+_43428843 | 1.92 |
ENSMUST00000223787.2
ENSMUST00000165311.3 |
Cnnm1
|
cyclin M1 |
| chr10_-_128237087 | 1.91 |
ENSMUST00000042666.13
|
Slc39a5
|
solute carrier family 39 (metal ion transporter), member 5 |
| chr5_-_147831610 | 1.91 |
ENSMUST00000118527.8
ENSMUST00000031655.4 ENSMUST00000138244.2 |
Slc46a3
|
solute carrier family 46, member 3 |
| chr9_+_45314436 | 1.89 |
ENSMUST00000041005.6
|
Fxyd2
|
FXYD domain-containing ion transport regulator 2 |
| chr5_+_136996713 | 1.89 |
ENSMUST00000001790.6
|
Cldn15
|
claudin 15 |
| chr6_-_124441731 | 1.89 |
ENSMUST00000008297.5
|
Clstn3
|
calsyntenin 3 |
| chr6_-_69037208 | 1.88 |
ENSMUST00000103343.4
|
Igkv4-78
|
immunoglobulin kappa variable 4-78 |
| chr5_-_24829395 | 1.88 |
ENSMUST00000195943.2
|
Smarcd3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr9_+_108685555 | 1.88 |
ENSMUST00000035218.9
ENSMUST00000195323.2 ENSMUST00000194819.2 |
Nckipsd
|
NCK interacting protein with SH3 domain |
| chr2_-_90410922 | 1.87 |
ENSMUST00000168621.3
|
Ptprj
|
protein tyrosine phosphatase, receptor type, J |
| chr2_+_30331839 | 1.86 |
ENSMUST00000131476.8
|
Ptpa
|
protein phosphatase 2 protein activator |
| chr6_-_69162381 | 1.85 |
ENSMUST00000103344.3
|
Igkv4-74
|
immunoglobulin kappa variable 4-74 |
| chr3_+_87704258 | 1.85 |
ENSMUST00000029711.9
ENSMUST00000107582.3 |
Insrr
|
insulin receptor-related receptor |
| chr12_+_109419575 | 1.85 |
ENSMUST00000173539.8
ENSMUST00000109841.9 |
Dlk1
|
delta like non-canonical Notch ligand 1 |
| chr4_-_155430153 | 1.84 |
ENSMUST00000103178.11
|
Prkcz
|
protein kinase C, zeta |
| chr9_+_108356935 | 1.83 |
ENSMUST00000194147.2
ENSMUST00000065014.10 ENSMUST00000195483.6 ENSMUST00000195058.2 |
Lamb2
|
laminin, beta 2 |
| chr11_+_94901104 | 1.83 |
ENSMUST00000124735.2
|
Samd14
|
sterile alpha motif domain containing 14 |
| chr4_+_155045372 | 1.82 |
ENSMUST00000049621.7
|
Hes5
|
hes family bHLH transcription factor 5 |
| chr12_+_109419454 | 1.81 |
ENSMUST00000109846.11
|
Dlk1
|
delta like non-canonical Notch ligand 1 |
| chr7_+_62026747 | 1.78 |
ENSMUST00000080403.7
|
Magel2
|
MAGE family member L2 |
| chr10_-_90959817 | 1.78 |
ENSMUST00000164505.2
|
Slc25a3
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 3 |
| chr2_+_178056302 | 1.77 |
ENSMUST00000094251.11
|
Fam217b
|
family with sequence similarity 217, member B |
| chr1_-_74788013 | 1.77 |
ENSMUST00000188073.7
|
Prkag3
|
protein kinase, AMP-activated, gamma 3 non-catalytic subunit |
| chr2_+_152596075 | 1.77 |
ENSMUST00000010020.12
|
Cox4i2
|
cytochrome c oxidase subunit 4I2 |
| chr6_+_67701864 | 1.75 |
ENSMUST00000103304.3
|
Igkv1-133
|
immunoglobulin kappa variable 1-133 |
| chr1_-_171108754 | 1.75 |
ENSMUST00000073120.11
|
Ppox
|
protoporphyrinogen oxidase |
| chr10_+_41352310 | 1.74 |
ENSMUST00000019967.16
ENSMUST00000119962.8 ENSMUST00000099934.11 |
Mical1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr4_-_137523659 | 1.74 |
ENSMUST00000030551.11
|
Alpl
|
alkaline phosphatase, liver/bone/kidney |
| chr9_-_58648826 | 1.74 |
ENSMUST00000098674.6
|
Rec114
|
REC114 meiotic recombination protein |
| chr1_-_154602102 | 1.72 |
ENSMUST00000187541.7
|
Cacna1e
|
calcium channel, voltage-dependent, R type, alpha 1E subunit |
| chr11_+_95733109 | 1.69 |
ENSMUST00000107714.9
ENSMUST00000107711.8 |
Gngt2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr10_+_80134917 | 1.69 |
ENSMUST00000154212.8
|
Apc2
|
APC regulator of WNT signaling pathway 2 |
| chr5_+_117919082 | 1.69 |
ENSMUST00000138579.3
|
Nos1
|
nitric oxide synthase 1, neuronal |
| chr7_-_4847673 | 1.67 |
ENSMUST00000066041.12
ENSMUST00000119433.4 |
Shisa7
|
shisa family member 7 |
| chr9_-_44231526 | 1.67 |
ENSMUST00000214602.2
ENSMUST00000065080.10 |
C2cd2l
|
C2 calcium-dependent domain containing 2-like |
| chr19_-_10847121 | 1.66 |
ENSMUST00000120524.2
ENSMUST00000025645.14 |
Tmem132a
|
transmembrane protein 132A |
| chr11_-_120520954 | 1.65 |
ENSMUST00000106180.2
|
Mafg
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) |
| chr2_+_170573727 | 1.65 |
ENSMUST00000029075.5
|
Dok5
|
docking protein 5 |
| chrX_+_81992467 | 1.64 |
ENSMUST00000114000.8
|
Dmd
|
dystrophin, muscular dystrophy |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 11.6 | GO:0031283 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 2.7 | 27.0 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 2.0 | 12.1 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 1.8 | 5.3 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 1.6 | 6.3 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 1.5 | 4.6 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 1.5 | 4.6 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 1.4 | 8.6 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 1.3 | 5.1 | GO:0072233 | ascending thin limb development(GO:0072021) thick ascending limb development(GO:0072023) metanephric ascending thin limb development(GO:0072218) metanephric thick ascending limb development(GO:0072233) |
| 1.1 | 5.5 | GO:0070543 | response to linoleic acid(GO:0070543) |
| 1.0 | 8.2 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 1.0 | 3.0 | GO:1902226 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 0.9 | 4.7 | GO:0061643 | chemorepulsion of axon(GO:0061643) |
| 0.9 | 3.7 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.9 | 4.6 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.9 | 3.6 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.9 | 3.6 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.9 | 3.5 | GO:1904616 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.9 | 2.6 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 0.9 | 6.9 | GO:0014004 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.8 | 5.6 | GO:1904717 | regulation of AMPA glutamate receptor clustering(GO:1904717) |
| 0.8 | 3.9 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.7 | 2.2 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.7 | 7.1 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.7 | 5.7 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.7 | 2.8 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.7 | 2.0 | GO:0051610 | serotonin uptake(GO:0051610) |
| 0.7 | 4.7 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.6 | 1.9 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.6 | 1.8 | GO:0072312 | metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.6 | 1.7 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.6 | 2.3 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.6 | 2.2 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.6 | 2.2 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.6 | 2.8 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.5 | 1.6 | GO:0021629 | olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 0.5 | 1.6 | GO:0036145 | dendritic cell homeostasis(GO:0036145) |
| 0.5 | 1.6 | GO:0070237 | positive regulation of activation-induced cell death of T cells(GO:0070237) |
| 0.5 | 2.1 | GO:0007522 | visceral muscle development(GO:0007522) |
| 0.5 | 3.0 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.5 | 2.0 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.5 | 19.7 | GO:0014823 | response to activity(GO:0014823) |
| 0.5 | 2.8 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.5 | 1.8 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.4 | 2.7 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.4 | 1.3 | GO:2001055 | positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 0.4 | 2.6 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.4 | 2.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.4 | 1.3 | GO:0021893 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.4 | 2.9 | GO:0051572 | negative regulation of histone H3-K4 methylation(GO:0051572) |
| 0.4 | 3.3 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.4 | 1.6 | GO:2000984 | regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.4 | 2.8 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.4 | 1.2 | GO:0070839 | divalent metal ion export(GO:0070839) |
| 0.4 | 1.2 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.4 | 1.2 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.4 | 1.9 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.4 | 4.0 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.4 | 1.8 | GO:0072086 | specification of loop of Henle identity(GO:0072086) |
| 0.4 | 2.2 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.4 | 2.2 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.4 | 3.2 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.4 | 1.1 | GO:0007386 | compartment pattern specification(GO:0007386) cerebellar cortex structural organization(GO:0021698) |
| 0.3 | 1.7 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.3 | 1.9 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.3 | 5.4 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.3 | 9.3 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.3 | 4.1 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.3 | 0.9 | GO:0033577 | protein glycosylation in endoplasmic reticulum(GO:0033577) |
| 0.3 | 6.0 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.3 | 0.5 | GO:0055095 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.3 | 2.7 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.3 | 1.8 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.3 | 1.0 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.2 | 1.0 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.2 | 1.0 | GO:1903944 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.2 | 0.2 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.2 | 2.4 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.2 | 1.9 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.2 | 2.3 | GO:0035878 | nail development(GO:0035878) |
| 0.2 | 32.1 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.2 | 1.3 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.2 | 1.5 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.2 | 1.0 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.2 | 0.4 | GO:2000412 | positive regulation of thymocyte migration(GO:2000412) |
| 0.2 | 7.2 | GO:1904376 | negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.2 | 0.8 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.2 | 1.7 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.2 | 2.5 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.2 | 6.2 | GO:0046852 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.2 | 1.9 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.2 | 0.6 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 0.2 | 2.3 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.2 | 3.3 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.2 | 2.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.2 | 3.1 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.2 | 0.5 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.2 | 3.5 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.2 | 0.6 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.2 | 0.5 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.1 | 0.4 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.1 | 5.0 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.1 | 12.1 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.1 | 0.4 | GO:1900620 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.1 | 5.3 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.1 | 1.4 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.1 | 7.1 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.1 | 0.3 | GO:0032831 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) |
| 0.1 | 2.3 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.1 | 0.8 | GO:0032226 | positive regulation of synaptic transmission, dopaminergic(GO:0032226) |
| 0.1 | 28.9 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 3.7 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.1 | 1.8 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 2.5 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 1.9 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.1 | 1.7 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 1.2 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.1 | 0.8 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.1 | 3.7 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.1 | 0.7 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.1 | 2.1 | GO:0007614 | short-term memory(GO:0007614) |
| 0.1 | 3.2 | GO:0000731 | DNA synthesis involved in DNA repair(GO:0000731) |
| 0.1 | 0.5 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 1.6 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.1 | 1.8 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.1 | 2.7 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 0.6 | GO:0033087 | negative regulation of immature T cell proliferation(GO:0033087) |
| 0.1 | 1.7 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 2.6 | GO:0048535 | lymph node development(GO:0048535) |
| 0.1 | 0.8 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 0.3 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.1 | 0.5 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 0.1 | 4.7 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 2.0 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.1 | 0.5 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.1 | 0.2 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.1 | 0.4 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 | 2.2 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.8 | GO:0021554 | optic nerve development(GO:0021554) |
| 0.0 | 4.5 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 3.3 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.8 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.4 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 1.5 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 1.3 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.0 | 1.7 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.0 | 1.5 | GO:0030517 | negative regulation of axon extension(GO:0030517) |
| 0.0 | 1.7 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 1.2 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.6 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 2.4 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.0 | 6.6 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 2.5 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 3.0 | GO:0042398 | cellular modified amino acid biosynthetic process(GO:0042398) |
| 0.0 | 2.9 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.1 | GO:1904306 | positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 0.0 | 0.8 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 3.6 | GO:0061726 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.2 | GO:0003011 | involuntary skeletal muscle contraction(GO:0003011) |
| 0.0 | 0.4 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.7 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 2.1 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.9 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 | 1.8 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.5 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 0.2 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 1.3 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 2.9 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 0.9 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.0 | 1.2 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 | 0.1 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.0 | 0.8 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.4 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.0 | 0.3 | GO:0048149 | adult feeding behavior(GO:0008343) behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.6 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.3 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.4 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 4.1 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 1.5 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 1.0 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.3 | GO:0033129 | positive regulation of histone phosphorylation(GO:0033129) |
| 0.0 | 0.2 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 2.9 | GO:0017157 | regulation of exocytosis(GO:0017157) |
| 0.0 | 0.0 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.0 | 0.3 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 1.2 | GO:1901655 | cellular response to ketone(GO:1901655) |
| 0.0 | 2.2 | GO:0009411 | response to UV(GO:0009411) |
| 0.0 | 1.6 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.6 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.3 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 1.5 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 1.6 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.0 | 1.7 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.2 | GO:1901016 | regulation of potassium ion transmembrane transporter activity(GO:1901016) |
| 0.0 | 0.4 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.0 | 1.6 | GO:0010506 | regulation of autophagy(GO:0010506) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 27.0 | GO:0045298 | tubulin complex(GO:0045298) |
| 1.7 | 12.1 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.9 | 5.7 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.8 | 3.4 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.8 | 3.0 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.7 | 7.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.7 | 10.2 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.6 | 1.9 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.6 | 3.2 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.6 | 3.0 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.6 | 8.7 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.6 | 2.8 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.5 | 1.8 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.4 | 10.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.4 | 3.5 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.3 | 2.2 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.3 | 4.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.3 | 2.0 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.3 | 32.1 | GO:0042571 | immunoglobulin complex(GO:0019814) immunoglobulin complex, circulating(GO:0042571) |
| 0.3 | 1.6 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.2 | 1.2 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.2 | 1.0 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.2 | 5.3 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.2 | 3.0 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 3.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 8.3 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.2 | 1.8 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 14.7 | GO:0032420 | stereocilium(GO:0032420) |
| 0.2 | 2.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.2 | 7.4 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 4.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 1.7 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 7.2 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 3.7 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 27.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 3.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 2.2 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 1.9 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 3.1 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.1 | 5.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 7.1 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 3.0 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 5.3 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 1.9 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.1 | 11.8 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 2.7 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.1 | 1.0 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 2.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 1.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 1.7 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.1 | 2.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 4.0 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 1.2 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 3.0 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 0.6 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 1.9 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 1.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 2.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 10.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 1.7 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 1.8 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 0.6 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 0.3 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 21.9 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 1.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 3.3 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 4.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.5 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 1.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 2.8 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 1.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 4.7 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 1.9 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.6 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 8.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 2.1 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.5 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.5 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 1.1 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 1.7 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 2.2 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 3.8 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 4.2 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 2.2 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 5.1 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 3.1 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 7.5 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 0.4 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.0 | 6.4 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 1.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 27.0 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 1.1 | 5.5 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 1.1 | 3.2 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 1.0 | 3.0 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.9 | 5.7 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.9 | 4.6 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.8 | 3.3 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.8 | 5.6 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.8 | 7.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.8 | 3.8 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.7 | 5.1 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.7 | 2.0 | GO:0005330 | dopamine:sodium symporter activity(GO:0005330) |
| 0.7 | 2.7 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.7 | 2.6 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.6 | 1.9 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.6 | 3.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.5 | 5.3 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.5 | 5.7 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.5 | 4.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.5 | 1.9 | GO:0070905 | serine binding(GO:0070905) |
| 0.5 | 2.4 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.5 | 2.4 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.5 | 1.9 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.4 | 9.8 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.4 | 3.0 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.4 | 3.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.4 | 5.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.4 | 7.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.4 | 11.9 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.3 | 1.7 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.3 | 2.0 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.3 | 3.0 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.3 | 2.1 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.3 | 0.9 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.3 | 0.8 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.3 | 3.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.3 | 1.9 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.3 | 2.7 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.3 | 2.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 1.7 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.2 | 32.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.2 | 1.2 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.2 | 2.3 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.2 | 5.8 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 4.7 | GO:0031005 | filamin binding(GO:0031005) |
| 0.2 | 0.6 | GO:0047223 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.2 | 2.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.2 | 9.3 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.2 | 5.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.2 | 10.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 4.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.2 | 1.7 | GO:0043559 | insulin binding(GO:0043559) |
| 0.2 | 4.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.2 | 3.6 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.2 | 3.8 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.2 | 0.8 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.2 | 3.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 1.6 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 1.8 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.4 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 12.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 1.9 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 6.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 1.0 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 1.0 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 4.9 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 1.8 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.1 | 1.2 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 1.7 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.1 | 1.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 2.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.6 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 1.0 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 1.0 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.1 | 3.9 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 2.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 3.2 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.5 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 3.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 2.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 2.1 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 2.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.8 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.1 | 0.6 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 1.0 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.1 | GO:0052642 | lysophosphatidic acid phosphatase activity(GO:0052642) |
| 0.1 | 1.8 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 3.3 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.1 | 1.0 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.1 | 1.9 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 1.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 1.8 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.7 | GO:0071949 | FAD binding(GO:0071949) |
| 0.1 | 1.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 4.3 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 6.3 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.1 | 1.7 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 2.1 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 2.8 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 1.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 6.0 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.1 | 0.4 | GO:0033265 | choline binding(GO:0033265) |
| 0.1 | 1.1 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.3 | GO:0019976 | interleukin-2 binding(GO:0019976) |
| 0.0 | 0.7 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 2.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.2 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 3.3 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.5 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 2.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.8 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 12.8 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.2 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.2 | GO:0008308 | voltage-gated anion channel activity(GO:0008308) |
| 0.0 | 2.5 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.0 | 4.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.4 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.6 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.8 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 2.0 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 4.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 3.3 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 3.0 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.2 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 3.8 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 0.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 1.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 2.6 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 1.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.2 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.5 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 1.3 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 3.6 | GO:0042277 | peptide binding(GO:0042277) |
| 0.0 | 5.5 | GO:0003779 | actin binding(GO:0003779) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 27.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.2 | 4.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 7.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.2 | 3.0 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 7.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 5.1 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 7.3 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 9.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 9.9 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 8.2 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 5.7 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 2.7 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 4.0 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 0.6 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 0.8 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 5.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 0.8 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 2.2 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 2.7 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.1 | 4.3 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 2.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 2.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.9 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 1.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 2.9 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 2.1 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 4.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 5.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 4.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.5 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.1 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.0 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 26.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.4 | 3.0 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.3 | 4.0 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 3.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.2 | 11.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 10.0 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.2 | 7.2 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.2 | 2.9 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.2 | 10.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 3.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.2 | 3.8 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.2 | 3.0 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 5.9 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 8.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 5.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 3.5 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 1.8 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 5.9 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 7.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 3.0 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 2.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 2.5 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 4.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 4.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.9 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 2.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 12.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 1.8 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 6.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 1.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.7 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 0.8 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 2.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.0 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 0.8 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 0.6 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 1.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 4.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 2.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 2.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 6.9 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 3.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.8 | REACTOME SIGNALING BY NOTCH1 | Genes involved in Signaling by NOTCH1 |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.3 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 1.1 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 1.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.9 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.5 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 4.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.3 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 3.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |