Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
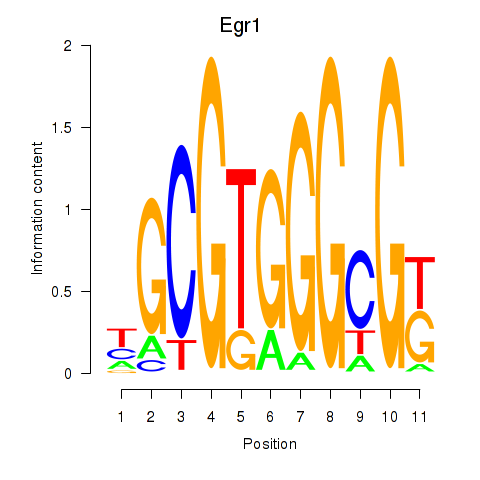
Results for Egr1
Z-value: 1.94
Transcription factors associated with Egr1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Egr1
|
ENSMUSG00000038418.8 | Egr1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Egr1 | mm39_v1_chr18_+_34994253_34994268 | 0.71 | 4.4e-12 | Click! |
Activity profile of Egr1 motif
Sorted Z-values of Egr1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Egr1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_6015538 | 22.10 |
ENSMUST00000101585.10
ENSMUST00000066431.14 ENSMUST00000109815.9 ENSMUST00000109812.9 ENSMUST00000101586.3 ENSMUST00000093355.12 ENSMUST00000019133.11 |
Camk2b
|
calcium/calmodulin-dependent protein kinase II, beta |
| chr10_-_108846816 | 19.90 |
ENSMUST00000105276.8
ENSMUST00000064054.14 |
Syt1
|
synaptotagmin I |
| chrX_-_20787150 | 19.84 |
ENSMUST00000081893.7
ENSMUST00000115345.8 |
Syn1
|
synapsin I |
| chr4_+_127062924 | 19.74 |
ENSMUST00000046659.14
|
Dlgap3
|
DLG associated protein 3 |
| chr9_-_37464200 | 19.65 |
ENSMUST00000065668.12
|
Nrgn
|
neurogranin |
| chrX_+_7504913 | 16.93 |
ENSMUST00000128890.2
|
Syp
|
synaptophysin |
| chr6_-_60805873 | 16.90 |
ENSMUST00000114268.5
|
Snca
|
synuclein, alpha |
| chr5_-_139115914 | 14.87 |
ENSMUST00000129851.8
|
Prkar1b
|
protein kinase, cAMP dependent regulatory, type I beta |
| chr5_+_137551774 | 14.49 |
ENSMUST00000136088.8
ENSMUST00000139395.8 |
Actl6b
|
actin-like 6B |
| chr5_-_139115417 | 14.02 |
ENSMUST00000026973.14
|
Prkar1b
|
protein kinase, cAMP dependent regulatory, type I beta |
| chrX_+_35459621 | 13.94 |
ENSMUST00000115256.2
|
Zcchc12
|
zinc finger, CCHC domain containing 12 |
| chr2_+_158452651 | 13.23 |
ENSMUST00000045738.5
|
Slc32a1
|
solute carrier family 32 (GABA vesicular transporter), member 1 |
| chr4_+_42917228 | 12.94 |
ENSMUST00000107976.9
ENSMUST00000069184.9 |
Phf24
|
PHD finger protein 24 |
| chr6_+_117988399 | 12.72 |
ENSMUST00000164960.4
|
Rasgef1a
|
RasGEF domain family, member 1A |
| chr16_-_34334314 | 12.31 |
ENSMUST00000151491.8
ENSMUST00000114960.9 |
Kalrn
|
kalirin, RhoGEF kinase |
| chr12_+_108300599 | 12.19 |
ENSMUST00000021684.6
|
Cyp46a1
|
cytochrome P450, family 46, subfamily a, polypeptide 1 |
| chr7_-_46782448 | 11.97 |
ENSMUST00000033142.13
|
Ptpn5
|
protein tyrosine phosphatase, non-receptor type 5 |
| chr15_-_44978223 | 11.81 |
ENSMUST00000022967.7
|
Kcnv1
|
potassium channel, subfamily V, member 1 |
| chr10_-_127099183 | 11.62 |
ENSMUST00000099172.5
|
Kif5a
|
kinesin family member 5A |
| chr5_+_137059127 | 11.58 |
ENSMUST00000041543.9
ENSMUST00000186451.2 |
Vgf
|
VGF nerve growth factor inducible |
| chr11_-_97464866 | 11.35 |
ENSMUST00000207653.2
ENSMUST00000107593.8 |
Srcin1
|
SRC kinase signaling inhibitor 1 |
| chr8_+_96404713 | 11.31 |
ENSMUST00000041318.14
|
Ndrg4
|
N-myc downstream regulated gene 4 |
| chr1_-_189075903 | 11.18 |
ENSMUST00000192723.2
ENSMUST00000110920.7 |
Kcnk2
|
potassium channel, subfamily K, member 2 |
| chr16_-_34334454 | 11.18 |
ENSMUST00000089655.12
|
Kalrn
|
kalirin, RhoGEF kinase |
| chr7_-_27095964 | 11.11 |
ENSMUST00000108363.8
|
Sptbn4
|
spectrin beta, non-erythrocytic 4 |
| chr5_+_137551790 | 10.78 |
ENSMUST00000136565.8
ENSMUST00000149292.8 ENSMUST00000125489.2 |
Actl6b
|
actin-like 6B |
| chr9_+_20943350 | 10.74 |
ENSMUST00000019616.6
|
Icam5
|
intercellular adhesion molecule 5, telencephalin |
| chr9_+_60620272 | 10.28 |
ENSMUST00000038407.6
|
Larp6
|
La ribonucleoprotein domain family, member 6 |
| chr11_+_69231274 | 10.11 |
ENSMUST00000129321.2
|
Rnf227
|
ring finger protein 227 |
| chr10_-_116309764 | 10.03 |
ENSMUST00000068233.11
|
Kcnmb4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4 |
| chr6_-_113911640 | 9.98 |
ENSMUST00000101044.9
|
Atp2b2
|
ATPase, Ca++ transporting, plasma membrane 2 |
| chr15_-_74544409 | 9.81 |
ENSMUST00000023268.14
ENSMUST00000110009.4 |
Arc
|
activity regulated cytoskeletal-associated protein |
| chrX_-_72703330 | 9.78 |
ENSMUST00000114473.8
ENSMUST00000002087.14 |
Pnck
|
pregnancy upregulated non-ubiquitously expressed CaM kinase |
| chr8_-_125161061 | 9.77 |
ENSMUST00000140012.8
|
Pgbd5
|
piggyBac transposable element derived 5 |
| chr11_+_80367839 | 9.72 |
ENSMUST00000053413.12
ENSMUST00000147694.2 |
Cdk5r1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
| chr4_-_126647156 | 9.56 |
ENSMUST00000030637.14
ENSMUST00000106116.2 |
Ncdn
|
neurochondrin |
| chrX_-_72703652 | 9.47 |
ENSMUST00000114472.8
|
Pnck
|
pregnancy upregulated non-ubiquitously expressed CaM kinase |
| chr15_-_66158445 | 9.38 |
ENSMUST00000070256.9
|
Kcnq3
|
potassium voltage-gated channel, subfamily Q, member 3 |
| chr7_-_45016138 | 9.34 |
ENSMUST00000211067.2
ENSMUST00000003961.16 |
Ppfia3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr1_-_189075515 | 9.26 |
ENSMUST00000193319.6
|
Kcnk2
|
potassium channel, subfamily K, member 2 |
| chr3_-_89230190 | 9.15 |
ENSMUST00000200436.2
ENSMUST00000029673.10 |
Efna3
|
ephrin A3 |
| chr10_-_79975181 | 9.09 |
ENSMUST00000105369.8
|
Cbarp
|
calcium channel, voltage-dependent, beta subunit associated regulatory protein |
| chr7_+_81824544 | 8.95 |
ENSMUST00000032874.14
|
Sh3gl3
|
SH3-domain GRB2-like 3 |
| chr10_+_79552421 | 8.93 |
ENSMUST00000099513.8
ENSMUST00000020581.3 |
Hcn2
|
hyperpolarization-activated, cyclic nucleotide-gated K+ 2 |
| chr7_+_29991101 | 8.92 |
ENSMUST00000150892.2
ENSMUST00000126216.2 ENSMUST00000014065.16 |
Clip3
|
CAP-GLY domain containing linker protein 3 |
| chr13_-_12121831 | 8.89 |
ENSMUST00000021750.15
ENSMUST00000170156.3 ENSMUST00000220597.2 |
Ryr2
|
ryanodine receptor 2, cardiac |
| chr2_+_157756535 | 8.88 |
ENSMUST00000109523.2
|
Vstm2l
|
V-set and transmembrane domain containing 2-like |
| chr11_-_118460736 | 8.88 |
ENSMUST00000136551.3
|
Rbfox3
|
RNA binding protein, fox-1 homolog (C. elegans) 3 |
| chr7_-_25374472 | 8.68 |
ENSMUST00000108404.8
ENSMUST00000108405.2 ENSMUST00000079439.10 |
Tmem91
|
transmembrane protein 91 |
| chr11_+_69231589 | 8.65 |
ENSMUST00000218008.2
ENSMUST00000151617.3 |
Rnf227
|
ring finger protein 227 |
| chr14_-_76794103 | 8.65 |
ENSMUST00000064517.9
ENSMUST00000228055.2 |
Serp2
|
stress-associated endoplasmic reticulum protein family member 2 |
| chr7_+_99876515 | 8.60 |
ENSMUST00000084935.11
|
Pgm2l1
|
phosphoglucomutase 2-like 1 |
| chrX_+_35459557 | 8.51 |
ENSMUST00000115258.9
|
Zcchc12
|
zinc finger, CCHC domain containing 12 |
| chr9_-_112016966 | 8.51 |
ENSMUST00000178410.2
ENSMUST00000172380.10 |
Arpp21
|
cyclic AMP-regulated phosphoprotein, 21 |
| chr2_-_167030706 | 8.49 |
ENSMUST00000207917.2
|
Kcnb1
|
potassium voltage gated channel, Shab-related subfamily, member 1 |
| chrX_+_35459589 | 8.46 |
ENSMUST00000048067.10
|
Zcchc12
|
zinc finger, CCHC domain containing 12 |
| chr14_-_70873385 | 8.43 |
ENSMUST00000228295.2
ENSMUST00000022695.16 |
Dmtn
|
dematin actin binding protein |
| chrX_+_35459601 | 8.26 |
ENSMUST00000115257.8
|
Zcchc12
|
zinc finger, CCHC domain containing 12 |
| chr12_+_108602008 | 8.23 |
ENSMUST00000172409.2
|
Evl
|
Ena-vasodilator stimulated phosphoprotein |
| chr5_-_115332343 | 8.21 |
ENSMUST00000112113.8
|
Cabp1
|
calcium binding protein 1 |
| chr9_-_89620461 | 8.21 |
ENSMUST00000060700.4
ENSMUST00000185470.3 |
Ankrd34c
|
ankyrin repeat domain 34C |
| chr9_+_58489523 | 8.07 |
ENSMUST00000177292.8
ENSMUST00000085651.12 ENSMUST00000176557.8 ENSMUST00000114121.11 ENSMUST00000177064.8 |
Nptn
|
neuroplastin |
| chr15_+_100768806 | 8.05 |
ENSMUST00000201549.4
ENSMUST00000108908.6 |
Scn8a
|
sodium channel, voltage-gated, type VIII, alpha |
| chr11_-_97464755 | 8.01 |
ENSMUST00000126287.2
ENSMUST00000107590.9 |
Srcin1
|
SRC kinase signaling inhibitor 1 |
| chr11_+_69909245 | 7.99 |
ENSMUST00000231415.2
ENSMUST00000108588.9 |
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr13_+_83672965 | 7.93 |
ENSMUST00000199432.5
ENSMUST00000198069.5 ENSMUST00000197681.5 ENSMUST00000197722.5 ENSMUST00000197938.5 |
Mef2c
|
myocyte enhancer factor 2C |
| chr3_+_96503944 | 7.89 |
ENSMUST00000058943.8
|
Ankrd34a
|
ankyrin repeat domain 34A |
| chr16_+_20408886 | 7.84 |
ENSMUST00000232279.2
ENSMUST00000232474.2 |
Vwa5b2
|
von Willebrand factor A domain containing 5B2 |
| chr2_+_156455583 | 7.83 |
ENSMUST00000109567.10
ENSMUST00000169464.9 |
Dlgap4
|
DLG associated protein 4 |
| chr5_-_108515740 | 7.83 |
ENSMUST00000197216.3
|
Gm42517
|
predicted gene 42517 |
| chr1_+_66507523 | 7.74 |
ENSMUST00000061620.17
ENSMUST00000212557.3 |
Unc80
|
unc-80, NALCN activator |
| chr12_+_108601963 | 7.73 |
ENSMUST00000223109.2
|
Evl
|
Ena-vasodilator stimulated phosphoprotein |
| chr15_-_78428865 | 7.73 |
ENSMUST00000053239.4
|
Sstr3
|
somatostatin receptor 3 |
| chr15_+_89383799 | 7.66 |
ENSMUST00000109309.9
|
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr9_-_102231884 | 7.66 |
ENSMUST00000035129.14
ENSMUST00000085169.12 ENSMUST00000149800.3 |
Ephb1
|
Eph receptor B1 |
| chr16_-_18448614 | 7.63 |
ENSMUST00000231956.2
ENSMUST00000096987.7 |
Septin5
|
septin 5 |
| chr5_-_5430172 | 7.62 |
ENSMUST00000030763.13
|
Cdk14
|
cyclin-dependent kinase 14 |
| chr11_-_75686874 | 7.61 |
ENSMUST00000021209.8
|
Doc2b
|
double C2, beta |
| chr16_+_94171477 | 7.60 |
ENSMUST00000117648.9
ENSMUST00000147352.8 ENSMUST00000150346.8 ENSMUST00000155692.8 ENSMUST00000153988.9 ENSMUST00000139513.9 ENSMUST00000141856.8 ENSMUST00000152117.8 ENSMUST00000150097.8 ENSMUST00000122895.8 ENSMUST00000151770.8 ENSMUST00000231569.2 ENSMUST00000147046.8 ENSMUST00000149885.8 ENSMUST00000127667.8 ENSMUST00000119131.3 ENSMUST00000145883.2 |
Ttc3
|
tetratricopeptide repeat domain 3 |
| chrX_-_7440480 | 7.52 |
ENSMUST00000115742.9
ENSMUST00000150787.8 |
Ppp1r3f
|
protein phosphatase 1, regulatory subunit 3F |
| chr10_-_81308693 | 7.42 |
ENSMUST00000147524.3
ENSMUST00000119060.8 |
Celf5
|
CUGBP, Elav-like family member 5 |
| chr5_+_137059522 | 7.41 |
ENSMUST00000187382.2
|
Vgf
|
VGF nerve growth factor inducible |
| chr16_-_20440005 | 7.39 |
ENSMUST00000052939.4
|
Camk2n2
|
calcium/calmodulin-dependent protein kinase II inhibitor 2 |
| chr3_-_86827640 | 7.32 |
ENSMUST00000195561.6
|
Dclk2
|
doublecortin-like kinase 2 |
| chr12_-_76756772 | 7.29 |
ENSMUST00000166101.2
|
Sptb
|
spectrin beta, erythrocytic |
| chr9_-_20657643 | 7.25 |
ENSMUST00000215999.2
|
Olfm2
|
olfactomedin 2 |
| chr5_+_9316097 | 7.14 |
ENSMUST00000134991.8
ENSMUST00000069538.14 ENSMUST00000115348.9 |
Elapor2
|
endosome-lysosome associated apoptosis and autophagy regulator family member 2 |
| chr2_+_143388062 | 7.12 |
ENSMUST00000028905.10
|
Pcsk2
|
proprotein convertase subtilisin/kexin type 2 |
| chr15_+_81119700 | 7.11 |
ENSMUST00000166855.3
|
Mchr1
|
melanin-concentrating hormone receptor 1 |
| chr15_+_100768776 | 7.08 |
ENSMUST00000108909.9
|
Scn8a
|
sodium channel, voltage-gated, type VIII, alpha |
| chr5_+_120787253 | 6.90 |
ENSMUST00000156722.2
|
Rasal1
|
RAS protein activator like 1 (GAP1 like) |
| chr7_+_122270599 | 6.78 |
ENSMUST00000182563.2
|
Cacng3
|
calcium channel, voltage-dependent, gamma subunit 3 |
| chr7_+_26958150 | 6.76 |
ENSMUST00000079258.7
|
Numbl
|
numb-like |
| chrX_+_5959507 | 6.72 |
ENSMUST00000103007.4
|
Nudt11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11 |
| chr19_+_5100475 | 6.69 |
ENSMUST00000225427.2
|
Rin1
|
Ras and Rab interactor 1 |
| chr1_-_79836344 | 6.64 |
ENSMUST00000027467.11
|
Serpine2
|
serine (or cysteine) peptidase inhibitor, clade E, member 2 |
| chr11_-_97944239 | 6.63 |
ENSMUST00000017544.9
|
Stac2
|
SH3 and cysteine rich domain 2 |
| chr13_+_55517545 | 6.57 |
ENSMUST00000063771.14
|
Rgs14
|
regulator of G-protein signaling 14 |
| chr14_-_20596580 | 6.48 |
ENSMUST00000022355.11
ENSMUST00000161445.8 ENSMUST00000159027.8 |
Ppp3cb
|
protein phosphatase 3, catalytic subunit, beta isoform |
| chr11_-_78388560 | 6.47 |
ENSMUST00000061174.7
|
Sarm1
|
sterile alpha and HEAT/Armadillo motif containing 1 |
| chr13_-_54897660 | 6.44 |
ENSMUST00000135343.2
|
Gprin1
|
G protein-regulated inducer of neurite outgrowth 1 |
| chr7_+_122270623 | 6.42 |
ENSMUST00000182095.2
|
Cacng3
|
calcium channel, voltage-dependent, gamma subunit 3 |
| chr11_-_103844870 | 6.41 |
ENSMUST00000103075.11
|
Nsf
|
N-ethylmaleimide sensitive fusion protein |
| chr13_-_54897425 | 6.41 |
ENSMUST00000099506.2
|
Gprin1
|
G protein-regulated inducer of neurite outgrowth 1 |
| chr13_-_69147639 | 6.39 |
ENSMUST00000022013.8
|
Adcy2
|
adenylate cyclase 2 |
| chr13_-_10410857 | 6.31 |
ENSMUST00000187510.7
|
Chrm3
|
cholinergic receptor, muscarinic 3, cardiac |
| chr12_+_44375665 | 6.28 |
ENSMUST00000110748.4
|
Nrcam
|
neuronal cell adhesion molecule |
| chr11_-_78388284 | 6.21 |
ENSMUST00000108287.10
|
Sarm1
|
sterile alpha and HEAT/Armadillo motif containing 1 |
| chr2_+_81883566 | 6.18 |
ENSMUST00000047527.8
|
Zfp804a
|
zinc finger protein 804A |
| chr11_-_4696778 | 6.17 |
ENSMUST00000009219.3
|
Cabp7
|
calcium binding protein 7 |
| chr2_-_25209107 | 6.17 |
ENSMUST00000114318.10
ENSMUST00000114310.10 ENSMUST00000114308.10 ENSMUST00000114317.10 ENSMUST00000028335.13 ENSMUST00000114314.10 ENSMUST00000114307.8 |
Grin1
|
glutamate receptor, ionotropic, NMDA1 (zeta 1) |
| chrX_+_7744535 | 6.13 |
ENSMUST00000033495.15
|
Pim2
|
proviral integration site 2 |
| chr16_-_18448454 | 6.12 |
ENSMUST00000231622.2
|
Septin5
|
septin 5 |
| chrX_-_58179754 | 6.01 |
ENSMUST00000033473.12
|
Fgf13
|
fibroblast growth factor 13 |
| chr7_+_126550009 | 6.01 |
ENSMUST00000106332.3
|
Sez6l2
|
seizure related 6 homolog like 2 |
| chr17_+_25946644 | 5.89 |
ENSMUST00000237183.2
ENSMUST00000237785.2 ENSMUST00000047273.3 |
Rpusd1
|
RNA pseudouridylate synthase domain containing 1 |
| chr2_-_24653059 | 5.84 |
ENSMUST00000100348.10
ENSMUST00000041342.12 ENSMUST00000114447.8 ENSMUST00000102939.9 ENSMUST00000070864.14 |
Cacna1b
|
calcium channel, voltage-dependent, N type, alpha 1B subunit |
| chr14_-_33169099 | 5.80 |
ENSMUST00000111944.10
ENSMUST00000022504.12 ENSMUST00000111945.9 |
Mapk8
|
mitogen-activated protein kinase 8 |
| chr11_+_69909659 | 5.78 |
ENSMUST00000232002.2
ENSMUST00000134376.10 ENSMUST00000231221.2 |
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr3_-_152373997 | 5.74 |
ENSMUST00000045262.11
|
Ak5
|
adenylate kinase 5 |
| chr3_+_136376440 | 5.71 |
ENSMUST00000056758.9
|
Ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isoform |
| chr16_+_80997580 | 5.71 |
ENSMUST00000037785.14
ENSMUST00000067602.5 |
Ncam2
|
neural cell adhesion molecule 2 |
| chr16_+_4964849 | 5.70 |
ENSMUST00000165810.2
ENSMUST00000230616.2 |
Sec14l5
|
SEC14-like lipid binding 5 |
| chr9_-_112016834 | 5.67 |
ENSMUST00000111872.9
ENSMUST00000164754.9 |
Arpp21
|
cyclic AMP-regulated phosphoprotein, 21 |
| chr18_+_86413077 | 5.63 |
ENSMUST00000058829.4
|
Neto1
|
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr13_+_83672708 | 5.58 |
ENSMUST00000199105.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr7_-_126303947 | 5.55 |
ENSMUST00000032949.14
|
Coro1a
|
coronin, actin binding protein 1A |
| chr15_+_100768551 | 5.49 |
ENSMUST00000082209.13
|
Scn8a
|
sodium channel, voltage-gated, type VIII, alpha |
| chr17_-_56447332 | 5.49 |
ENSMUST00000001256.11
|
Sema6b
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B |
| chr11_-_115258508 | 5.45 |
ENSMUST00000044152.13
ENSMUST00000106542.9 |
Hid1
|
HID1 domain containing |
| chr6_+_85164420 | 5.41 |
ENSMUST00000045942.9
|
Emx1
|
empty spiracles homeobox 1 |
| chr6_-_126621751 | 5.40 |
ENSMUST00000055168.5
|
Kcna1
|
potassium voltage-gated channel, shaker-related subfamily, member 1 |
| chr9_+_21077010 | 5.40 |
ENSMUST00000039413.15
|
Pde4a
|
phosphodiesterase 4A, cAMP specific |
| chr7_+_29991366 | 5.36 |
ENSMUST00000144508.2
|
Clip3
|
CAP-GLY domain containing linker protein 3 |
| chr2_-_118534444 | 5.35 |
ENSMUST00000104937.2
|
Ankrd63
|
ankyrin repeat domain 63 |
| chr11_-_100246209 | 5.26 |
ENSMUST00000146878.3
|
Hap1
|
huntingtin-associated protein 1 |
| chr6_-_126621770 | 5.24 |
ENSMUST00000203094.2
|
Kcna1
|
potassium voltage-gated channel, shaker-related subfamily, member 1 |
| chr4_+_42949814 | 5.23 |
ENSMUST00000037872.10
ENSMUST00000098112.9 |
Dnajb5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr15_+_99122742 | 5.17 |
ENSMUST00000041415.5
|
Kcnh3
|
potassium voltage-gated channel, subfamily H (eag-related), member 3 |
| chr11_+_119833589 | 5.14 |
ENSMUST00000106231.8
ENSMUST00000075180.12 ENSMUST00000103021.10 ENSMUST00000026436.10 ENSMUST00000106233.2 |
Baiap2
|
brain-specific angiogenesis inhibitor 1-associated protein 2 |
| chr12_-_32111214 | 5.09 |
ENSMUST00000003079.12
ENSMUST00000036497.16 |
Prkar2b
|
protein kinase, cAMP dependent regulatory, type II beta |
| chr7_+_28151370 | 5.09 |
ENSMUST00000190954.7
|
Lrfn1
|
leucine rich repeat and fibronectin type III domain containing 1 |
| chr7_-_62862261 | 5.05 |
ENSMUST00000032738.7
|
Chrna7
|
cholinergic receptor, nicotinic, alpha polypeptide 7 |
| chr12_+_102915102 | 5.01 |
ENSMUST00000101099.12
|
Unc79
|
unc-79 homolog |
| chr7_-_126303351 | 4.96 |
ENSMUST00000106364.8
|
Coro1a
|
coronin, actin binding protein 1A |
| chr11_+_68979308 | 4.83 |
ENSMUST00000021273.13
|
Vamp2
|
vesicle-associated membrane protein 2 |
| chr2_+_164328763 | 4.80 |
ENSMUST00000109349.9
|
Dbndd2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr11_-_97520511 | 4.80 |
ENSMUST00000052281.6
|
Epop
|
elongin BC and polycomb repressive complex 2 associated protein |
| chr4_-_47474283 | 4.77 |
ENSMUST00000044148.3
|
Alg2
|
asparagine-linked glycosylation 2 (alpha-1,3-mannosyltransferase) |
| chr5_-_31453206 | 4.74 |
ENSMUST00000041266.11
ENSMUST00000172435.8 ENSMUST00000201417.2 |
Fndc4
|
fibronectin type III domain containing 4 |
| chr5_+_100187844 | 4.69 |
ENSMUST00000169390.8
ENSMUST00000031268.8 |
Enoph1
|
enolase-phosphatase 1 |
| chr7_-_37806912 | 4.63 |
ENSMUST00000108023.10
|
Ccne1
|
cyclin E1 |
| chr7_+_43474819 | 4.63 |
ENSMUST00000107967.3
|
Klk6
|
kallikrein related-peptidase 6 |
| chr2_+_145009625 | 4.61 |
ENSMUST00000110007.8
|
Slc24a3
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 |
| chr3_-_89000591 | 4.59 |
ENSMUST00000090929.12
ENSMUST00000052539.13 |
Rusc1
|
RUN and SH3 domain containing 1 |
| chr13_+_83672654 | 4.52 |
ENSMUST00000199019.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr7_-_142213219 | 4.50 |
ENSMUST00000121128.8
|
Igf2
|
insulin-like growth factor 2 |
| chr3_+_28317570 | 4.47 |
ENSMUST00000160307.9
ENSMUST00000159680.9 ENSMUST00000160518.8 ENSMUST00000162485.8 ENSMUST00000159308.8 ENSMUST00000162777.8 ENSMUST00000161964.2 |
Tnik
|
TRAF2 and NCK interacting kinase |
| chr2_+_149672708 | 4.45 |
ENSMUST00000109935.8
|
Syndig1
|
synapse differentiation inducing 1 |
| chr6_-_120470768 | 4.42 |
ENSMUST00000178687.2
|
Tmem121b
|
transmembrane protein 121B |
| chr18_-_20879461 | 4.40 |
ENSMUST00000070080.6
|
B4galt6
|
UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 6 |
| chr19_-_50667079 | 4.39 |
ENSMUST00000209413.2
ENSMUST00000072685.13 ENSMUST00000164039.9 |
Sorcs1
|
sortilin-related VPS10 domain containing receptor 1 |
| chr6_+_85408953 | 4.36 |
ENSMUST00000045693.8
|
Smyd5
|
SET and MYND domain containing 5 |
| chr12_+_80509978 | 4.35 |
ENSMUST00000219272.2
|
Exd2
|
exonuclease 3'-5' domain containing 2 |
| chr7_+_128290204 | 4.33 |
ENSMUST00000118605.2
|
Inpp5f
|
inositol polyphosphate-5-phosphatase F |
| chr13_-_105430932 | 4.30 |
ENSMUST00000224662.2
|
Rnf180
|
ring finger protein 180 |
| chr15_-_64184485 | 4.30 |
ENSMUST00000177083.8
ENSMUST00000177371.8 |
Asap1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
| chr8_-_69187708 | 4.26 |
ENSMUST00000136060.8
ENSMUST00000130214.8 ENSMUST00000078350.13 |
Csgalnact1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr4_-_151946219 | 4.26 |
ENSMUST00000097774.9
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr8_-_122305486 | 4.14 |
ENSMUST00000212985.2
ENSMUST00000059018.14 |
Fbxo31
|
F-box protein 31 |
| chr15_+_87509413 | 4.12 |
ENSMUST00000068088.8
|
Tafa5
|
TAFA chemokine like family member 5 |
| chr6_+_4747298 | 4.11 |
ENSMUST00000166678.2
ENSMUST00000176204.8 |
Peg10
|
paternally expressed 10 |
| chr2_+_149672814 | 4.08 |
ENSMUST00000137280.2
ENSMUST00000149705.2 |
Syndig1
|
synapse differentiation inducing 1 |
| chr19_+_23736205 | 4.06 |
ENSMUST00000025830.9
|
Apba1
|
amyloid beta (A4) precursor protein binding, family A, member 1 |
| chr12_+_102915709 | 4.05 |
ENSMUST00000179002.8
|
Unc79
|
unc-79 homolog |
| chr7_-_16007542 | 4.01 |
ENSMUST00000169612.3
|
Inafm1
|
InaF motif containing 1 |
| chr19_-_5135510 | 4.00 |
ENSMUST00000140389.8
ENSMUST00000151413.2 ENSMUST00000077066.8 |
Tmem151a
|
transmembrane protein 151A |
| chr11_-_72026547 | 3.99 |
ENSMUST00000108508.3
ENSMUST00000075258.13 |
Pitpnm3
|
PITPNM family member 3 |
| chr13_-_105430889 | 3.99 |
ENSMUST00000226044.2
|
Rnf180
|
ring finger protein 180 |
| chr13_-_43457626 | 3.99 |
ENSMUST00000055341.7
|
Gfod1
|
glucose-fructose oxidoreductase domain containing 1 |
| chr2_+_116951855 | 3.98 |
ENSMUST00000028829.13
|
Spred1
|
sprouty protein with EVH-1 domain 1, related sequence |
| chr7_+_44240310 | 3.85 |
ENSMUST00000107906.6
|
Kcnc3
|
potassium voltage gated channel, Shaw-related subfamily, member 3 |
| chr14_+_84680993 | 3.82 |
ENSMUST00000071370.7
|
Pcdh17
|
protocadherin 17 |
| chr14_+_54713557 | 3.82 |
ENSMUST00000164766.8
|
Rem2
|
rad and gem related GTP binding protein 2 |
| chr10_+_83558729 | 3.77 |
ENSMUST00000150459.3
|
1500009L16Rik
|
RIKEN cDNA 1500009L16 gene |
| chr1_+_60448813 | 3.77 |
ENSMUST00000188594.7
ENSMUST00000188618.7 ENSMUST00000189980.7 |
Abi2
|
abl interactor 2 |
| chr1_-_74544946 | 3.72 |
ENSMUST00000044260.11
ENSMUST00000186282.7 |
Usp37
|
ubiquitin specific peptidase 37 |
| chr12_+_105302853 | 3.71 |
ENSMUST00000180458.9
|
Tunar
|
Tcl1 upstream neural differentiation associated RNA |
| chr17_+_8744201 | 3.69 |
ENSMUST00000115715.8
|
Pde10a
|
phosphodiesterase 10A |
| chr2_-_25209199 | 3.66 |
ENSMUST00000114312.2
|
Grin1
|
glutamate receptor, ionotropic, NMDA1 (zeta 1) |
| chr8_+_40876827 | 3.64 |
ENSMUST00000049389.11
ENSMUST00000128166.8 ENSMUST00000167766.2 |
Zdhhc2
|
zinc finger, DHHC domain containing 2 |
| chr1_-_52272370 | 3.64 |
ENSMUST00000114513.9
ENSMUST00000114510.8 |
Gls
|
glutaminase |
| chr6_+_99669640 | 3.63 |
ENSMUST00000101122.3
|
Gpr27
|
G protein-coupled receptor 27 |
| chr11_-_86648309 | 3.63 |
ENSMUST00000060766.16
ENSMUST00000103186.11 |
Cltc
|
clathrin, heavy polypeptide (Hc) |
| chr7_-_81104423 | 3.63 |
ENSMUST00000178892.3
ENSMUST00000098331.10 |
Cpeb1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr1_-_52271455 | 3.62 |
ENSMUST00000114512.8
|
Gls
|
glutaminase |
| chr2_+_29855572 | 3.58 |
ENSMUST00000113719.9
ENSMUST00000113717.8 ENSMUST00000113741.8 ENSMUST00000100225.9 ENSMUST00000095083.11 ENSMUST00000046257.14 |
Sptan1
|
spectrin alpha, non-erythrocytic 1 |
| chr1_+_75526225 | 3.58 |
ENSMUST00000154101.8
|
Slc4a3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr15_-_100393369 | 3.53 |
ENSMUST00000061457.7
|
Csrnp2
|
cysteine-serine-rich nuclear protein 2 |
| chr3_+_84859453 | 3.49 |
ENSMUST00000029727.8
|
Fbxw7
|
F-box and WD-40 domain protein 7 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.4 | 22.1 | GO:0060466 | activation of meiosis involved in egg activation(GO:0060466) |
| 6.8 | 20.4 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 5.7 | 34.2 | GO:2000327 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 5.6 | 16.9 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 5.6 | 16.9 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 4.9 | 19.6 | GO:0021586 | pons maturation(GO:0021586) |
| 4.2 | 37.8 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 3.0 | 12.0 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 2.9 | 23.5 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 2.4 | 7.1 | GO:0030070 | insulin processing(GO:0030070) |
| 2.3 | 11.6 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 2.2 | 6.6 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 2.1 | 10.6 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 2.0 | 8.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 1.9 | 17.5 | GO:2000969 | positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 1.9 | 5.7 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 1.8 | 14.3 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 1.8 | 21.4 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 1.7 | 17.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 1.7 | 5.0 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) |
| 1.7 | 3.3 | GO:0072054 | renal outer medulla development(GO:0072054) |
| 1.6 | 4.8 | GO:0033577 | protein glycosylation in endoplasmic reticulum(GO:0033577) |
| 1.5 | 32.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 1.4 | 10.0 | GO:0048840 | otolith development(GO:0048840) |
| 1.4 | 5.6 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 1.4 | 8.4 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 1.4 | 8.2 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 1.3 | 16.0 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 1.3 | 13.2 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 1.3 | 6.6 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 1.3 | 10.5 | GO:0032796 | uropod organization(GO:0032796) early endosome to recycling endosome transport(GO:0061502) |
| 1.3 | 3.9 | GO:0072708 | response to sorbitol(GO:0072708) response to dithiothreitol(GO:0072720) |
| 1.3 | 8.9 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 1.3 | 29.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 1.3 | 6.3 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 1.2 | 6.2 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 1.2 | 7.3 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 1.2 | 3.6 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 1.2 | 7.3 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 1.2 | 15.6 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 1.2 | 4.7 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 1.2 | 5.8 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 1.1 | 3.4 | GO:0071874 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) cellular response to norepinephrine stimulus(GO:0071874) |
| 1.1 | 4.4 | GO:1903061 | positive regulation of protein lipidation(GO:1903061) negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 1.1 | 16.0 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 1.1 | 13.8 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 1.1 | 5.3 | GO:0032901 | positive regulation of neurotrophin production(GO:0032901) |
| 1.0 | 9.1 | GO:1905245 | regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902959) positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) regulation of aspartic-type peptidase activity(GO:1905245) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 1.0 | 6.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 1.0 | 4.9 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 1.0 | 15.4 | GO:0043084 | penile erection(GO:0043084) |
| 1.0 | 6.7 | GO:1901909 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 1.0 | 4.8 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.9 | 9.2 | GO:0014719 | skeletal muscle satellite cell activation(GO:0014719) |
| 0.9 | 2.8 | GO:0098885 | modification of postsynaptic actin cytoskeleton(GO:0098885) |
| 0.9 | 2.7 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.9 | 7.1 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.9 | 2.6 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.8 | 12.5 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.8 | 12.4 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.8 | 3.3 | GO:1905077 | negative regulation of interleukin-17 secretion(GO:1905077) |
| 0.8 | 5.6 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.8 | 2.4 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.8 | 12.2 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.8 | 6.0 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.7 | 9.0 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.7 | 19.6 | GO:1900273 | positive regulation of long-term synaptic potentiation(GO:1900273) |
| 0.7 | 3.6 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.7 | 3.6 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.7 | 4.9 | GO:0002447 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.7 | 9.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.7 | 3.5 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.7 | 3.4 | GO:0070428 | regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.6 | 9.1 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.6 | 4.3 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.6 | 3.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.6 | 7.3 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.6 | 10.8 | GO:0042428 | serotonin metabolic process(GO:0042428) |
| 0.6 | 5.9 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.6 | 6.3 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.6 | 2.8 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.5 | 13.2 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.5 | 2.7 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.5 | 2.1 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.5 | 1.6 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.5 | 4.0 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.5 | 7.2 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.5 | 6.8 | GO:0021873 | forebrain neuroblast division(GO:0021873) |
| 0.5 | 9.4 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.5 | 3.2 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.5 | 6.4 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.4 | 1.8 | GO:1901026 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.4 | 2.2 | GO:0072368 | regulation of lipid transport by negative regulation of transcription from RNA polymerase II promoter(GO:0072368) |
| 0.4 | 1.7 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.4 | 22.2 | GO:0061098 | positive regulation of protein tyrosine kinase activity(GO:0061098) |
| 0.4 | 4.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.4 | 7.6 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.4 | 1.2 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 0.4 | 3.2 | GO:0019336 | phenol-containing compound catabolic process(GO:0019336) |
| 0.4 | 12.9 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.4 | 1.6 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.4 | 3.2 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.4 | 3.1 | GO:0060313 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.4 | 1.2 | GO:0048003 | synaptic vesicle recycling via endosome(GO:0036466) antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.4 | 5.1 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.4 | 2.7 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.4 | 5.4 | GO:0070444 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.4 | 1.1 | GO:0021664 | rhombomere formation(GO:0021594) rhombomere 3 formation(GO:0021660) rhombomere 5 morphogenesis(GO:0021664) rhombomere 5 formation(GO:0021666) |
| 0.4 | 5.8 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.4 | 3.2 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.3 | 2.4 | GO:0015871 | choline transport(GO:0015871) |
| 0.3 | 8.1 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.3 | 2.3 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.3 | 23.1 | GO:0007628 | adult walking behavior(GO:0007628) |
| 0.3 | 1.0 | GO:0060618 | nipple development(GO:0060618) mammary gland bud morphogenesis(GO:0060648) |
| 0.3 | 5.8 | GO:0033574 | positive regulation of neurotransmitter secretion(GO:0001956) response to testosterone(GO:0033574) |
| 0.3 | 7.3 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.3 | 6.0 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.3 | 2.5 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.3 | 3.7 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.3 | 0.9 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.3 | 1.5 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.3 | 5.1 | GO:2000463 | positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.3 | 0.3 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.3 | 1.5 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.3 | 2.3 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.3 | 9.6 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.3 | 3.1 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.3 | 1.9 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.3 | 1.1 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.3 | 1.6 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.3 | 9.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.3 | 6.9 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.2 | 2.9 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.2 | 4.1 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.2 | 1.6 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.2 | 6.0 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.2 | 4.4 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.2 | 0.7 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.2 | 1.7 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.2 | 2.1 | GO:0032536 | regulation of microvillus length(GO:0032532) regulation of cell projection size(GO:0032536) |
| 0.2 | 1.9 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.2 | 6.5 | GO:0003299 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.2 | 2.3 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.2 | 0.6 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) |
| 0.2 | 4.0 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.2 | 14.8 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.2 | 3.0 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.2 | 1.3 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.2 | 1.3 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.2 | 6.4 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.2 | 10.9 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.2 | 0.9 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.2 | 1.4 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.2 | 1.6 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.2 | 4.6 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.2 | 6.2 | GO:0051703 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.2 | 1.6 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.2 | 3.7 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.2 | 2.7 | GO:0046473 | phosphatidic acid metabolic process(GO:0046473) |
| 0.1 | 2.1 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.1 | 6.1 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 1.4 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 | 1.3 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.1 | 3.9 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.1 | 4.8 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.1 | 3.9 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 2.0 | GO:0030834 | regulation of actin filament depolymerization(GO:0030834) |
| 0.1 | 1.6 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.1 | 8.6 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.1 | 1.4 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 0.6 | GO:0009173 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.1 | 2.2 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 3.9 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.1 | 1.6 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 9.8 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.1 | 5.3 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 0.8 | GO:0034651 | cortisol biosynthetic process(GO:0034651) |
| 0.1 | 3.3 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.1 | 3.2 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.1 | 0.5 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.1 | 14.5 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.1 | 2.6 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.1 | 2.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 3.2 | GO:0007616 | long-term memory(GO:0007616) |
| 0.1 | 1.1 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 8.5 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.1 | 4.6 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.1 | 7.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 6.0 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 4.1 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.1 | 0.5 | GO:1900038 | negative regulation of cellular response to hypoxia(GO:1900038) |
| 0.1 | 0.2 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.1 | 0.3 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.1 | 1.1 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 0.5 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.1 | 0.5 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 2.5 | GO:0071158 | positive regulation of cell cycle arrest(GO:0071158) |
| 0.1 | 0.7 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 3.9 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 0.8 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.1 | 2.1 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 1.1 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 0.3 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.1 | 1.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.1 | 2.5 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 1.5 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.1 | 1.0 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.1 | 2.7 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 2.1 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 1.4 | GO:0006356 | regulation of transcription from RNA polymerase I promoter(GO:0006356) |
| 0.1 | 0.2 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.1 | 1.3 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 5.7 | GO:0006165 | nucleoside diphosphate phosphorylation(GO:0006165) |
| 0.0 | 0.7 | GO:1904869 | protein localization to nuclear body(GO:1903405) positive regulation of establishment of protein localization to telomere(GO:1904851) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.0 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.8 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 3.4 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.0 | 0.8 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.9 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.8 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.0 | 0.7 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 1.8 | GO:0032435 | negative regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032435) |
| 0.0 | 1.3 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 1.8 | GO:0031623 | receptor internalization(GO:0031623) |
| 0.0 | 0.3 | GO:0043247 | telomere maintenance in response to DNA damage(GO:0043247) |
| 0.0 | 2.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.8 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.7 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.1 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 1.4 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.4 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 1.4 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 | 0.1 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.0 | 0.4 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.6 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 1.1 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 2.3 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 1.7 | GO:0006338 | chromatin remodeling(GO:0006338) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 16.9 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 3.4 | 20.4 | GO:0044305 | calyx of Held(GO:0044305) |
| 3.0 | 8.9 | GO:0098855 | HCN channel complex(GO:0098855) |
| 2.6 | 13.2 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 2.4 | 9.7 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 2.3 | 18.4 | GO:0008091 | spectrin(GO:0008091) |
| 2.2 | 35.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 2.0 | 13.9 | GO:0005955 | calcineurin complex(GO:0005955) |
| 2.0 | 9.8 | GO:0044307 | dendritic branch(GO:0044307) |
| 1.5 | 41.7 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 1.5 | 7.6 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 1.4 | 20.4 | GO:0044327 | dendritic spine head(GO:0044327) |
| 1.3 | 4.0 | GO:1903095 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 1.3 | 7.9 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 1.2 | 8.4 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 1.2 | 29.4 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 1.1 | 19.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 1.0 | 12.7 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.9 | 28.0 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.9 | 3.6 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.9 | 39.7 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.9 | 3.5 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.8 | 4.8 | GO:0070449 | elongin complex(GO:0070449) |
| 0.7 | 7.7 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.7 | 13.8 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.7 | 22.1 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.6 | 3.6 | GO:0031523 | Myb complex(GO:0031523) |
| 0.6 | 11.4 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.6 | 20.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.6 | 29.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.5 | 4.4 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.5 | 20.5 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.5 | 25.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.5 | 3.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.5 | 3.5 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.5 | 6.6 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.5 | 0.9 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.5 | 12.6 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.4 | 1.7 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.4 | 7.7 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.4 | 6.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.4 | 2.8 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.4 | 1.2 | GO:0098830 | presynaptic endosome(GO:0098830) |
| 0.4 | 6.2 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.4 | 22.5 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.4 | 9.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.4 | 11.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.3 | 8.0 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.3 | 2.6 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.3 | 6.9 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.3 | 3.6 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.3 | 25.8 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.3 | 2.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.3 | 4.4 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.3 | 3.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.2 | 4.3 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.2 | 1.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.2 | 6.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.2 | 1.8 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.2 | 20.5 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.2 | 1.5 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.2 | 19.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.2 | 8.9 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.2 | 0.9 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.2 | 20.8 | GO:0030175 | filopodium(GO:0030175) |
| 0.2 | 1.5 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.2 | 23.4 | GO:0043204 | perikaryon(GO:0043204) |
| 0.2 | 1.8 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.2 | 1.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 2.6 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.1 | 2.4 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 2.6 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 0.5 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 2.9 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 4.7 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 0.2 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) cyclin K-CDK13 complex(GO:0002945) |
| 0.1 | 15.2 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.1 | 3.2 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 0.3 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.1 | 15.9 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 20.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 17.2 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 41.1 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 1.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.4 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 8.0 | GO:0016234 | inclusion body(GO:0016234) |
| 0.1 | 44.0 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 1.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 0.8 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.8 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 2.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 1.6 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 4.9 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 0.7 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 1.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 0.7 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 1.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.4 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 1.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 1.0 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 0.2 | GO:0070722 | Tle3-Aes complex(GO:0070722) |
| 0.0 | 3.9 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.7 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.2 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.7 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.7 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.8 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.8 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 1.9 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 1.3 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.6 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.6 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.3 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.3 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 2.1 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.2 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 3.3 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 1.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.8 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.8 | GO:0030139 | endocytic vesicle(GO:0030139) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.6 | 19.9 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 3.4 | 13.6 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 2.9 | 14.3 | GO:0097001 | ceramide binding(GO:0097001) |
| 2.6 | 13.2 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 2.3 | 13.9 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 2.3 | 34.0 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 2.0 | 13.8 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 1.8 | 5.3 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 1.6 | 4.8 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 1.5 | 16.9 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 1.5 | 7.5 | GO:2001069 | glycogen binding(GO:2001069) |
| 1.5 | 8.9 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 1.5 | 7.4 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 1.4 | 8.6 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 1.4 | 10.0 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 1.4 | 28.9 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 1.3 | 7.7 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 1.3 | 6.3 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 1.2 | 6.2 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) D-glucose transmembrane transporter activity(GO:0055056) |
| 1.2 | 9.8 | GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 1.2 | 7.3 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 1.2 | 7.3 | GO:0004359 | glutaminase activity(GO:0004359) |
| 1.2 | 5.8 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 1.1 | 7.7 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 1.1 | 38.0 | GO:0032183 | SUMO binding(GO:0032183) |
| 1.1 | 4.3 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 1.0 | 30.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 1.0 | 6.9 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 1.0 | 6.7 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 1.0 | 28.6 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.9 | 24.0 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.9 | 4.3 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.9 | 2.6 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.8 | 3.4 | GO:0031762 | alpha-1A adrenergic receptor binding(GO:0031691) alpha-1B adrenergic receptor binding(GO:0031692) follicle-stimulating hormone receptor binding(GO:0031762) |
| 0.8 | 3.2 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.8 | 4.0 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.8 | 4.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.8 | 2.4 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.8 | 21.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.8 | 7.8 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.8 | 2.3 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.7 | 9.6 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.6 | 1.9 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.6 | 23.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.6 | 5.1 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.6 | 18.0 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.6 | 3.7 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.6 | 11.0 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.6 | 3.5 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.6 | 9.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.6 | 6.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.5 | 19.0 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.5 | 3.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.5 | 2.6 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.5 | 2.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.5 | 5.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.5 | 10.5 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.5 | 1.9 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.5 | 5.9 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.4 | 21.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.4 | 2.7 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.4 | 3.0 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.4 | 3.4 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.4 | 2.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.4 | 2.5 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.4 | 5.7 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.4 | 28.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.4 | 7.1 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.4 | 11.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.4 | 7.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.3 | 22.3 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.3 | 10.0 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.3 | 1.6 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.3 | 14.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.3 | 24.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.3 | 3.6 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.3 | 6.3 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.3 | 6.6 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.3 | 26.8 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.3 | 22.6 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.3 | 3.6 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.3 | 4.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.2 | 1.7 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.2 | 11.0 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.2 | 2.7 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.2 | 1.8 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.2 | 1.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.2 | 2.7 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.2 | 1.5 | GO:0044547 | rRNA primary transcript binding(GO:0042134) DNA topoisomerase binding(GO:0044547) |
| 0.2 | 2.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.2 | 2.1 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.2 | 0.6 | GO:0001132 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.2 | 0.6 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.2 | 5.4 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.2 | 1.7 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.2 | 3.3 | GO:0004970 | ionotropic glutamate receptor activity(GO:0004970) extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.2 | 18.1 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.2 | 8.6 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.2 | 2.6 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.2 | 2.7 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 3.1 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.2 | 1.6 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.2 | 3.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.2 | 1.4 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.2 | 8.0 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.2 | 2.8 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 4.4 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.1 | 3.4 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.1 | 4.8 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 1.6 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 7.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 3.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 2.0 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.1 | 4.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 3.7 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 1.3 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 6.4 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 6.0 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 0.8 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 2.2 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 1.0 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 0.3 | GO:0016890 | site-specific endodeoxyribonuclease activity, specific for altered base(GO:0016890) |
| 0.1 | 1.9 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 4.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.6 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 1.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 6.6 | GO:0004620 | phospholipase activity(GO:0004620) |
| 0.1 | 1.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 1.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 1.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 2.3 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 1.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 1.5 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.1 | 12.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 7.8 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.1 | 0.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 6.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 6.2 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.5 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 1.0 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 3.3 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.5 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.3 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.9 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 1.4 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 4.0 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 2.8 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.0 | 1.4 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 11.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.7 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 1.0 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.3 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) mRNA CDS binding(GO:1990715) |
| 0.0 | 1.3 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 6.0 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 2.6 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 1.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.1 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.7 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 0.7 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 3.1 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 4.8 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 2.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 9.1 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.4 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 1.3 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.8 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 39.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.6 | 20.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.6 | 34.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.6 | 19.6 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.6 | 11.1 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.5 | 23.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.5 | 22.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.4 | 2.9 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.4 | 20.8 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.3 | 14.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.3 | 7.7 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.3 | 3.4 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.3 | 10.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.3 | 9.8 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.2 | 4.6 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 9.1 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.2 | 32.5 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.2 | 2.3 | PID MYC PATHWAY | C-MYC pathway |
| 0.2 | 5.9 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.2 | 5.7 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.2 | 1.7 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.2 | 3.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.2 | 3.4 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 8.9 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 14.5 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 2.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 1.1 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.1 | 4.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 4.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 4.2 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.1 | 1.4 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 1.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 2.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 3.1 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 3.4 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 0.5 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 3.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 0.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.0 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.2 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 2.7 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.1 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.0 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.8 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.7 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 3.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 44.6 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 1.5 | 45.1 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 1.1 | 18.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 1.1 | 56.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 1.1 | 31.9 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.8 | 13.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.8 | 12.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.7 | 40.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.7 | 8.6 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.6 | 7.3 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.5 | 19.6 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.5 | 13.0 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.5 | 11.8 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.4 | 8.0 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.4 | 25.5 | REACTOME MAPK TARGETS NUCLEAR EVENTS MEDIATED BY MAP KINASES | Genes involved in MAPK targets/ Nuclear events mediated by MAP kinases |
| 0.4 | 13.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.4 | 16.0 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.4 | 12.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.4 | 8.7 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.4 | 2.6 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.4 | 6.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.4 | 11.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.4 | 7.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.3 | 3.7 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.3 | 26.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.3 | 3.6 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.2 | 1.8 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.2 | 3.1 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.2 | 4.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 4.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.2 | 4.6 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.2 | 5.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 2.7 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 2.7 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.1 | 4.0 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 3.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 3.9 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 4.8 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 4.8 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 2.7 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 1.1 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 2.9 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 3.3 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 0.9 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 2.8 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.7 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 5.1 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 1.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 2.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 0.8 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 2.6 | REACTOME RNA POL I TRANSCRIPTION | Genes involved in RNA Polymerase I Transcription |
| 0.1 | 5.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 1.0 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 2.2 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.1 | 1.1 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 2.0 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 12.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 1.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 0.7 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 8.2 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.9 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.6 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.0 | 0.5 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.9 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.6 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.7 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 1.7 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.3 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.3 | REACTOME HOST INTERACTIONS OF HIV FACTORS | Genes involved in Host Interactions of HIV factors |
| 0.0 | 0.3 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.4 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.6 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 2.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |