Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
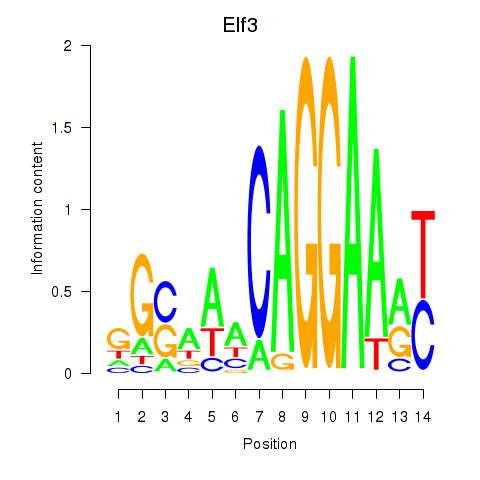
Results for Elf3
Z-value: 1.34
Transcription factors associated with Elf3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Elf3
|
ENSMUSG00000003051.14 | Elf3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Elf3 | mm39_v1_chr1_-_135186176_135186306 | 0.70 | 6.2e-12 | Click! |
Activity profile of Elf3 motif
Sorted Z-values of Elf3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Elf3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_165957784 | 11.52 |
ENSMUST00000060833.14
|
Gpa33
|
glycoprotein A33 (transmembrane) |
| chr7_-_3680530 | 10.68 |
ENSMUST00000038743.15
|
Tmc4
|
transmembrane channel-like gene family 4 |
| chr8_-_65582206 | 10.62 |
ENSMUST00000098713.5
|
Smim31
|
small integral membrane protein 31 |
| chr19_-_11058452 | 9.90 |
ENSMUST00000025636.8
|
Ms4a8a
|
membrane-spanning 4-domains, subfamily A, member 8A |
| chr7_+_130633776 | 9.27 |
ENSMUST00000084509.7
ENSMUST00000213064.3 ENSMUST00000208311.4 |
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr17_+_35268942 | 8.57 |
ENSMUST00000007257.10
|
Clic1
|
chloride intracellular channel 1 |
| chr2_+_151414524 | 8.52 |
ENSMUST00000028950.9
|
Sdcbp2
|
syndecan binding protein (syntenin) 2 |
| chr11_+_115778427 | 8.35 |
ENSMUST00000167507.3
|
Myo15b
|
myosin XVB |
| chr9_-_76474374 | 8.13 |
ENSMUST00000183437.8
|
Fam83b
|
family with sequence similarity 83, member B |
| chr6_+_60921456 | 7.73 |
ENSMUST00000129603.4
ENSMUST00000204333.2 |
Mmrn1
|
multimerin 1 |
| chr1_+_165957909 | 7.47 |
ENSMUST00000166159.2
|
Gpa33
|
glycoprotein A33 (transmembrane) |
| chr1_-_87029312 | 6.62 |
ENSMUST00000113270.3
|
Alpi
|
alkaline phosphatase, intestinal |
| chr10_-_117128763 | 6.42 |
ENSMUST00000092162.7
|
Lyz1
|
lysozyme 1 |
| chr10_+_34359395 | 6.36 |
ENSMUST00000019913.15
|
Frk
|
fyn-related kinase |
| chr7_+_30463175 | 6.25 |
ENSMUST00000165887.8
ENSMUST00000085691.11 ENSMUST00000054427.13 ENSMUST00000085688.11 |
Dmkn
|
dermokine |
| chr3_-_92833537 | 6.17 |
ENSMUST00000067318.6
|
Lce3a
|
late cornified envelope 3A |
| chr10_+_34359513 | 6.08 |
ENSMUST00000170771.3
|
Frk
|
fyn-related kinase |
| chr7_+_24476597 | 6.07 |
ENSMUST00000038069.9
ENSMUST00000206847.2 |
Ceacam10
|
carcinoembryonic antigen-related cell adhesion molecule 10 |
| chr19_+_34044745 | 5.78 |
ENSMUST00000025682.12
ENSMUST00000126710.8 |
Lipn
|
lipase, family member N |
| chr2_-_119060366 | 5.65 |
ENSMUST00000076084.6
|
Ppp1r14d
|
protein phosphatase 1, regulatory inhibitor subunit 14D |
| chr7_+_30475819 | 5.47 |
ENSMUST00000041703.10
|
Dmkn
|
dermokine |
| chr6_-_86770504 | 5.45 |
ENSMUST00000204441.3
ENSMUST00000204398.2 ENSMUST00000001187.15 |
Anxa4
|
annexin A4 |
| chr1_-_136158027 | 5.38 |
ENSMUST00000150163.8
ENSMUST00000144464.7 |
Inava
|
innate immunity activator |
| chr16_+_92295009 | 5.28 |
ENSMUST00000023670.4
|
Clic6
|
chloride intracellular channel 6 |
| chr10_-_117074501 | 5.18 |
ENSMUST00000159193.8
ENSMUST00000020392.5 |
9530003J23Rik
|
RIKEN cDNA 9530003J23 gene |
| chr13_+_120085355 | 5.08 |
ENSMUST00000099241.4
|
Ccl28
|
chemokine (C-C motif) ligand 28 |
| chrX_+_162922317 | 4.96 |
ENSMUST00000112271.10
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr8_+_75599801 | 4.90 |
ENSMUST00000034034.10
|
Isx
|
intestine specific homeobox |
| chr2_-_119060330 | 4.90 |
ENSMUST00000110820.3
|
Ppp1r14d
|
protein phosphatase 1, regulatory inhibitor subunit 14D |
| chr3_+_92874366 | 4.86 |
ENSMUST00000098886.5
|
Lce3e
|
late cornified envelope 3E |
| chr7_+_141988714 | 4.77 |
ENSMUST00000118276.8
ENSMUST00000105976.8 ENSMUST00000097939.9 |
Syt8
|
synaptotagmin VIII |
| chr14_+_80237691 | 4.65 |
ENSMUST00000228749.2
ENSMUST00000088735.4 |
Olfm4
|
olfactomedin 4 |
| chr3_-_30067537 | 4.64 |
ENSMUST00000108270.10
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr16_+_96001650 | 4.52 |
ENSMUST00000048770.16
|
Sh3bgr
|
SH3-binding domain glutamic acid-rich protein |
| chr5_+_37892863 | 4.49 |
ENSMUST00000073554.4
|
Cytl1
|
cytokine-like 1 |
| chr4_+_135413593 | 4.47 |
ENSMUST00000074408.7
|
Ifnlr1
|
interferon lambda receptor 1 |
| chr6_+_113448388 | 4.44 |
ENSMUST00000058300.14
|
Il17rc
|
interleukin 17 receptor C |
| chr10_-_62067026 | 4.41 |
ENSMUST00000047883.11
|
Tspan15
|
tetraspanin 15 |
| chr15_+_79400597 | 4.36 |
ENSMUST00000010974.9
|
Kdelr3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chr18_-_9619460 | 4.36 |
ENSMUST00000234003.2
ENSMUST00000062769.7 |
Cetn1
|
centrin 1 |
| chr2_+_24105430 | 4.30 |
ENSMUST00000028361.5
|
Il1f6
|
interleukin 1 family, member 6 |
| chr16_+_92282593 | 4.04 |
ENSMUST00000162181.8
|
Clic6
|
chloride intracellular channel 6 |
| chr5_+_17779273 | 4.01 |
ENSMUST00000030568.14
|
Sema3c
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr11_-_103235475 | 3.97 |
ENSMUST00000041385.14
|
Arhgap27
|
Rho GTPase activating protein 27 |
| chr14_-_70761507 | 3.80 |
ENSMUST00000022692.5
|
Sftpc
|
surfactant associated protein C |
| chr1_-_133834790 | 3.62 |
ENSMUST00000149380.8
ENSMUST00000124051.9 |
Optc
|
opticin |
| chr11_-_69576363 | 3.56 |
ENSMUST00000018896.14
|
Tnfsf13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr3_-_92366022 | 3.46 |
ENSMUST00000058142.4
|
Sprr3
|
small proline-rich protein 3 |
| chr1_-_133617824 | 3.36 |
ENSMUST00000189524.2
ENSMUST00000169295.8 |
Lax1
|
lymphocyte transmembrane adaptor 1 |
| chr5_+_115680964 | 3.35 |
ENSMUST00000137716.8
|
Pxn
|
paxillin |
| chr19_-_40982576 | 3.32 |
ENSMUST00000117695.8
|
Blnk
|
B cell linker |
| chr4_-_14621805 | 3.30 |
ENSMUST00000042221.14
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr7_-_101582987 | 3.27 |
ENSMUST00000106964.8
ENSMUST00000106963.2 ENSMUST00000078448.11 ENSMUST00000106966.8 |
Lrrc51
|
leucine rich repeat containing 51 |
| chr2_+_164404499 | 3.26 |
ENSMUST00000017867.10
ENSMUST00000109344.9 ENSMUST00000109345.9 |
Wfdc2
|
WAP four-disulfide core domain 2 |
| chr3_+_90511068 | 3.24 |
ENSMUST00000001046.7
|
S100a4
|
S100 calcium binding protein A4 |
| chr11_+_73068063 | 3.18 |
ENSMUST00000108477.2
|
Tax1bp3
|
Tax1 (human T cell leukemia virus type I) binding protein 3 |
| chr7_+_126808016 | 3.16 |
ENSMUST00000206204.2
ENSMUST00000206772.2 |
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chrX_+_106860083 | 3.15 |
ENSMUST00000143975.8
ENSMUST00000144695.8 ENSMUST00000167154.2 |
Tent5d
|
terminal nucleotidyltransferase 5D |
| chr3_+_92864693 | 3.07 |
ENSMUST00000059053.11
|
Lce3d
|
late cornified envelope 3D |
| chr3_+_84573499 | 2.98 |
ENSMUST00000107682.2
|
Tmem154
|
transmembrane protein 154 |
| chr11_+_78811613 | 2.91 |
ENSMUST00000018610.7
|
Nos2
|
nitric oxide synthase 2, inducible |
| chr11_-_120971954 | 2.87 |
ENSMUST00000106120.3
ENSMUST00000100126.9 ENSMUST00000106119.9 |
Sectm1a
|
secreted and transmembrane 1A |
| chr16_+_9988080 | 2.73 |
ENSMUST00000121292.8
ENSMUST00000044103.6 |
Rpl39l
|
ribosomal protein L39-like |
| chr3_+_146205562 | 2.73 |
ENSMUST00000090031.12
ENSMUST00000118280.2 |
Gng5
|
guanine nucleotide binding protein (G protein), gamma 5 |
| chr8_-_123302187 | 2.68 |
ENSMUST00000213062.2
|
Aprt
|
adenine phosphoribosyl transferase |
| chr13_-_74956924 | 2.66 |
ENSMUST00000223206.2
|
Cast
|
calpastatin |
| chr3_-_84489783 | 2.64 |
ENSMUST00000107687.9
ENSMUST00000098990.10 |
Arfip1
|
ADP-ribosylation factor interacting protein 1 |
| chr1_+_40363701 | 2.63 |
ENSMUST00000095020.9
ENSMUST00000194296.6 |
Il1rl2
|
interleukin 1 receptor-like 2 |
| chrX_+_138511360 | 2.59 |
ENSMUST00000113026.2
|
Rnf128
|
ring finger protein 128 |
| chr7_-_25176959 | 2.44 |
ENSMUST00000098668.3
ENSMUST00000206687.2 ENSMUST00000206676.2 ENSMUST00000205308.2 ENSMUST00000098669.8 ENSMUST00000206171.2 ENSMUST00000098666.9 |
Ceacam1
|
carcinoembryonic antigen-related cell adhesion molecule 1 |
| chr12_-_65219328 | 2.41 |
ENSMUST00000124201.2
ENSMUST00000052201.9 ENSMUST00000222244.2 ENSMUST00000221296.2 |
Mis18bp1
|
MIS18 binding protein 1 |
| chr7_+_141040988 | 2.33 |
ENSMUST00000053670.12
|
Cracr2b
|
calcium release activated channel regulator 2B |
| chr16_+_48692976 | 2.31 |
ENSMUST00000065666.6
|
Retnlg
|
resistin like gamma |
| chr3_-_89325594 | 2.31 |
ENSMUST00000029679.4
|
Cks1b
|
CDC28 protein kinase 1b |
| chr9_-_114393406 | 2.30 |
ENSMUST00000111816.3
|
Trim71
|
tripartite motif-containing 71 |
| chr4_-_14621669 | 2.19 |
ENSMUST00000143105.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr3_-_52012462 | 2.19 |
ENSMUST00000121440.4
|
Maml3
|
mastermind like transcriptional coactivator 3 |
| chr7_-_25239229 | 2.17 |
ENSMUST00000044547.10
ENSMUST00000066503.14 ENSMUST00000064862.13 |
Ceacam2
|
carcinoembryonic antigen-related cell adhesion molecule 2 |
| chr4_-_156312996 | 2.16 |
ENSMUST00000105571.4
|
Plekhn1
|
pleckstrin homology domain containing, family N member 1 |
| chr11_+_32483290 | 2.14 |
ENSMUST00000102821.4
|
Stk10
|
serine/threonine kinase 10 |
| chr17_+_25690538 | 2.14 |
ENSMUST00000234449.2
ENSMUST00000025002.3 ENSMUST00000235033.2 |
Tekt4
|
tektin 4 |
| chr13_+_13612136 | 2.11 |
ENSMUST00000005532.9
|
Nid1
|
nidogen 1 |
| chr10_-_23662948 | 2.05 |
ENSMUST00000220070.2
|
Rps12
|
ribosomal protein S12 |
| chr11_-_120971984 | 2.00 |
ENSMUST00000026162.12
|
Sectm1a
|
secreted and transmembrane 1A |
| chrX_+_11184495 | 1.99 |
ENSMUST00000179859.2
|
H2al1g
|
H2A histone family member L1G |
| chr11_-_117670430 | 1.90 |
ENSMUST00000143406.8
|
Tmc6
|
transmembrane channel-like gene family 6 |
| chr7_+_143027473 | 1.90 |
ENSMUST00000052348.12
|
Slc22a18
|
solute carrier family 22 (organic cation transporter), member 18 |
| chr6_+_70648743 | 1.90 |
ENSMUST00000103401.3
|
Igkv3-4
|
immunoglobulin kappa variable 3-4 |
| chr16_-_43836681 | 1.86 |
ENSMUST00000036174.10
|
Gramd1c
|
GRAM domain containing 1C |
| chr5_+_143166759 | 1.86 |
ENSMUST00000031574.10
|
Spdye4b
|
speedy/RINGO cell cycle regulator family, member E4B |
| chr6_+_83214357 | 1.85 |
ENSMUST00000039212.8
ENSMUST00000113899.8 |
Slc4a5
|
solute carrier family 4, sodium bicarbonate cotransporter, member 5 |
| chr9_+_110867807 | 1.83 |
ENSMUST00000197575.2
|
Ltf
|
lactotransferrin |
| chr4_-_156312961 | 1.83 |
ENSMUST00000217885.2
|
Plekhn1
|
pleckstrin homology domain containing, family N member 1 |
| chr1_+_58841650 | 1.83 |
ENSMUST00000165549.8
|
Casp8
|
caspase 8 |
| chr3_-_146205429 | 1.82 |
ENSMUST00000029839.11
|
Spata1
|
spermatogenesis associated 1 |
| chr11_+_119205613 | 1.80 |
ENSMUST00000053245.7
|
Card14
|
caspase recruitment domain family, member 14 |
| chr11_+_53660834 | 1.79 |
ENSMUST00000108920.10
ENSMUST00000140866.9 ENSMUST00000108922.9 |
Irf1
|
interferon regulatory factor 1 |
| chr8_+_126721878 | 1.79 |
ENSMUST00000046765.10
|
Kcnk1
|
potassium channel, subfamily K, member 1 |
| chr2_+_144665576 | 1.78 |
ENSMUST00000028918.4
|
Scp2d1
|
SCP2 sterol-binding domain containing 1 |
| chr2_+_153621851 | 1.78 |
ENSMUST00000126656.4
|
Efcab8
|
EF-hand calcium binding domain 8 |
| chr7_-_143294051 | 1.78 |
ENSMUST00000119499.8
|
Osbpl5
|
oxysterol binding protein-like 5 |
| chr18_+_35347983 | 1.78 |
ENSMUST00000235449.2
ENSMUST00000235269.2 |
Ctnna1
|
catenin (cadherin associated protein), alpha 1 |
| chr9_-_108140925 | 1.77 |
ENSMUST00000171412.7
ENSMUST00000195429.6 ENSMUST00000080435.9 |
Dag1
|
dystroglycan 1 |
| chr1_-_130557349 | 1.77 |
ENSMUST00000142416.2
ENSMUST00000039862.11 ENSMUST00000128128.8 |
Zp3r
|
zona pellucida 3 receptor |
| chr9_-_119019450 | 1.76 |
ENSMUST00000093775.12
|
Slc22a14
|
solute carrier family 22 (organic cation transporter), member 14 |
| chr2_-_29641647 | 1.76 |
ENSMUST00000129574.3
|
Gm13420
|
predicted gene 13420 |
| chr2_-_84545504 | 1.76 |
ENSMUST00000035840.6
|
Zdhhc5
|
zinc finger, DHHC domain containing 5 |
| chr3_+_85946145 | 1.75 |
ENSMUST00000238331.2
|
Sh3d19
|
SH3 domain protein D19 |
| chr11_+_77656414 | 1.75 |
ENSMUST00000164315.2
|
Myo18a
|
myosin XVIIIA |
| chr8_-_89362745 | 1.72 |
ENSMUST00000034087.9
|
Snx20
|
sorting nexin 20 |
| chr1_-_36578439 | 1.72 |
ENSMUST00000191849.6
|
Gm42417
|
predicted gene, 42417 |
| chr1_-_186947618 | 1.71 |
ENSMUST00000110945.4
ENSMUST00000183931.8 ENSMUST00000027908.13 |
Spata17
|
spermatogenesis associated 17 |
| chr17_+_28491085 | 1.70 |
ENSMUST00000169040.3
|
Ppard
|
peroxisome proliferator activator receptor delta |
| chr11_+_49554430 | 1.69 |
ENSMUST00000043873.10
ENSMUST00000076006.5 |
Scgb3a1
|
secretoglobin, family 3A, member 1 |
| chr6_+_48963795 | 1.68 |
ENSMUST00000037696.6
|
Svs1
|
seminal vesicle secretory protein 1 |
| chr6_+_17636979 | 1.68 |
ENSMUST00000015877.14
ENSMUST00000152005.3 |
Capza2
|
capping protein (actin filament) muscle Z-line, alpha 2 |
| chr1_+_58834532 | 1.66 |
ENSMUST00000027189.15
|
Casp8
|
caspase 8 |
| chr2_+_68966125 | 1.65 |
ENSMUST00000041865.8
|
Nostrin
|
nitric oxide synthase trafficker |
| chr4_-_14621497 | 1.65 |
ENSMUST00000149633.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr8_-_83129832 | 1.65 |
ENSMUST00000034148.7
|
Il15
|
interleukin 15 |
| chr9_-_50657800 | 1.64 |
ENSMUST00000239417.2
ENSMUST00000034564.4 |
2310030G06Rik
|
RIKEN cDNA 2310030G06 gene |
| chr5_+_115681003 | 1.64 |
ENSMUST00000157050.8
|
Pxn
|
paxillin |
| chr3_+_89325750 | 1.64 |
ENSMUST00000039110.12
ENSMUST00000125036.8 ENSMUST00000191485.7 ENSMUST00000154791.8 |
Shc1
|
src homology 2 domain-containing transforming protein C1 |
| chr16_+_48637219 | 1.63 |
ENSMUST00000023328.8
|
Retnlb
|
resistin like beta |
| chr19_-_57227742 | 1.62 |
ENSMUST00000111559.8
|
Ablim1
|
actin-binding LIM protein 1 |
| chr8_+_126722113 | 1.61 |
ENSMUST00000212831.2
|
Kcnk1
|
potassium channel, subfamily K, member 1 |
| chr1_+_58841808 | 1.60 |
ENSMUST00000190213.2
|
Casp8
|
caspase 8 |
| chr6_+_21986445 | 1.60 |
ENSMUST00000115382.8
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr9_+_44990447 | 1.59 |
ENSMUST00000050020.8
|
Jaml
|
junction adhesion molecule like |
| chr5_+_145063568 | 1.56 |
ENSMUST00000138922.2
|
Arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr1_-_156936197 | 1.56 |
ENSMUST00000187546.7
ENSMUST00000118207.8 ENSMUST00000027884.13 ENSMUST00000121911.8 |
Tex35
|
testis expressed 35 |
| chr11_+_109253598 | 1.55 |
ENSMUST00000106702.4
ENSMUST00000020930.14 |
Gna13
|
guanine nucleotide binding protein, alpha 13 |
| chr4_-_92035446 | 1.55 |
ENSMUST00000107108.8
ENSMUST00000143542.2 |
Izumo3
|
IZUMO family member 3 |
| chr11_+_53661251 | 1.53 |
ENSMUST00000138913.8
ENSMUST00000123376.8 ENSMUST00000019043.13 ENSMUST00000133291.3 |
Irf1
|
interferon regulatory factor 1 |
| chr11_-_94673526 | 1.53 |
ENSMUST00000100554.8
|
Tmem92
|
transmembrane protein 92 |
| chr3_-_85909798 | 1.53 |
ENSMUST00000061343.4
|
Prss48
|
protease, serine 48 |
| chr14_+_32713349 | 1.52 |
ENSMUST00000120866.8
ENSMUST00000120588.8 |
Lrrc18
|
leucine rich repeat containing 18 |
| chr17_+_88976088 | 1.51 |
ENSMUST00000024970.11
ENSMUST00000161481.2 |
Gtf2a1l
|
general transcription factor IIA, 1-like |
| chr10_+_79824418 | 1.51 |
ENSMUST00000004784.11
ENSMUST00000105374.2 |
Cnn2
|
calponin 2 |
| chr9_-_67739607 | 1.49 |
ENSMUST00000054500.7
|
C2cd4a
|
C2 calcium-dependent domain containing 4A |
| chr7_-_79882228 | 1.46 |
ENSMUST00000123279.8
|
Cib1
|
calcium and integrin binding 1 (calmyrin) |
| chr17_+_13574834 | 1.44 |
ENSMUST00000233944.2
ENSMUST00000097403.4 |
Tcp10c
|
t-complex protein 10c |
| chr9_+_45014092 | 1.43 |
ENSMUST00000217074.2
|
Jaml
|
junction adhesion molecule like |
| chr18_+_50411431 | 1.42 |
ENSMUST00000039121.4
ENSMUST00000238078.2 |
Fam170a
|
family with sequence similarity 170, member A |
| chr1_-_186947651 | 1.40 |
ENSMUST00000183819.8
|
Spata17
|
spermatogenesis associated 17 |
| chr7_+_119393210 | 1.40 |
ENSMUST00000033218.15
ENSMUST00000106520.9 |
Rexo5
|
RNA exonuclease 5 |
| chr8_+_72943455 | 1.40 |
ENSMUST00000072097.14
|
Hsh2d
|
hematopoietic SH2 domain containing |
| chr1_+_136395673 | 1.39 |
ENSMUST00000189413.7
ENSMUST00000047817.12 |
Kif14
|
kinesin family member 14 |
| chr1_-_22386016 | 1.39 |
ENSMUST00000164877.8
|
Rims1
|
regulating synaptic membrane exocytosis 1 |
| chr17_+_7592045 | 1.39 |
ENSMUST00000095726.11
ENSMUST00000128533.8 ENSMUST00000129709.8 ENSMUST00000147803.8 ENSMUST00000140192.8 ENSMUST00000138222.8 ENSMUST00000144861.2 |
Tcp10a
|
t-complex protein 10a |
| chr19_+_29923182 | 1.36 |
ENSMUST00000025724.9
|
Il33
|
interleukin 33 |
| chr14_+_32713387 | 1.35 |
ENSMUST00000123822.8
ENSMUST00000120951.2 |
Lrrc18
|
leucine rich repeat containing 18 |
| chr12_-_113649535 | 1.35 |
ENSMUST00000103449.4
ENSMUST00000195707.3 |
Ighv2-5
|
immunoglobulin heavy variable 2-5 |
| chr3_-_122778052 | 1.33 |
ENSMUST00000199401.2
ENSMUST00000197314.5 ENSMUST00000197934.5 ENSMUST00000090379.7 |
Usp53
|
ubiquitin specific peptidase 53 |
| chr12_+_105651643 | 1.33 |
ENSMUST00000051934.7
|
Gskip
|
GSK3B interacting protein |
| chr15_-_83316995 | 1.33 |
ENSMUST00000165095.9
|
Pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr6_-_128277701 | 1.32 |
ENSMUST00000143004.2
ENSMUST00000006311.13 ENSMUST00000112157.4 ENSMUST00000133118.2 |
Tead4
|
TEA domain family member 4 |
| chr16_+_19578981 | 1.32 |
ENSMUST00000079780.10
ENSMUST00000164397.8 |
B3gnt5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr11_-_55310724 | 1.31 |
ENSMUST00000108858.8
ENSMUST00000141530.2 |
Sparc
|
secreted acidic cysteine rich glycoprotein |
| chr15_-_83317020 | 1.31 |
ENSMUST00000231184.2
|
Pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr11_-_83540175 | 1.31 |
ENSMUST00000001008.6
|
Ccl3
|
chemokine (C-C motif) ligand 3 |
| chr5_+_124577952 | 1.31 |
ENSMUST00000059580.11
|
Kmt5a
|
lysine methyltransferase 5A |
| chr7_-_25089522 | 1.31 |
ENSMUST00000054301.14
|
Lipe
|
lipase, hormone sensitive |
| chr15_-_76079891 | 1.30 |
ENSMUST00000023226.13
|
Plec
|
plectin |
| chr19_+_4198615 | 1.30 |
ENSMUST00000123874.8
ENSMUST00000008893.9 ENSMUST00000237125.2 |
Coro1b
|
coronin, actin binding protein 1B |
| chr1_+_84817547 | 1.29 |
ENSMUST00000097672.4
|
Fbxo36
|
F-box protein 36 |
| chr7_+_130179063 | 1.27 |
ENSMUST00000207918.2
ENSMUST00000215492.2 ENSMUST00000084513.12 ENSMUST00000059145.14 |
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr17_-_40630096 | 1.26 |
ENSMUST00000026498.5
|
Crisp1
|
cysteine-rich secretory protein 1 |
| chrX_+_11165496 | 1.26 |
ENSMUST00000188439.2
|
H2al1a
|
H2A histone family member L1A |
| chr3_+_108444837 | 1.26 |
ENSMUST00000029485.6
|
1700013F07Rik
|
RIKEN cDNA 1700013F07 gene |
| chr1_+_16758629 | 1.26 |
ENSMUST00000026881.11
|
Ly96
|
lymphocyte antigen 96 |
| chr2_-_25086810 | 1.25 |
ENSMUST00000081869.7
|
Tor4a
|
torsin family 4, member A |
| chr8_+_93687561 | 1.25 |
ENSMUST00000072939.8
|
Slc6a2
|
solute carrier family 6 (neurotransmitter transporter, noradrenalin), member 2 |
| chr19_+_40648182 | 1.24 |
ENSMUST00000112231.9
ENSMUST00000127828.8 |
Entpd1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr18_+_4165832 | 1.24 |
ENSMUST00000025076.10
|
Lyzl1
|
lysozyme-like 1 |
| chr2_-_120119544 | 1.24 |
ENSMUST00000094665.5
|
Pla2g4d
|
phospholipase A2, group IVD |
| chr11_-_121009503 | 1.23 |
ENSMUST00000039146.4
|
Tex19.2
|
testis expressed gene 19.2 |
| chr16_+_19578945 | 1.22 |
ENSMUST00000121344.8
|
B3gnt5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chrX_+_9150003 | 1.21 |
ENSMUST00000073949.4
|
H2al1m
|
H2A histone family member L1M |
| chr5_-_124018055 | 1.18 |
ENSMUST00000164267.2
|
Hcar1
|
hydrocarboxylic acid receptor 1 |
| chr1_+_58834621 | 1.16 |
ENSMUST00000191201.7
|
Casp8
|
caspase 8 |
| chr9_-_119169053 | 1.15 |
ENSMUST00000035092.7
|
Myd88
|
myeloid differentiation primary response gene 88 |
| chrX_+_11178173 | 1.12 |
ENSMUST00000178979.2
|
H2al1e
|
H2A histone family member L1E |
| chr2_-_72810782 | 1.12 |
ENSMUST00000102689.10
|
Sp3
|
trans-acting transcription factor 3 |
| chr5_+_115567644 | 1.11 |
ENSMUST00000150779.8
|
Msi1
|
musashi RNA-binding protein 1 |
| chr14_-_54491365 | 1.09 |
ENSMUST00000128231.2
|
Dad1
|
defender against cell death 1 |
| chr3_+_89325901 | 1.09 |
ENSMUST00000128238.8
ENSMUST00000107417.9 |
Shc1
|
src homology 2 domain-containing transforming protein C1 |
| chr2_+_153816842 | 1.08 |
ENSMUST00000109753.3
|
Bpifa6
|
BPI fold containing family A, member 6 |
| chr14_+_32713336 | 1.08 |
ENSMUST00000038956.12
|
Lrrc18
|
leucine rich repeat containing 18 |
| chr14_+_54701594 | 1.07 |
ENSMUST00000022782.10
|
Lrp10
|
low-density lipoprotein receptor-related protein 10 |
| chr5_+_14075281 | 1.04 |
ENSMUST00000073957.8
|
Sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr4_-_34050038 | 1.03 |
ENSMUST00000084734.11
|
Spaca1
|
sperm acrosome associated 1 |
| chr18_+_56565188 | 1.02 |
ENSMUST00000070166.6
|
Gramd3
|
GRAM domain containing 3 |
| chr7_+_119393312 | 1.01 |
ENSMUST00000084644.3
|
Rexo5
|
RNA exonuclease 5 |
| chr7_+_130467564 | 1.00 |
ENSMUST00000075181.11
ENSMUST00000151119.9 ENSMUST00000048180.12 |
Plekha1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1 |
| chr8_-_44152524 | 1.00 |
ENSMUST00000212185.2
|
Adam34
|
a disintegrin and metallopeptidase domain 34 |
| chr2_-_25086770 | 1.00 |
ENSMUST00000142857.2
ENSMUST00000137920.2 |
Tor4a
|
torsin family 4, member A |
| chr3_+_87826834 | 1.00 |
ENSMUST00000137775.2
|
Mrpl24
|
mitochondrial ribosomal protein L24 |
| chr2_-_60793536 | 0.99 |
ENSMUST00000028347.13
|
Rbms1
|
RNA binding motif, single stranded interacting protein 1 |
| chr11_-_59118988 | 0.99 |
ENSMUST00000163300.8
ENSMUST00000061242.8 |
Arf1
|
ADP-ribosylation factor 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 11.7 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 1.6 | 4.9 | GO:1901738 | regulation of vitamin A metabolic process(GO:1901738) |
| 1.3 | 5.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 1.2 | 5.0 | GO:0015827 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 1.2 | 3.6 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.9 | 2.8 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.8 | 3.3 | GO:2000564 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.8 | 14.8 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.8 | 7.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.7 | 5.4 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.7 | 2.7 | GO:0046083 | adenine salvage(GO:0006168) adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.7 | 2.7 | GO:1900191 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.6 | 4.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.6 | 1.8 | GO:0021682 | nerve maturation(GO:0021682) |
| 0.6 | 1.7 | GO:1903028 | regulation of opsonization(GO:1903027) positive regulation of opsonization(GO:1903028) |
| 0.6 | 2.3 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.6 | 1.7 | GO:0006429 | leucyl-tRNA aminoacylation(GO:0006429) |
| 0.5 | 2.1 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.5 | 4.0 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.5 | 2.9 | GO:1990839 | response to endothelin(GO:1990839) |
| 0.5 | 4.4 | GO:0035696 | monocyte extravasation(GO:0035696) |
| 0.5 | 3.4 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.5 | 1.4 | GO:0033624 | cerebellar cortex structural organization(GO:0021698) regulation of Rap protein signal transduction(GO:0032487) negative regulation of integrin activation(GO:0033624) |
| 0.4 | 6.2 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.4 | 1.3 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.4 | 2.9 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.4 | 1.9 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.4 | 0.7 | GO:0071640 | regulation of macrophage inflammatory protein 1 alpha production(GO:0071640) |
| 0.4 | 1.5 | GO:0038163 | endomitotic cell cycle(GO:0007113) thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.3 | 1.0 | GO:0097212 | cleavage furrow ingression(GO:0036090) lysosomal membrane organization(GO:0097212) |
| 0.3 | 1.6 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-15(GO:0071350) |
| 0.3 | 1.3 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) diacylglycerol catabolic process(GO:0046340) |
| 0.3 | 8.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.3 | 1.8 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.3 | 1.8 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.3 | 1.5 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.3 | 0.9 | GO:0099526 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.3 | 4.6 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.3 | 1.7 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.3 | 1.1 | GO:0072566 | chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.3 | 1.4 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.3 | 1.3 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.3 | 1.8 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.2 | 2.7 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.2 | 17.0 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.2 | 2.2 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.2 | 0.7 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.2 | 1.4 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.2 | 17.9 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.2 | 4.1 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.2 | 0.7 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.2 | 4.7 | GO:0002385 | mucosal immune response(GO:0002385) |
| 0.2 | 1.2 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.2 | 3.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.2 | 0.6 | GO:1902460 | transforming growth factor beta activation(GO:0036363) regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.2 | 0.6 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 0.2 | 0.7 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.2 | 2.3 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.2 | 1.2 | GO:1900045 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.2 | 0.5 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.2 | 2.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.2 | 2.6 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.2 | 4.5 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.2 | 0.5 | GO:0046087 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.2 | 1.4 | GO:0061517 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) |
| 0.2 | 0.5 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.1 | 0.6 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.1 | 2.7 | GO:0007343 | egg activation(GO:0007343) |
| 0.1 | 5.0 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 0.7 | GO:0002268 | follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
| 0.1 | 0.7 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 | 1.6 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.1 | 2.1 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 1.0 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.1 | 0.4 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.1 | 0.6 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.1 | 2.3 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.3 | GO:0045212 | neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.1 | 0.8 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.1 | 1.5 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) embryonic process involved in female pregnancy(GO:0060136) |
| 0.1 | 1.3 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 4.8 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.3 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) |
| 0.1 | 0.2 | GO:2000019 | negative regulation of male gonad development(GO:2000019) |
| 0.1 | 0.3 | GO:0060722 | spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) |
| 0.1 | 1.3 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.1 | 2.2 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.1 | 1.0 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.1 | 2.6 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.1 | 5.3 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 0.4 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.1 | 1.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.6 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.1 | 0.6 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.5 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.1 | 0.9 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.1 | 0.2 | GO:0002148 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 0.1 | 0.5 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.1 | 0.5 | GO:0038109 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Fc-epsilon receptor signaling pathway(GO:0038095) Kit signaling pathway(GO:0038109) |
| 0.1 | 1.7 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 6.9 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.1 | 2.1 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 1.5 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.1 | 3.3 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 1.7 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 7.6 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.1 | 1.9 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.1 | 0.9 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.1 | 6.1 | GO:0030317 | sperm motility(GO:0030317) |
| 0.1 | 0.6 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.1 | 1.3 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.1 | 0.2 | GO:0009955 | adaxial/abaxial pattern specification(GO:0009955) regulation of adaxial/abaxial pattern formation(GO:2000011) |
| 0.1 | 0.8 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.4 | GO:1990009 | retinal cell apoptotic process(GO:1990009) |
| 0.1 | 1.2 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.1 | 1.0 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 1.2 | GO:0015874 | norepinephrine transport(GO:0015874) |
| 0.0 | 1.3 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.0 | 0.4 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 1.8 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.0 | 0.5 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.2 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 2.2 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 1.4 | GO:0001953 | negative regulation of cell-matrix adhesion(GO:0001953) |
| 0.0 | 1.0 | GO:0002360 | T cell lineage commitment(GO:0002360) |
| 0.0 | 1.0 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 1.6 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 1.0 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.2 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 1.3 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 6.7 | GO:0035304 | regulation of protein dephosphorylation(GO:0035304) |
| 0.0 | 1.8 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 2.7 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 2.5 | GO:0030148 | sphingolipid biosynthetic process(GO:0030148) |
| 0.0 | 0.4 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.2 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.7 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.5 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 0.2 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 2.7 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 0.7 | GO:0006672 | ceramide metabolic process(GO:0006672) |
| 0.0 | 4.2 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 6.2 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.0 | 0.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.9 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.1 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 | 0.1 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.4 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.5 | GO:0046326 | positive regulation of glucose import(GO:0046326) |
| 0.0 | 1.0 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 2.5 | GO:0071773 | response to BMP(GO:0071772) cellular response to BMP stimulus(GO:0071773) |
| 0.0 | 0.0 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.2 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.0 | 0.2 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 1.6 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.1 | GO:0032532 | regulation of microvillus length(GO:0032532) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 6.2 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 0.9 | 6.4 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.9 | 2.7 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.8 | 9.9 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.7 | 2.7 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.7 | 3.4 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.4 | 1.8 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.4 | 1.6 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 0.3 | 3.8 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.3 | 1.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.3 | 6.5 | GO:0042581 | specific granule(GO:0042581) |
| 0.3 | 17.0 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.3 | 1.5 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.2 | 2.8 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.2 | 1.4 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.2 | 18.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.2 | 1.8 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.2 | 0.6 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.2 | 2.4 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.2 | 1.0 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 1.8 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.1 | 0.7 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.1 | 1.9 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 3.4 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 0.6 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.1 | 0.5 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 1.0 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 1.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.5 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 2.4 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 6.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 2.6 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 5.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.9 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 1.0 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 1.5 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 10.1 | GO:0042641 | actomyosin(GO:0042641) |
| 0.1 | 0.8 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 3.2 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 1.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.0 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.9 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 8.1 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 3.4 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 1.7 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.0 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 1.0 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 2.4 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 2.3 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 2.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.2 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 2.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 7.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.1 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.6 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.2 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 3.9 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 1.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 5.7 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.7 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 11.6 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.2 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.6 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 1.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.7 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.2 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.7 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 5.9 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 54.1 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.8 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 1.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 9.3 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 3.1 | 9.3 | GO:0035375 | zymogen binding(GO:0035375) |
| 2.0 | 8.1 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 1.6 | 6.2 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 1.2 | 5.0 | GO:0051435 | BH4 domain binding(GO:0051435) |
| 0.9 | 6.6 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.9 | 2.7 | GO:0002055 | adenine binding(GO:0002055) adenine phosphoribosyltransferase activity(GO:0003999) |
| 0.9 | 4.4 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.8 | 2.5 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.8 | 5.0 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.8 | 14.8 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.8 | 3.3 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.7 | 2.9 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.7 | 2.6 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.6 | 1.7 | GO:0004823 | leucine-tRNA ligase activity(GO:0004823) |
| 0.6 | 1.7 | GO:0016501 | prostacyclin receptor activity(GO:0016501) |
| 0.5 | 1.6 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.5 | 4.4 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.5 | 3.9 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.5 | 4.4 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.5 | 2.7 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.4 | 1.2 | GO:0005333 | norepinephrine transmembrane transporter activity(GO:0005333) |
| 0.4 | 7.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.4 | 2.3 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.4 | 2.7 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.3 | 1.8 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.3 | 1.5 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.3 | 1.1 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.3 | 5.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 3.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.2 | 4.6 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.2 | 0.4 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.2 | 10.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.2 | 0.6 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.2 | 1.3 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.2 | 1.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.2 | 4.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.2 | 3.4 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.2 | 0.8 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.2 | 6.4 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.2 | 1.8 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 11.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 1.2 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.1 | 0.4 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.1 | 0.7 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.1 | 0.8 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 0.3 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.1 | 4.4 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 1.1 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 1.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 1.0 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 0.9 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 1.8 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.2 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.1 | 9.1 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.1 | 3.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 2.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 2.3 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 0.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 4.4 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 0.7 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.3 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 1.8 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 1.6 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 2.1 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 1.9 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 0.1 | 1.1 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 0.5 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.1 | 0.9 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 0.5 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.5 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.3 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.8 | GO:0001164 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 1.9 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 1.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.4 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.5 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.7 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 1.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 1.7 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.4 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 2.9 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.6 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 1.0 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.9 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 2.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 9.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.4 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.8 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.2 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 6.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.8 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 1.8 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.7 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.8 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.6 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.5 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0052894 | norspermine:oxygen oxidoreductase activity(GO:0052894) N1-acetylspermine:oxygen oxidoreductase (N1-acetylspermidine-forming) activity(GO:0052895) |
| 0.0 | 0.4 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 1.3 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 8.5 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 1.5 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.6 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.2 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 1.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 3.6 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 11.1 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.2 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.7 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.2 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 1.2 | GO:0032934 | sterol binding(GO:0032934) |
| 0.0 | 0.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 1.6 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 5.1 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 1.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.4 | GO:0101005 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 1.7 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 1.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.2 | 1.8 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.2 | 7.9 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 5.2 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 1.6 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 4.0 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 20.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 3.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 2.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 1.0 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 0.9 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 1.0 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 3.0 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 2.0 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 2.3 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 2.0 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 5.3 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 1.0 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.9 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 1.6 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.5 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.6 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.3 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.3 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 2.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.7 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.2 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.2 | 1.8 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.2 | 3.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 3.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 3.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 2.9 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 2.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 1.9 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 2.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 1.4 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 1.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 4.1 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.1 | 3.3 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 1.0 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 3.9 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 4.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 10.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 1.3 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 3.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 8.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 0.9 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 0.7 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 1.6 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 3.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 0.7 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 1.6 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 1.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.5 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 2.3 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 1.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.9 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.9 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 0.6 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 3.0 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.7 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.8 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 1.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 1.3 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.5 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.5 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |