Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Emx1_Emx2
Z-value: 1.22
Transcription factors associated with Emx1_Emx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
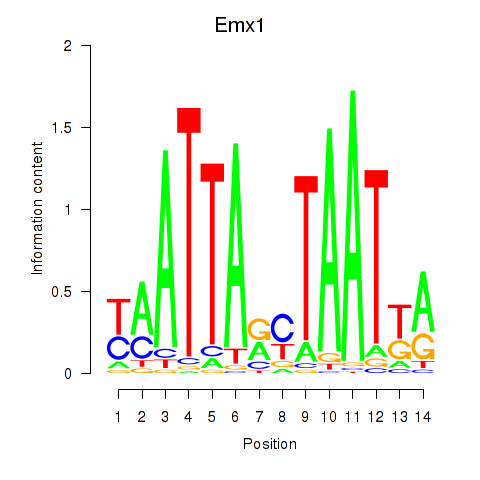
Emx1
|
ENSMUSG00000033726.9 | Emx1 |
|
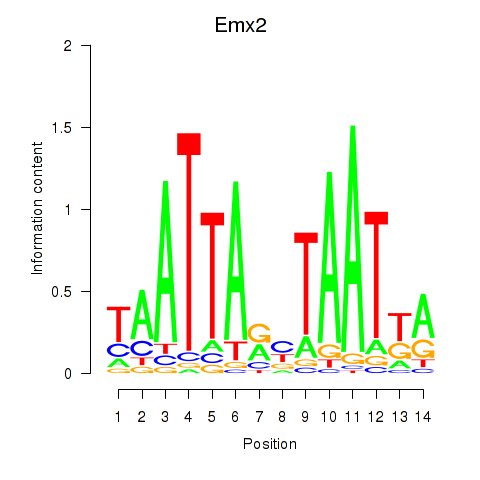
Emx2
|
ENSMUSG00000043969.5 | Emx2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Emx1 | mm39_v1_chr6_+_85164420_85164505 | 0.15 | 2.0e-01 | Click! |
| Emx2 | mm39_v1_chr19_+_59447950_59448025 | -0.04 | 7.7e-01 | Click! |
Activity profile of Emx1_Emx2 motif
Sorted Z-values of Emx1_Emx2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Emx1_Emx2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_39801188 | 15.42 |
ENSMUST00000162507.2
ENSMUST00000160476.9 ENSMUST00000239028.2 |
Cyp2c40
|
cytochrome P450, family 2, subfamily c, polypeptide 40 |
| chr5_-_89605622 | 13.61 |
ENSMUST00000049209.13
|
Gc
|
vitamin D binding protein |
| chr12_-_11485639 | 11.46 |
ENSMUST00000220506.2
|
Vsnl1
|
visinin-like 1 |
| chr4_-_61972348 | 9.90 |
ENSMUST00000074018.4
|
Mup20
|
major urinary protein 20 |
| chr19_+_39275518 | 8.95 |
ENSMUST00000003137.15
|
Cyp2c29
|
cytochrome P450, family 2, subfamily c, polypeptide 29 |
| chr13_+_4486105 | 8.81 |
ENSMUST00000156277.2
|
Akr1c6
|
aldo-keto reductase family 1, member C6 |
| chr16_-_26190578 | 8.42 |
ENSMUST00000023154.3
|
Cldn1
|
claudin 1 |
| chr4_-_60070411 | 8.18 |
ENSMUST00000079697.10
ENSMUST00000125282.2 ENSMUST00000166098.8 |
Mup7
|
major urinary protein 7 |
| chr16_+_22737128 | 7.73 |
ENSMUST00000170805.9
|
Fetub
|
fetuin beta |
| chr16_+_22737050 | 7.52 |
ENSMUST00000231768.2
|
Fetub
|
fetuin beta |
| chr16_+_22737227 | 7.27 |
ENSMUST00000231880.2
|
Fetub
|
fetuin beta |
| chr19_-_39637489 | 6.92 |
ENSMUST00000067328.7
|
Cyp2c67
|
cytochrome P450, family 2, subfamily c, polypeptide 67 |
| chr19_-_39875192 | 6.65 |
ENSMUST00000168838.3
|
Cyp2c69
|
cytochrome P450, family 2, subfamily c, polypeptide 69 |
| chr5_-_87240405 | 6.06 |
ENSMUST00000132667.2
ENSMUST00000145617.8 ENSMUST00000094649.11 |
Ugt2b36
|
UDP glucuronosyltransferase 2 family, polypeptide B36 |
| chr12_+_74044435 | 5.94 |
ENSMUST00000221220.2
|
Syt16
|
synaptotagmin XVI |
| chr14_-_57371041 | 5.65 |
ENSMUST00000039380.9
|
Gjb6
|
gap junction protein, beta 6 |
| chr1_-_139786421 | 5.45 |
ENSMUST00000194186.6
ENSMUST00000094489.5 ENSMUST00000239380.2 |
Cfhr2
|
complement factor H-related 2 |
| chr1_+_58152295 | 5.27 |
ENSMUST00000040999.14
ENSMUST00000162011.3 |
Aox3
|
aldehyde oxidase 3 |
| chr1_-_106980033 | 5.09 |
ENSMUST00000112717.3
|
Serpinb3a
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 3A |
| chr4_-_61357980 | 4.71 |
ENSMUST00000095049.5
|
Mup15
|
major urinary protein 15 |
| chr3_+_57332735 | 4.59 |
ENSMUST00000029377.8
|
Tm4sf4
|
transmembrane 4 superfamily member 4 |
| chr1_-_158183894 | 4.46 |
ENSMUST00000004133.11
|
Brinp2
|
bone morphogenic protein/retinoic acid inducible neural-specific 2 |
| chr19_-_39451509 | 4.36 |
ENSMUST00000035488.3
|
Cyp2c38
|
cytochrome P450, family 2, subfamily c, polypeptide 38 |
| chr10_-_85847697 | 4.04 |
ENSMUST00000105304.2
ENSMUST00000061699.12 |
Bpifc
|
BPI fold containing family C |
| chr2_-_17465410 | 4.02 |
ENSMUST00000145492.2
|
Nebl
|
nebulette |
| chr10_+_81093110 | 4.01 |
ENSMUST00000117488.8
ENSMUST00000105328.10 ENSMUST00000121205.8 |
Matk
|
megakaryocyte-associated tyrosine kinase |
| chr14_+_14210932 | 3.96 |
ENSMUST00000022271.14
|
Acox2
|
acyl-Coenzyme A oxidase 2, branched chain |
| chr1_-_139708906 | 3.74 |
ENSMUST00000111986.8
ENSMUST00000027612.11 ENSMUST00000111989.9 |
Cfhr4
|
complement factor H-related 4 |
| chr1_-_174749379 | 3.48 |
ENSMUST00000055294.4
|
Grem2
|
gremlin 2, DAN family BMP antagonist |
| chr3_+_92899521 | 3.24 |
ENSMUST00000090863.5
|
Lce3f
|
late cornified envelope 3F |
| chr19_-_39729431 | 3.23 |
ENSMUST00000099472.4
|
Cyp2c68
|
cytochrome P450, family 2, subfamily c, polypeptide 68 |
| chrX_-_159777661 | 3.09 |
ENSMUST00000087104.11
|
Cdkl5
|
cyclin-dependent kinase-like 5 |
| chr2_+_110551927 | 3.06 |
ENSMUST00000111017.9
|
Muc15
|
mucin 15 |
| chr6_-_57512355 | 3.00 |
ENSMUST00000042766.6
|
Ppm1k
|
protein phosphatase 1K (PP2C domain containing) |
| chrX_+_158242121 | 2.99 |
ENSMUST00000112470.3
ENSMUST00000043151.12 ENSMUST00000156172.3 |
Map7d2
|
MAP7 domain containing 2 |
| chr2_+_110551685 | 2.95 |
ENSMUST00000111016.9
|
Muc15
|
mucin 15 |
| chr14_+_65504067 | 2.86 |
ENSMUST00000224629.2
|
Fbxo16
|
F-box protein 16 |
| chr19_+_34078333 | 2.84 |
ENSMUST00000025685.8
|
Lipm
|
lipase, family member M |
| chr9_+_21634779 | 2.77 |
ENSMUST00000034713.9
|
Ldlr
|
low density lipoprotein receptor |
| chr18_-_38999755 | 2.72 |
ENSMUST00000115582.8
ENSMUST00000236060.2 |
Fgf1
|
fibroblast growth factor 1 |
| chr2_+_110551976 | 2.69 |
ENSMUST00000090332.5
|
Muc15
|
mucin 15 |
| chr10_-_67748461 | 2.60 |
ENSMUST00000064656.8
|
Zfp365
|
zinc finger protein 365 |
| chr17_+_44337566 | 2.59 |
ENSMUST00000229939.2
|
Rcan2
|
regulator of calcineurin 2 |
| chr6_+_129510331 | 2.55 |
ENSMUST00000204956.2
ENSMUST00000204639.2 |
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr6_+_129510117 | 2.53 |
ENSMUST00000032264.9
|
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr5_+_104350475 | 2.53 |
ENSMUST00000066708.7
|
Dmp1
|
dentin matrix protein 1 |
| chr6_+_129510145 | 2.46 |
ENSMUST00000204487.3
|
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr7_+_24335969 | 2.43 |
ENSMUST00000080718.6
|
Lypd3
|
Ly6/Plaur domain containing 3 |
| chr16_-_43836681 | 2.35 |
ENSMUST00000036174.10
|
Gramd1c
|
GRAM domain containing 1C |
| chr6_-_144155197 | 2.32 |
ENSMUST00000038815.14
ENSMUST00000111749.8 ENSMUST00000170367.9 |
Sox5
|
SRY (sex determining region Y)-box 5 |
| chr13_+_16189041 | 2.31 |
ENSMUST00000164993.2
|
Inhba
|
inhibin beta-A |
| chr3_+_92864693 | 2.29 |
ENSMUST00000059053.11
|
Lce3d
|
late cornified envelope 3D |
| chr17_+_79919267 | 2.27 |
ENSMUST00000223924.2
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr12_+_37930305 | 2.27 |
ENSMUST00000220990.2
|
Dgkb
|
diacylglycerol kinase, beta |
| chr4_-_82423944 | 2.24 |
ENSMUST00000107248.8
ENSMUST00000107247.8 |
Nfib
|
nuclear factor I/B |
| chr8_-_126625029 | 2.21 |
ENSMUST00000047239.13
ENSMUST00000131127.3 |
Pcnx2
|
pecanex homolog 2 |
| chr10_-_49659355 | 2.21 |
ENSMUST00000105484.10
ENSMUST00000218598.2 ENSMUST00000079751.9 ENSMUST00000218441.2 |
Grik2
|
glutamate receptor, ionotropic, kainate 2 (beta 2) |
| chr4_-_112632013 | 2.19 |
ENSMUST00000060327.4
|
Skint10
|
selection and upkeep of intraepithelial T cells 10 |
| chr9_+_78099229 | 2.18 |
ENSMUST00000034903.7
|
Gsta4
|
glutathione S-transferase, alpha 4 |
| chr6_-_58884038 | 2.14 |
ENSMUST00000059539.5
|
Nap1l5
|
nucleosome assembly protein 1-like 5 |
| chr14_+_27598021 | 2.10 |
ENSMUST00000211684.2
ENSMUST00000210924.2 |
Erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chrX_+_151922936 | 2.05 |
ENSMUST00000039720.11
ENSMUST00000144175.3 |
Rragb
|
Ras-related GTP binding B |
| chr11_+_109434519 | 1.96 |
ENSMUST00000106696.2
|
Arsg
|
arylsulfatase G |
| chr17_+_56312672 | 1.94 |
ENSMUST00000133998.8
|
Mpnd
|
MPN domain containing |
| chr11_-_41891111 | 1.92 |
ENSMUST00000109290.2
|
Gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 2 |
| chr1_+_107288928 | 1.89 |
ENSMUST00000191425.7
|
Serpinb11
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 11 |
| chr5_-_118382926 | 1.83 |
ENSMUST00000117177.8
ENSMUST00000133372.2 ENSMUST00000154786.8 ENSMUST00000121369.8 |
Rnft2
|
ring finger protein, transmembrane 2 |
| chr2_-_157408239 | 1.80 |
ENSMUST00000109528.9
ENSMUST00000088494.3 |
Blcap
|
bladder cancer associated protein |
| chr8_+_31579499 | 1.78 |
ENSMUST00000036631.14
|
Dusp26
|
dual specificity phosphatase 26 (putative) |
| chr1_-_37996838 | 1.78 |
ENSMUST00000027254.10
ENSMUST00000114894.2 |
Lyg1
|
lysozyme G-like 1 |
| chr13_-_99653045 | 1.77 |
ENSMUST00000064762.6
|
Map1b
|
microtubule-associated protein 1B |
| chr8_-_123087485 | 1.75 |
ENSMUST00000044123.2
|
Trhr2
|
thyrotropin releasing hormone receptor 2 |
| chr3_+_106020545 | 1.74 |
ENSMUST00000079132.12
ENSMUST00000139086.2 |
Chia1
|
chitinase, acidic 1 |
| chr9_-_73876182 | 1.73 |
ENSMUST00000184666.8
|
Unc13c
|
unc-13 homolog C |
| chr15_+_16778187 | 1.72 |
ENSMUST00000026432.8
|
Cdh9
|
cadherin 9 |
| chr7_-_97387429 | 1.65 |
ENSMUST00000206389.2
|
Aqp11
|
aquaporin 11 |
| chr8_+_31579633 | 1.62 |
ENSMUST00000170204.8
|
Dusp26
|
dual specificity phosphatase 26 (putative) |
| chr18_-_34712123 | 1.53 |
ENSMUST00000079287.12
|
Nme5
|
NME/NM23 family member 5 |
| chr4_-_4077510 | 1.53 |
ENSMUST00000108383.2
|
Sdr16c6
|
short chain dehydrogenase/reductase family 16C, member 6 |
| chr18_-_39000056 | 1.53 |
ENSMUST00000236630.2
ENSMUST00000237356.2 |
Fgf1
|
fibroblast growth factor 1 |
| chr13_-_4659120 | 1.46 |
ENSMUST00000091848.7
ENSMUST00000110691.10 |
Akr1e1
|
aldo-keto reductase family 1, member E1 |
| chr18_+_57275854 | 1.43 |
ENSMUST00000139892.2
|
Megf10
|
multiple EGF-like-domains 10 |
| chr4_+_126503611 | 1.41 |
ENSMUST00000097886.4
ENSMUST00000164362.2 |
5730409E04Rik
|
RIKEN cDNA 5730409E04Rik gene |
| chr12_+_37930169 | 1.41 |
ENSMUST00000221176.2
|
Dgkb
|
diacylglycerol kinase, beta |
| chr9_-_117843228 | 1.37 |
ENSMUST00000187803.3
|
Zcwpw2
|
zinc finger, CW type with PWWP domain 2 |
| chr16_-_59138611 | 1.37 |
ENSMUST00000216261.2
|
Olfr204
|
olfactory receptor 204 |
| chr16_-_19341016 | 1.37 |
ENSMUST00000214315.2
|
Olfr167
|
olfactory receptor 167 |
| chr4_+_41966058 | 1.36 |
ENSMUST00000108026.3
|
Fam205a4
|
family with sequence similarity 205, member A4 |
| chr3_-_86827664 | 1.35 |
ENSMUST00000194452.2
ENSMUST00000191752.6 |
Dclk2
|
doublecortin-like kinase 2 |
| chr4_+_52596266 | 1.35 |
ENSMUST00000029995.6
|
Toporsl
|
topoisomerase I binding, arginine/serine-rich like |
| chr14_-_96756503 | 1.35 |
ENSMUST00000022666.9
|
Klhl1
|
kelch-like 1 |
| chr4_+_110254858 | 1.34 |
ENSMUST00000106589.9
ENSMUST00000106587.9 ENSMUST00000106591.8 ENSMUST00000106592.8 |
Agbl4
|
ATP/GTP binding protein-like 4 |
| chr5_-_5315968 | 1.33 |
ENSMUST00000115451.8
ENSMUST00000115452.8 ENSMUST00000131392.8 |
Cdk14
|
cyclin-dependent kinase 14 |
| chr1_+_128079543 | 1.32 |
ENSMUST00000189317.3
|
R3hdm1
|
R3H domain containing 1 |
| chr15_+_100202021 | 1.30 |
ENSMUST00000230472.2
|
Mettl7a1
|
methyltransferase like 7A1 |
| chr9_+_50466127 | 1.27 |
ENSMUST00000213916.2
|
Il18
|
interleukin 18 |
| chr15_-_102419115 | 1.24 |
ENSMUST00000171565.8
|
Map3k12
|
mitogen-activated protein kinase kinase kinase 12 |
| chr8_+_45960931 | 1.17 |
ENSMUST00000171337.10
ENSMUST00000067107.15 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr15_+_100202079 | 1.14 |
ENSMUST00000230252.2
ENSMUST00000231166.2 |
Mettl7a1
|
methyltransferase like 7A1 |
| chr16_-_16647139 | 1.14 |
ENSMUST00000023468.6
|
Spag6l
|
sperm associated antigen 6-like |
| chr8_+_94537910 | 1.14 |
ENSMUST00000138659.9
|
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chr4_-_86587728 | 1.14 |
ENSMUST00000149700.8
|
Plin2
|
perilipin 2 |
| chr4_-_149184259 | 1.09 |
ENSMUST00000103217.11
|
Pex14
|
peroxisomal biogenesis factor 14 |
| chr8_-_96615138 | 1.09 |
ENSMUST00000034097.8
|
Got2
|
glutamatic-oxaloacetic transaminase 2, mitochondrial |
| chr1_+_179788675 | 1.07 |
ENSMUST00000076687.12
ENSMUST00000097450.10 ENSMUST00000212756.2 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr19_-_34143437 | 1.06 |
ENSMUST00000025686.9
|
Ankrd22
|
ankyrin repeat domain 22 |
| chr1_-_157084252 | 1.05 |
ENSMUST00000134543.8
|
Rasal2
|
RAS protein activator like 2 |
| chr15_+_100202061 | 1.04 |
ENSMUST00000229574.2
ENSMUST00000229217.2 |
Mettl7a1
|
methyltransferase like 7A1 |
| chr11_+_6510167 | 1.03 |
ENSMUST00000109722.9
|
Ccm2
|
cerebral cavernous malformation 2 |
| chr10_+_89906956 | 1.03 |
ENSMUST00000183109.2
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr13_-_24118139 | 1.01 |
ENSMUST00000052776.4
|
H2bc1
|
H2B clustered histone 1 |
| chr4_+_110254907 | 1.00 |
ENSMUST00000097920.9
ENSMUST00000080744.13 |
Agbl4
|
ATP/GTP binding protein-like 4 |
| chr7_-_101494472 | 1.00 |
ENSMUST00000211566.2
ENSMUST00000094141.7 ENSMUST00000209329.2 |
Folr2
|
folate receptor 2 (fetal) |
| chr8_+_55053809 | 0.97 |
ENSMUST00000033917.7
|
Spata4
|
spermatogenesis associated 4 |
| chr8_+_45960804 | 0.96 |
ENSMUST00000067065.14
ENSMUST00000124544.8 ENSMUST00000138049.9 ENSMUST00000132139.9 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr4_-_112632057 | 0.94 |
ENSMUST00000068851.6
|
Skint10
|
selection and upkeep of intraepithelial T cells 10 |
| chr4_+_102446883 | 0.94 |
ENSMUST00000097949.11
ENSMUST00000106901.2 |
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chrX_-_58211440 | 0.93 |
ENSMUST00000119306.2
|
Fgf13
|
fibroblast growth factor 13 |
| chr3_-_86827640 | 0.93 |
ENSMUST00000195561.6
|
Dclk2
|
doublecortin-like kinase 2 |
| chr4_+_150321142 | 0.92 |
ENSMUST00000150175.8
|
Eno1
|
enolase 1, alpha non-neuron |
| chr15_+_99192968 | 0.90 |
ENSMUST00000128352.8
ENSMUST00000145482.8 |
Prpf40b
|
pre-mRNA processing factor 40B |
| chr4_-_107975723 | 0.89 |
ENSMUST00000030340.15
|
Scp2
|
sterol carrier protein 2, liver |
| chr6_-_144155167 | 0.88 |
ENSMUST00000077160.12
|
Sox5
|
SRY (sex determining region Y)-box 5 |
| chr14_-_8798841 | 0.88 |
ENSMUST00000061045.3
|
Sntn
|
sentan, cilia apical structure protein |
| chr1_+_163607143 | 0.86 |
ENSMUST00000077642.12
ENSMUST00000027877.7 |
Kifap3
|
kinesin-associated protein 3 |
| chr4_+_147576874 | 0.84 |
ENSMUST00000105721.9
|
Zfp982
|
zinc finger protein 982 |
| chr17_-_40630096 | 0.84 |
ENSMUST00000026498.5
|
Crisp1
|
cysteine-rich secretory protein 1 |
| chr1_+_179788037 | 0.83 |
ENSMUST00000097453.9
ENSMUST00000111117.8 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr2_-_12424212 | 0.82 |
ENSMUST00000124603.8
ENSMUST00000129993.3 ENSMUST00000028105.13 |
Mindy3
|
MINDY lysine 48 deubiquitinase 3 |
| chr2_-_34716083 | 0.79 |
ENSMUST00000113077.8
ENSMUST00000028220.10 |
Fbxw2
|
F-box and WD-40 domain protein 2 |
| chr2_-_86906161 | 0.79 |
ENSMUST00000214049.2
|
Olfr1107
|
olfactory receptor 1107 |
| chr7_-_29553079 | 0.78 |
ENSMUST00000108223.8
|
Zfp940
|
zinc finger protein 940 |
| chr9_-_103569984 | 0.78 |
ENSMUST00000049452.15
|
Tmem108
|
transmembrane protein 108 |
| chrX_-_8059597 | 0.77 |
ENSMUST00000143223.2
ENSMUST00000033509.15 |
Ebp
|
phenylalkylamine Ca2+ antagonist (emopamil) binding protein |
| chr6_+_17749169 | 0.76 |
ENSMUST00000053148.14
ENSMUST00000115417.4 |
St7
|
suppression of tumorigenicity 7 |
| chr15_-_36496880 | 0.75 |
ENSMUST00000228601.2
ENSMUST00000057486.9 |
Ankrd46
|
ankyrin repeat domain 46 |
| chr18_+_37488174 | 0.75 |
ENSMUST00000192867.2
ENSMUST00000051163.3 |
Pcdhb8
|
protocadherin beta 8 |
| chr7_-_12819142 | 0.74 |
ENSMUST00000094829.2
|
Vmn1r85
|
vomeronasal 1 receptor 85 |
| chr14_+_118374511 | 0.74 |
ENSMUST00000022728.4
|
Gpr180
|
G protein-coupled receptor 180 |
| chr17_-_15163362 | 0.74 |
ENSMUST00000238668.2
ENSMUST00000228330.2 |
Wdr27
|
WD repeat domain 27 |
| chr2_-_86180622 | 0.74 |
ENSMUST00000099894.5
ENSMUST00000213564.3 |
Olfr1055
|
olfactory receptor 1055 |
| chr11_-_49004584 | 0.73 |
ENSMUST00000203007.2
|
Olfr1396
|
olfactory receptor 1396 |
| chr7_+_45271229 | 0.71 |
ENSMUST00000033100.5
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr17_-_45910529 | 0.70 |
ENSMUST00000171847.8
ENSMUST00000166633.8 ENSMUST00000169729.8 |
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr7_+_130936227 | 0.70 |
ENSMUST00000207784.2
|
Fam24a
|
family with sequence similarity 24, member A |
| chr2_+_85876205 | 0.69 |
ENSMUST00000213496.2
|
Olfr1034
|
olfactory receptor 1034 |
| chr16_-_63684477 | 0.68 |
ENSMUST00000232654.2
ENSMUST00000064405.8 |
Epha3
|
Eph receptor A3 |
| chr15_+_65682066 | 0.68 |
ENSMUST00000211878.2
|
Efr3a
|
EFR3 homolog A |
| chr17_+_64203017 | 0.67 |
ENSMUST00000000129.14
|
Fer
|
fer (fms/fps related) protein kinase |
| chr4_-_150085722 | 0.66 |
ENSMUST00000153394.2
|
H6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr2_+_36263531 | 0.66 |
ENSMUST00000072114.4
ENSMUST00000217511.2 |
Olfr338
|
olfactory receptor 338 |
| chr17_+_38485977 | 0.66 |
ENSMUST00000074883.2
|
Olfr134
|
olfactory receptor 134 |
| chr7_-_142215595 | 0.65 |
ENSMUST00000145896.3
|
Igf2
|
insulin-like growth factor 2 |
| chrX_+_151909893 | 0.63 |
ENSMUST00000163801.2
|
Foxr2
|
forkhead box R2 |
| chr3_+_146558114 | 0.63 |
ENSMUST00000170055.8
ENSMUST00000037942.11 |
Ttll7
|
tubulin tyrosine ligase-like family, member 7 |
| chr14_+_25980463 | 0.63 |
ENSMUST00000173155.8
|
Duxbl1
|
double homeobox B-like 1 |
| chr6_+_114435480 | 0.63 |
ENSMUST00000160780.2
|
Hrh1
|
histamine receptor H1 |
| chr6_-_144993451 | 0.62 |
ENSMUST00000123930.8
|
Bcat1
|
branched chain aminotransferase 1, cytosolic |
| chr19_-_55229668 | 0.62 |
ENSMUST00000069183.8
|
Gucy2g
|
guanylate cyclase 2g |
| chr13_-_21257469 | 0.62 |
ENSMUST00000058168.2
|
Olfr1370
|
olfactory receptor 1370 |
| chr2_-_89678487 | 0.59 |
ENSMUST00000214428.3
|
Olfr48
|
olfactory receptor 48 |
| chr12_-_55045887 | 0.57 |
ENSMUST00000173529.2
|
Baz1a
|
bromodomain adjacent to zinc finger domain 1A |
| chr7_-_11414074 | 0.57 |
ENSMUST00000227010.2
ENSMUST00000209638.3 |
Vmn1r72
|
vomeronasal 1 receptor 72 |
| chr7_+_129193581 | 0.57 |
ENSMUST00000084519.7
|
Wdr11
|
WD repeat domain 11 |
| chr3_-_75072319 | 0.57 |
ENSMUST00000124618.2
|
Zbbx
|
zinc finger, B-box domain containing |
| chr2_-_12424189 | 0.57 |
ENSMUST00000124515.2
|
Mindy3
|
MINDY lysine 48 deubiquitinase 3 |
| chr4_+_126156118 | 0.57 |
ENSMUST00000030660.9
|
Trappc3
|
trafficking protein particle complex 3 |
| chr16_+_88525719 | 0.57 |
ENSMUST00000060494.8
|
Krtap13-1
|
keratin associated protein 13-1 |
| chr7_-_5325456 | 0.57 |
ENSMUST00000207520.2
|
Nlrp2
|
NLR family, pyrin domain containing 2 |
| chr7_+_26456567 | 0.57 |
ENSMUST00000077855.8
|
Cyp2b19
|
cytochrome P450, family 2, subfamily b, polypeptide 19 |
| chr2_+_85868891 | 0.56 |
ENSMUST00000218397.2
|
Olfr1033
|
olfactory receptor 1033 |
| chr2_-_92876398 | 0.56 |
ENSMUST00000111272.3
ENSMUST00000178666.8 ENSMUST00000147339.3 |
Prdm11
|
PR domain containing 11 |
| chr11_+_120499295 | 0.54 |
ENSMUST00000106194.8
ENSMUST00000106195.3 ENSMUST00000061309.5 |
Npb
|
neuropeptide B |
| chr9_-_19799300 | 0.54 |
ENSMUST00000079660.5
|
Olfr862
|
olfactory receptor 862 |
| chr14_+_25979825 | 0.54 |
ENSMUST00000173580.8
|
Duxbl1
|
double homeobox B-like 1 |
| chr2_+_111329683 | 0.53 |
ENSMUST00000219064.3
|
Olfr1291-ps1
|
olfactory receptor 1291, pseudogene 1 |
| chr12_+_71021395 | 0.53 |
ENSMUST00000160027.8
ENSMUST00000160864.8 |
Psma3
|
proteasome subunit alpha 3 |
| chr6_+_134617903 | 0.53 |
ENSMUST00000062755.10
|
Borcs5
|
BLOC-1 related complex subunit 5 |
| chr14_+_55797468 | 0.52 |
ENSMUST00000147981.2
ENSMUST00000133256.8 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr4_+_150321272 | 0.52 |
ENSMUST00000080926.13
|
Eno1
|
enolase 1, alpha non-neuron |
| chr11_+_5738480 | 0.51 |
ENSMUST00000109845.8
ENSMUST00000020769.14 ENSMUST00000102928.5 |
Dbnl
|
drebrin-like |
| chr2_+_144435974 | 0.51 |
ENSMUST00000136628.2
|
Smim26
|
small integral membrane protein 26 |
| chr13_-_21823691 | 0.51 |
ENSMUST00000043081.3
|
Olfr11
|
olfactory receptor 11 |
| chr6_-_89572629 | 0.51 |
ENSMUST00000113550.6
ENSMUST00000032172.14 |
Chchd6
|
coiled-coil-helix-coiled-coil-helix domain containing 6 |
| chr2_-_34716199 | 0.51 |
ENSMUST00000113075.8
ENSMUST00000113080.9 ENSMUST00000091020.10 |
Fbxw2
|
F-box and WD-40 domain protein 2 |
| chr14_+_7030760 | 0.50 |
ENSMUST00000055211.6
|
Lrrc3b
|
leucine rich repeat containing 3B |
| chr2_-_85966272 | 0.48 |
ENSMUST00000216566.3
ENSMUST00000214364.2 |
Olfr1039
|
olfactory receptor 1039 |
| chr11_+_99755302 | 0.48 |
ENSMUST00000092694.4
|
Gm11559
|
predicted gene 11559 |
| chr11_+_73489420 | 0.47 |
ENSMUST00000214228.2
|
Olfr384
|
olfactory receptor 384 |
| chr7_-_103191924 | 0.47 |
ENSMUST00000214269.3
|
Olfr612
|
olfactory receptor 612 |
| chr19_-_33602652 | 0.45 |
ENSMUST00000124230.3
|
Gm8978
|
predicted gene 8978 |
| chr6_+_132869974 | 0.44 |
ENSMUST00000079035.3
|
Tas2r113
|
taste receptor, type 2, member 113 |
| chr4_-_123507494 | 0.43 |
ENSMUST00000238866.2
|
Macf1
|
microtubule-actin crosslinking factor 1 |
| chr4_-_155729865 | 0.43 |
ENSMUST00000115821.3
|
Gm10563
|
predicted gene 10563 |
| chr12_+_80509978 | 0.43 |
ENSMUST00000219272.2
|
Exd2
|
exonuclease 3'-5' domain containing 2 |
| chr2_+_85551751 | 0.43 |
ENSMUST00000055517.3
|
Olfr1009
|
olfactory receptor 1009 |
| chr11_-_12362136 | 0.42 |
ENSMUST00000174874.8
|
Cobl
|
cordon-bleu WH2 repeat |
| chr2_+_85780781 | 0.42 |
ENSMUST00000080698.3
|
Olfr1028
|
olfactory receptor 1028 |
| chr9_-_14411778 | 0.41 |
ENSMUST00000058796.7
|
Kdm4d
|
lysine (K)-specific demethylase 4D |
| chr17_+_37978659 | 0.41 |
ENSMUST00000216551.2
|
Olfr118
|
olfactory receptor 118 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.8 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 2.8 | 8.4 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 1.6 | 9.9 | GO:0008355 | olfactory learning(GO:0008355) |
| 1.4 | 46.1 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 1.2 | 3.5 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 1.1 | 5.3 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.7 | 2.8 | GO:0010899 | regulation of phosphatidylcholine catabolic process(GO:0010899) positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.7 | 3.4 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.7 | 4.0 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.7 | 2.6 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.6 | 13.6 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.6 | 4.2 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.6 | 2.2 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.5 | 2.5 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.5 | 7.5 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.5 | 2.3 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.5 | 2.3 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.5 | 3.2 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.4 | 1.7 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.4 | 22.5 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.4 | 1.1 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.4 | 1.1 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) |
| 0.3 | 1.3 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.3 | 0.9 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.3 | 0.8 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.3 | 2.1 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.3 | 2.0 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.2 | 1.4 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.2 | 11.7 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.2 | 0.9 | GO:1901373 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) |
| 0.2 | 2.0 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.1 | 2.4 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 0.7 | GO:0035036 | sperm-egg recognition(GO:0035036) |
| 0.1 | 0.9 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 | 1.2 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 1.7 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 1.0 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.1 | 1.5 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 0.8 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.1 | 2.5 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 0.3 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.1 | 2.6 | GO:0031987 | short-term memory(GO:0007614) locomotion involved in locomotory behavior(GO:0031987) |
| 0.1 | 1.3 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 | 0.7 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 0.8 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.1 | 1.0 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 0.3 | GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) |
| 0.1 | 3.4 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 1.8 | GO:0061162 | establishment of monopolar cell polarity(GO:0061162) |
| 0.1 | 0.3 | GO:0050712 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.1 | 1.0 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 1.1 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.1 | 0.9 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 4.9 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 0.4 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 1.7 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.1 | 1.0 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.1 | 1.4 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 1.9 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.1 | 4.5 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 0.2 | GO:0042245 | tRNA wobble cytosine modification(GO:0002101) RNA repair(GO:0042245) |
| 0.0 | 1.8 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.3 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 0.3 | GO:0048496 | determination of pancreatic left/right asymmetry(GO:0035469) maintenance of organ identity(GO:0048496) |
| 0.0 | 0.2 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.4 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 2.5 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 3.1 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 4.0 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.7 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 1.4 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 1.3 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 0.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 5.7 | GO:0042471 | ear morphogenesis(GO:0042471) |
| 0.0 | 0.5 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.4 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.2 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 | 0.5 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.7 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.3 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 2.1 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.3 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.9 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.5 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 1.1 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.6 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 0.5 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.4 | GO:0031497 | chromatin assembly(GO:0031497) |
| 0.0 | 0.8 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.0 | 5.7 | GO:0006887 | exocytosis(GO:0006887) |
| 0.0 | 1.1 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 | 1.1 | GO:0019915 | lipid storage(GO:0019915) |
| 0.0 | 0.3 | GO:0034587 | piRNA metabolic process(GO:0034587) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.8 | 2.3 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.4 | 2.0 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.3 | 1.7 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.3 | 0.9 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.3 | 12.8 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.3 | 1.1 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.3 | 1.1 | GO:1990429 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.3 | 1.3 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.2 | 5.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 2.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 0.6 | GO:0008623 | CHRAC(GO:0008623) |
| 0.1 | 3.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 6.2 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 1.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 9.2 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 0.3 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.1 | 1.9 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.9 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.1 | 1.8 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 0.5 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.1 | 0.5 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 1.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.4 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.3 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.0 | 0.5 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.9 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.3 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 4.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.4 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 2.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.7 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.5 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.6 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.2 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.3 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 4.2 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 15.5 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.8 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 4.1 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.5 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 1.8 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 1.9 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 28.0 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.4 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 0.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 1.1 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 5.1 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 1.6 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 13.6 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 2.9 | 8.8 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 2.2 | 13.3 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 1.1 | 5.3 | GO:0016726 | aldehyde oxidase activity(GO:0004031) xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) molybdenum ion binding(GO:0030151) |
| 1.0 | 32.8 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 1.0 | 22.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 1.0 | 9.9 | GO:0005186 | pheromone activity(GO:0005186) |
| 0.6 | 1.7 | GO:0001565 | phorbol ester receptor activity(GO:0001565) non-kinase phorbol ester receptor activity(GO:0001566) |
| 0.6 | 3.4 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.5 | 4.0 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.4 | 7.5 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.4 | 2.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.4 | 1.1 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.3 | 3.3 | GO:0036122 | BMP binding(GO:0036122) |
| 0.3 | 8.4 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.3 | 2.3 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.3 | 2.6 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.3 | 1.0 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.2 | 1.9 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.2 | 1.4 | GO:0070004 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 0.2 | 2.0 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.2 | 0.9 | GO:0033814 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.2 | 0.7 | GO:0047936 | glucose 1-dehydrogenase [NAD(P)] activity(GO:0047936) |
| 0.2 | 3.7 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 1.8 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.2 | 4.2 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.2 | 0.6 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.2 | 1.4 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.2 | 0.6 | GO:0051381 | histamine binding(GO:0051381) |
| 0.1 | 6.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 1.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 1.0 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 0.8 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.1 | 2.0 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 1.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.9 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.1 | 1.0 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.1 | 0.3 | GO:0016005 | phospholipase A2 activator activity(GO:0016005) |
| 0.1 | 0.3 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.1 | 2.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.3 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.1 | 5.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 4.4 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 2.2 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.1 | 0.7 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 0.7 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 1.5 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 5.0 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 1.1 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 1.9 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.2 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.0 | 4.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 7.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.8 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 2.0 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 1.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 3.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.8 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.5 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.4 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.2 | GO:0043734 | DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 1.8 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.3 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.6 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.7 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.5 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 1.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.7 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.3 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 11.0 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.4 | GO:0003785 | actin monomer binding(GO:0003785) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 8.4 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 2.2 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 4.2 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 1.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 0.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 2.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 1.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 4.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.1 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 2.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.9 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 5.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.9 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 2.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.7 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.9 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 5.7 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.0 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.3 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.3 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.5 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 8.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.3 | 13.3 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.3 | 2.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.2 | 5.6 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 2.0 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.2 | 4.9 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.2 | 8.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 4.2 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 2.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 3.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 2.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 3.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 2.2 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 1.9 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 1.7 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 2.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 2.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.0 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 3.8 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 0.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.1 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 1.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.5 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.3 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.3 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.7 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 0.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |