Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Ehf
Z-value: 0.39
Transcription factors associated with Ehf
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ehf
|
ENSMUSG00000012350.16 | Ehf |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ehf | mm39_v1_chr2_-_103114105_103114147 | 0.02 | 8.4e-01 | Click! |
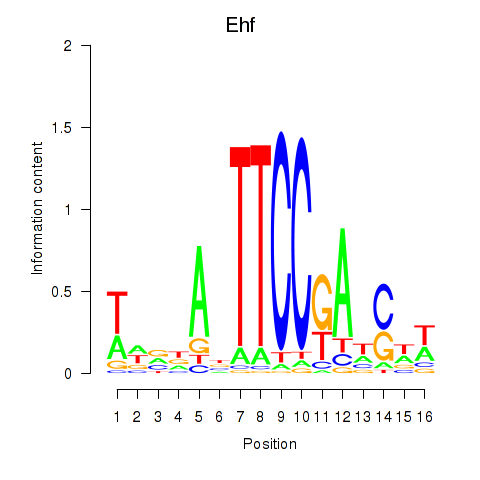
Activity profile of Ehf motif
Sorted Z-values of Ehf motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Ehf
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_41104954 | 2.00 |
ENSMUST00000131699.8
ENSMUST00000024724.14 ENSMUST00000232709.2 ENSMUST00000144243.8 |
Crisp2
|
cysteine-rich secretory protein 2 |
| chr9_+_32027335 | 1.98 |
ENSMUST00000174641.8
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr11_+_3464861 | 1.93 |
ENSMUST00000094469.6
|
Selenom
|
selenoprotein M |
| chr2_+_164647002 | 1.86 |
ENSMUST00000052107.5
|
Zswim3
|
zinc finger SWIM-type containing 3 |
| chr9_-_106324642 | 1.85 |
ENSMUST00000185334.7
ENSMUST00000187001.2 ENSMUST00000171678.9 ENSMUST00000190798.7 ENSMUST00000048685.13 ENSMUST00000171925.8 |
Abhd14a
|
abhydrolase domain containing 14A |
| chrX_+_152341587 | 1.73 |
ENSMUST00000112573.8
ENSMUST00000056754.4 |
Cypt3
|
cysteine-rich perinuclear theca 3 |
| chr18_+_37610858 | 1.57 |
ENSMUST00000051442.7
|
Pcdhb16
|
protocadherin beta 16 |
| chr6_+_54249817 | 1.57 |
ENSMUST00000204921.3
ENSMUST00000203091.3 ENSMUST00000204115.3 ENSMUST00000203941.3 ENSMUST00000204746.2 |
Chn2
|
chimerin 2 |
| chr8_-_106198112 | 1.51 |
ENSMUST00000014990.13
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr18_-_24736521 | 1.39 |
ENSMUST00000154205.2
|
Slc39a6
|
solute carrier family 39 (metal ion transporter), member 6 |
| chr18_-_24736848 | 1.36 |
ENSMUST00000070726.10
|
Slc39a6
|
solute carrier family 39 (metal ion transporter), member 6 |
| chr15_+_4404965 | 1.13 |
ENSMUST00000061925.5
|
Plcxd3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr4_+_48049080 | 0.99 |
ENSMUST00000153369.2
|
Nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr7_+_12246415 | 0.96 |
ENSMUST00000032541.5
|
2900092C05Rik
|
RIKEN cDNA 2900092C05 gene |
| chr11_-_54751738 | 0.93 |
ENSMUST00000144164.9
|
Lyrm7
|
LYR motif containing 7 |
| chr6_-_30892508 | 0.83 |
ENSMUST00000048580.2
|
Tsga13
|
testis specific gene A13 |
| chr16_-_14109219 | 0.77 |
ENSMUST00000230397.2
ENSMUST00000231567.2 ENSMUST00000090287.5 |
Myh11
|
myosin, heavy polypeptide 11, smooth muscle |
| chr13_-_8921027 | 0.74 |
ENSMUST00000177404.2
ENSMUST00000176922.8 ENSMUST00000021572.11 |
Wdr37
|
WD repeat domain 37 |
| chr3_-_141874955 | 0.72 |
ENSMUST00000098568.8
|
Bmpr1b
|
bone morphogenetic protein receptor, type 1B |
| chr15_+_41694317 | 0.69 |
ENSMUST00000166917.3
ENSMUST00000230127.2 ENSMUST00000230131.2 |
Oxr1
|
oxidation resistance 1 |
| chr2_-_164646794 | 0.66 |
ENSMUST00000103094.11
ENSMUST00000017451.7 |
Acot8
|
acyl-CoA thioesterase 8 |
| chr12_-_104831266 | 0.65 |
ENSMUST00000109937.9
|
Clmn
|
calmin |
| chr5_+_90942389 | 0.63 |
ENSMUST00000031322.7
|
Cxcl15
|
chemokine (C-X-C motif) ligand 15 |
| chr17_+_34596098 | 0.57 |
ENSMUST00000080254.7
|
Btnl1
|
butyrophilin-like 1 |
| chr11_-_95966477 | 0.53 |
ENSMUST00000090541.12
|
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 (subunit 9) |
| chr9_+_107419416 | 0.50 |
ENSMUST00000010201.9
|
Nprl2
|
NPR2 like, GATOR1 complex subunit |
| chr8_-_105368298 | 0.49 |
ENSMUST00000093234.5
|
Ciao2b
|
cytosolic iron-sulfur assembly component 2B |
| chr18_+_24737009 | 0.48 |
ENSMUST00000234266.2
ENSMUST00000025120.8 |
Elp2
|
elongator acetyltransferase complex subunit 2 |
| chr6_+_132928065 | 0.46 |
ENSMUST00000070991.5
|
Tas2r129
|
taste receptor, type 2, member 129 |
| chr13_-_8921000 | 0.45 |
ENSMUST00000164183.9
|
Wdr37
|
WD repeat domain 37 |
| chr4_-_120672900 | 0.44 |
ENSMUST00000120779.8
|
Nfyc
|
nuclear transcription factor-Y gamma |
| chr3_+_116388600 | 0.43 |
ENSMUST00000198386.5
ENSMUST00000198311.5 ENSMUST00000197335.2 |
Sass6
|
SAS-6 centriolar assembly protein |
| chr5_-_143133260 | 0.37 |
ENSMUST00000215102.2
ENSMUST00000213631.2 ENSMUST00000164536.5 |
Olfr718-ps1
|
olfactory receptor 718, pseudogene 1 |
| chr11_-_69286159 | 0.35 |
ENSMUST00000108660.8
ENSMUST00000051620.5 |
Cyb5d1
|
cytochrome b5 domain containing 1 |
| chr11_-_100653754 | 0.34 |
ENSMUST00000107360.3
ENSMUST00000055083.4 |
Hcrt
|
hypocretin |
| chr14_-_8798841 | 0.26 |
ENSMUST00000061045.3
|
Sntn
|
sentan, cilia apical structure protein |
| chr14_+_53186347 | 0.26 |
ENSMUST00000103597.3
|
Trav14d-2
|
T cell receptor alpha variable 14D-2 |
| chr11_+_69286473 | 0.25 |
ENSMUST00000144531.2
|
Naa38
|
N(alpha)-acetyltransferase 38, NatC auxiliary subunit |
| chr13_+_109397184 | 0.23 |
ENSMUST00000153234.8
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr9_+_77543776 | 0.18 |
ENSMUST00000057781.8
|
Klhl31
|
kelch-like 31 |
| chr11_-_95966407 | 0.14 |
ENSMUST00000107686.8
ENSMUST00000107684.2 |
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 (subunit 9) |
| chr6_+_132869974 | 0.14 |
ENSMUST00000079035.3
|
Tas2r113
|
taste receptor, type 2, member 113 |
| chr10_-_129577771 | 0.14 |
ENSMUST00000215142.3
ENSMUST00000213239.2 |
Olfr806
|
olfactory receptor 806 |
| chr3_-_116388334 | 0.10 |
ENSMUST00000197190.5
ENSMUST00000198454.2 |
Trmt13
|
tRNA methyltransferase 13 |
| chr13_-_91372072 | 0.09 |
ENSMUST00000022119.6
|
Atg10
|
autophagy related 10 |
| chr1_+_127796508 | 0.02 |
ENSMUST00000037649.6
ENSMUST00000212506.2 |
Rab3gap1
|
RAB3 GTPase activating protein subunit 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.2 | 1.0 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 2.8 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 0.7 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 0.6 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 0.1 | 0.9 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.1 | 0.8 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.7 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.0 | 1.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.3 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.7 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 1.2 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.5 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.7 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.2 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 1.5 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 0.4 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 0.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.8 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 1.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 1.5 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 0.5 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 0.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 0.8 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 0.5 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.3 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 0.4 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.7 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 1.6 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.7 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 2.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.7 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.7 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 1.0 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.8 | GO:0008307 | structural constituent of muscle(GO:0008307) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 1.0 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 2.0 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.6 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.7 | PID BMP PATHWAY | BMP receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.7 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.8 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |