Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for En1
Z-value: 1.22
Transcription factors associated with En1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
En1
|
ENSMUSG00000058665.9 | En1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| En1 | mm39_v1_chr1_+_120530134_120530147 | -0.34 | 3.7e-03 | Click! |
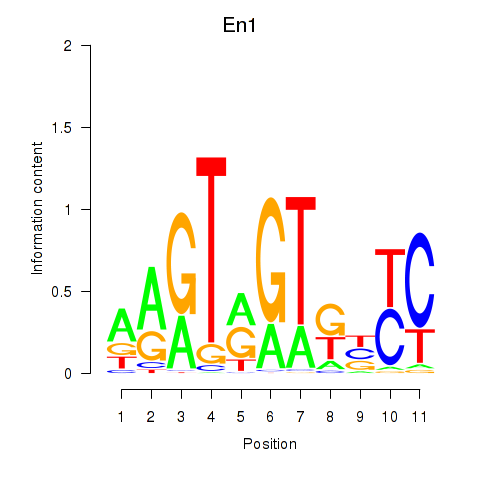
Activity profile of En1 motif
Sorted Z-values of En1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of En1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_144738526 | 12.42 |
ENSMUST00000029919.7
|
Clca1
|
chloride channel accessory 1 |
| chr12_-_76842263 | 9.33 |
ENSMUST00000082431.6
|
Gpx2
|
glutathione peroxidase 2 |
| chr6_-_70149254 | 8.19 |
ENSMUST00000197272.2
|
Igkv8-27
|
immunoglobulin kappa chain variable 8-27 |
| chr5_-_87716882 | 7.16 |
ENSMUST00000113314.3
|
Sult1d1
|
sulfotransferase family 1D, member 1 |
| chr6_+_70703409 | 5.97 |
ENSMUST00000103410.3
|
Igkc
|
immunoglobulin kappa constant |
| chr12_+_8062331 | 5.77 |
ENSMUST00000171239.2
|
Apob
|
apolipoprotein B |
| chr6_-_69415741 | 5.74 |
ENSMUST00000103354.3
|
Igkv4-59
|
immunoglobulin kappa variable 4-59 |
| chr9_-_31375497 | 5.39 |
ENSMUST00000217007.2
ENSMUST00000213807.2 |
Tmem45b
|
transmembrane protein 45b |
| chr6_-_70292451 | 4.93 |
ENSMUST00000103387.3
|
Igkv8-21
|
immunoglobulin kappa variable 8-21 |
| chr14_-_51384236 | 4.84 |
ENSMUST00000080126.4
|
Rnase1
|
ribonuclease, RNase A family, 1 (pancreatic) |
| chr6_-_68887957 | 4.53 |
ENSMUST00000200454.2
|
Igkv4-86
|
immunoglobulin kappa variable 4-86 |
| chr4_-_129121676 | 4.45 |
ENSMUST00000106051.8
|
C77080
|
expressed sequence C77080 |
| chr6_+_68247469 | 4.31 |
ENSMUST00000103321.3
|
Igkv1-110
|
immunoglobulin kappa variable 1-110 |
| chr6_-_68994064 | 4.20 |
ENSMUST00000103341.4
|
Igkv4-80
|
immunoglobulin kappa variable 4-80 |
| chr17_-_84990360 | 4.19 |
ENSMUST00000066175.10
|
Abcg5
|
ATP binding cassette subfamily G member 5 |
| chr6_+_68279392 | 4.16 |
ENSMUST00000103322.3
|
Igkv2-109
|
immunoglobulin kappa variable 2-109 |
| chr2_-_103114105 | 4.09 |
ENSMUST00000111174.8
|
Ehf
|
ets homologous factor |
| chr3_-_14843512 | 3.97 |
ENSMUST00000094365.11
|
Car1
|
carbonic anhydrase 1 |
| chr11_-_69696428 | 3.90 |
ENSMUST00000051025.5
|
Tmem102
|
transmembrane protein 102 |
| chr6_-_69626340 | 3.88 |
ENSMUST00000198328.2
|
Igkv4-53
|
immunoglobulin kappa variable 4-53 |
| chr6_-_68746087 | 3.86 |
ENSMUST00000103333.4
|
Igkv4-91
|
immunoglobulin kappa chain variable 4-91 |
| chr6_+_135042649 | 3.82 |
ENSMUST00000050104.8
|
Gprc5a
|
G protein-coupled receptor, family C, group 5, member A |
| chr6_-_69355456 | 3.80 |
ENSMUST00000196595.2
|
Igkv4-63
|
immunoglobulin kappa variable 4-63 |
| chr13_+_3974686 | 3.70 |
ENSMUST00000021639.8
|
Tubal3
|
tubulin, alpha-like 3 |
| chr1_+_88139678 | 3.68 |
ENSMUST00000073049.7
|
Ugt1a1
|
UDP glucuronosyltransferase 1 family, polypeptide A1 |
| chr6_+_41928559 | 3.60 |
ENSMUST00000031898.5
|
Sval1
|
seminal vesicle antigen-like 1 |
| chr6_-_70120881 | 3.60 |
ENSMUST00000103380.3
|
Igkv8-28
|
immunoglobulin kappa variable 8-28 |
| chr6_+_78347844 | 3.59 |
ENSMUST00000096904.6
ENSMUST00000203266.2 |
Reg3b
|
regenerating islet-derived 3 beta |
| chrX_+_111404963 | 3.51 |
ENSMUST00000026602.9
ENSMUST00000113412.3 |
2010106E10Rik
|
RIKEN cDNA 2010106E10 gene |
| chr2_-_92290791 | 3.51 |
ENSMUST00000125276.2
|
Slc35c1
|
solute carrier family 35, member C1 |
| chr19_+_31846154 | 3.48 |
ENSMUST00000224564.2
ENSMUST00000224304.2 ENSMUST00000075838.8 ENSMUST00000224400.2 |
A1cf
|
APOBEC1 complementation factor |
| chr17_-_73706284 | 3.48 |
ENSMUST00000095208.4
|
Capn13
|
calpain 13 |
| chr6_-_70094604 | 3.38 |
ENSMUST00000103378.3
|
Igkv8-30
|
immunoglobulin kappa chain variable 8-30 |
| chr6_-_70318437 | 3.38 |
ENSMUST00000196599.2
|
Igkv8-19
|
immunoglobulin kappa variable 8-19 |
| chr2_+_72306503 | 3.37 |
ENSMUST00000102691.11
ENSMUST00000157019.2 |
Cdca7
|
cell division cycle associated 7 |
| chr10_-_128796834 | 3.34 |
ENSMUST00000026398.5
|
Mettl7b
|
methyltransferase like 7B |
| chr12_-_113649535 | 3.32 |
ENSMUST00000103449.4
ENSMUST00000195707.3 |
Ighv2-5
|
immunoglobulin heavy variable 2-5 |
| chr3_-_90150393 | 3.28 |
ENSMUST00000107369.2
|
Creb3l4
|
cAMP responsive element binding protein 3-like 4 |
| chr12_-_113928438 | 3.22 |
ENSMUST00000103478.4
|
Ighv3-1
|
immunoglobulin heavy variable 3-1 |
| chr11_-_98915005 | 3.17 |
ENSMUST00000068031.8
|
Top2a
|
topoisomerase (DNA) II alpha |
| chr4_-_127222891 | 3.16 |
ENSMUST00000106091.9
|
Gjb3
|
gap junction protein, beta 3 |
| chr5_+_43976218 | 3.11 |
ENSMUST00000101237.8
|
Bst1
|
bone marrow stromal cell antigen 1 |
| chr8_+_75599801 | 3.09 |
ENSMUST00000034034.10
|
Isx
|
intestine specific homeobox |
| chr5_-_87054796 | 3.08 |
ENSMUST00000031181.16
ENSMUST00000113333.2 |
Ugt2b34
|
UDP glucuronosyltransferase 2 family, polypeptide B34 |
| chr6_-_69204417 | 3.07 |
ENSMUST00000103346.3
|
Igkv4-72
|
immunoglobulin kappa chain variable 4-72 |
| chr6_-_70121150 | 3.06 |
ENSMUST00000197525.2
|
Igkv8-28
|
immunoglobulin kappa variable 8-28 |
| chr6_+_129568745 | 3.02 |
ENSMUST00000032268.14
ENSMUST00000112063.9 ENSMUST00000119520.8 |
Klrd1
|
killer cell lectin-like receptor, subfamily D, member 1 |
| chr6_+_78347636 | 2.99 |
ENSMUST00000204873.3
|
Reg3b
|
regenerating islet-derived 3 beta |
| chr1_+_88066086 | 2.92 |
ENSMUST00000014263.6
|
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A |
| chr6_+_113435716 | 2.92 |
ENSMUST00000203661.3
ENSMUST00000204774.3 ENSMUST00000053569.7 ENSMUST00000101065.8 |
Il17re
|
interleukin 17 receptor E |
| chr9_+_75139295 | 2.84 |
ENSMUST00000036555.8
|
Myo5c
|
myosin VC |
| chr19_+_4203603 | 2.80 |
ENSMUST00000236632.2
|
Ptprcap
|
protein tyrosine phosphatase, receptor type, C polypeptide-associated protein |
| chr15_-_76004395 | 2.78 |
ENSMUST00000239552.1
|
EPPK1
|
epiplakin 1 |
| chr8_-_27664651 | 2.75 |
ENSMUST00000054212.7
ENSMUST00000033878.14 ENSMUST00000209377.2 |
Rab11fip1
|
RAB11 family interacting protein 1 (class I) |
| chr3_-_144514386 | 2.71 |
ENSMUST00000197013.2
|
Clca3a2
|
chloride channel accessory 3A2 |
| chr4_+_129000600 | 2.71 |
ENSMUST00000148979.2
|
Tmem54
|
transmembrane protein 54 |
| chr9_-_44891626 | 2.71 |
ENSMUST00000002101.12
ENSMUST00000160886.2 |
Cd3g
|
CD3 antigen, gamma polypeptide |
| chr3_+_10431961 | 2.71 |
ENSMUST00000029049.7
|
Chmp4c
|
charged multivesicular body protein 4C |
| chr5_-_72910106 | 2.70 |
ENSMUST00000197313.5
ENSMUST00000198464.3 ENSMUST00000113604.10 |
Txk
|
TXK tyrosine kinase |
| chr1_+_86230931 | 2.68 |
ENSMUST00000113306.4
|
B3gnt7
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7 |
| chr19_-_11243530 | 2.64 |
ENSMUST00000169159.3
|
Ms4a1
|
membrane-spanning 4-domains, subfamily A, member 1 |
| chr15_-_99717956 | 2.64 |
ENSMUST00000109024.9
|
Lima1
|
LIM domain and actin binding 1 |
| chr7_-_48531344 | 2.63 |
ENSMUST00000119223.2
|
E2f8
|
E2F transcription factor 8 |
| chr17_+_48761916 | 2.62 |
ENSMUST00000074574.13
|
Unc5cl
|
unc-5 family C-terminal like |
| chr6_+_68402550 | 2.57 |
ENSMUST00000103323.3
|
Igkv16-104
|
immunoglobulin kappa variable 16-104 |
| chr1_-_171434882 | 2.54 |
ENSMUST00000111277.2
ENSMUST00000004827.14 |
Ly9
|
lymphocyte antigen 9 |
| chr16_-_16687119 | 2.54 |
ENSMUST00000075017.5
|
Vpreb1
|
pre-B lymphocyte gene 1 |
| chr12_-_114057841 | 2.48 |
ENSMUST00000103471.2
ENSMUST00000195884.2 |
Ighv9-1
|
immunoglobulin heavy variable 9-1 |
| chr6_-_125213911 | 2.47 |
ENSMUST00000112282.3
ENSMUST00000112281.8 ENSMUST00000032486.13 |
Cd27
|
CD27 antigen |
| chr5_+_18167547 | 2.46 |
ENSMUST00000030561.9
|
Gnat3
|
guanine nucleotide binding protein, alpha transducing 3 |
| chr3_+_69129745 | 2.43 |
ENSMUST00000183126.2
|
Arl14
|
ADP-ribosylation factor-like 14 |
| chr1_+_132973724 | 2.42 |
ENSMUST00000077730.7
|
Pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta |
| chr12_-_113589576 | 2.39 |
ENSMUST00000103446.2
|
Ighv5-6
|
immunoglobulin heavy variable 5-6 |
| chr1_+_67162176 | 2.36 |
ENSMUST00000027144.8
|
Cps1
|
carbamoyl-phosphate synthetase 1 |
| chr1_+_86231208 | 2.32 |
ENSMUST00000188695.2
|
B3gnt7
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7 |
| chr6_+_113448388 | 2.32 |
ENSMUST00000058300.14
|
Il17rc
|
interleukin 17 receptor C |
| chr6_-_68840015 | 2.32 |
ENSMUST00000103336.2
|
Igkv1-88
|
immunoglobulin kappa chain variable 1-88 |
| chr5_-_66309244 | 2.31 |
ENSMUST00000167950.8
|
Rbm47
|
RNA binding motif protein 47 |
| chr17_+_34416707 | 2.31 |
ENSMUST00000025196.9
|
Psmb8
|
proteasome (prosome, macropain) subunit, beta type 8 (large multifunctional peptidase 7) |
| chr9_+_44951075 | 2.31 |
ENSMUST00000217097.2
|
Mpzl2
|
myelin protein zero-like 2 |
| chr6_-_68887922 | 2.29 |
ENSMUST00000103337.3
|
Igkv4-86
|
immunoglobulin kappa variable 4-86 |
| chr2_-_34848148 | 2.29 |
ENSMUST00000201690.2
ENSMUST00000172159.8 |
Traf1
|
TNF receptor-associated factor 1 |
| chr6_-_128803182 | 2.28 |
ENSMUST00000204756.3
ENSMUST00000204394.3 ENSMUST00000204423.3 ENSMUST00000204677.2 ENSMUST00000205130.3 ENSMUST00000174544.2 ENSMUST00000172887.8 ENSMUST00000032472.11 |
Gm44511
Klrb1b
|
predicted gene 44511 killer cell lectin-like receptor subfamily B member 1B |
| chr1_-_171434944 | 2.28 |
ENSMUST00000068878.14
|
Ly9
|
lymphocyte antigen 9 |
| chr2_+_121188195 | 2.27 |
ENSMUST00000125812.8
ENSMUST00000078222.9 ENSMUST00000125221.3 ENSMUST00000150271.8 |
Ckmt1
|
creatine kinase, mitochondrial 1, ubiquitous |
| chr12_-_113392174 | 2.24 |
ENSMUST00000103427.2
|
Ighj4
|
immunoglobulin heavy joining 4 |
| chr7_-_24997393 | 2.24 |
ENSMUST00000005583.12
|
Pafah1b3
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 3 |
| chr9_+_65494469 | 2.20 |
ENSMUST00000239405.2
ENSMUST00000047099.13 ENSMUST00000131483.3 ENSMUST00000141046.3 |
Pif1
|
PIF1 5'-to-3' DNA helicase |
| chr5_-_121045568 | 2.20 |
ENSMUST00000080322.8
|
Oas1a
|
2'-5' oligoadenylate synthetase 1A |
| chr11_-_80670815 | 2.19 |
ENSMUST00000041065.14
ENSMUST00000070997.6 |
Myo1d
|
myosin ID |
| chr9_+_92131797 | 2.17 |
ENSMUST00000093801.10
|
Plscr1
|
phospholipid scramblase 1 |
| chr5_-_41921834 | 2.16 |
ENSMUST00000060820.8
|
Nkx3-2
|
NK3 homeobox 2 |
| chr17_+_34416689 | 2.16 |
ENSMUST00000173441.9
|
Psmb8
|
proteasome (prosome, macropain) subunit, beta type 8 (large multifunctional peptidase 7) |
| chr3_-_88368489 | 2.12 |
ENSMUST00000166237.8
|
Sema4a
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr6_+_142244145 | 2.12 |
ENSMUST00000041993.3
|
Iapp
|
islet amyloid polypeptide |
| chr1_+_133237516 | 2.11 |
ENSMUST00000094557.7
ENSMUST00000192465.2 ENSMUST00000193888.6 ENSMUST00000194044.6 ENSMUST00000184603.8 |
Golt1a
Gm28040
Gm28040
|
golgi transport 1A predicted gene, 28040 predicted gene, 28040 |
| chr19_+_38085510 | 2.09 |
ENSMUST00000067098.8
|
Ffar4
|
free fatty acid receptor 4 |
| chr19_+_55240357 | 2.09 |
ENSMUST00000225551.2
|
Acsl5
|
acyl-CoA synthetase long-chain family member 5 |
| chr11_+_72098363 | 2.07 |
ENSMUST00000021158.4
|
Txndc17
|
thioredoxin domain containing 17 |
| chr5_-_87288177 | 2.06 |
ENSMUST00000067790.7
|
Ugt2b5
|
UDP glucuronosyltransferase 2 family, polypeptide B5 |
| chr4_-_111759951 | 2.04 |
ENSMUST00000102719.8
ENSMUST00000102721.8 |
Slc5a9
|
solute carrier family 5 (sodium/glucose cotransporter), member 9 |
| chr6_-_124698805 | 2.02 |
ENSMUST00000173315.8
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr13_-_100453124 | 2.00 |
ENSMUST00000042220.3
|
Naip6
|
NLR family, apoptosis inhibitory protein 6 |
| chr3_+_29136172 | 1.97 |
ENSMUST00000124809.8
|
Egfem1
|
EGF-like and EMI domain containing 1 |
| chr9_-_45847344 | 1.97 |
ENSMUST00000034590.4
|
Tagln
|
transgelin |
| chr3_+_94600863 | 1.96 |
ENSMUST00000090848.10
ENSMUST00000173981.8 ENSMUST00000173849.8 ENSMUST00000174223.2 |
Selenbp2
|
selenium binding protein 2 |
| chrX_-_133012457 | 1.95 |
ENSMUST00000159259.3
ENSMUST00000113275.10 |
Nox1
|
NADPH oxidase 1 |
| chr6_+_123206802 | 1.95 |
ENSMUST00000112554.9
ENSMUST00000024118.11 ENSMUST00000117130.8 |
Clec4n
|
C-type lectin domain family 4, member n |
| chr1_+_88062508 | 1.94 |
ENSMUST00000113134.8
ENSMUST00000140092.8 |
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A |
| chr4_-_129155185 | 1.93 |
ENSMUST00000145261.8
|
C77080
|
expressed sequence C77080 |
| chr9_-_106768601 | 1.92 |
ENSMUST00000069036.14
|
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr13_+_75987987 | 1.90 |
ENSMUST00000022082.8
ENSMUST00000223120.2 ENSMUST00000220523.2 |
Glrx
|
glutaredoxin |
| chr6_+_70640233 | 1.90 |
ENSMUST00000103400.3
|
Igkv3-5
|
immunoglobulin kappa chain variable 3-5 |
| chr1_+_72863641 | 1.87 |
ENSMUST00000047328.11
|
Igfbp2
|
insulin-like growth factor binding protein 2 |
| chr6_-_70021662 | 1.86 |
ENSMUST00000196959.2
|
Igkv8-34
|
immunoglobulin kappa variable 8-34 |
| chr3_+_138121245 | 1.86 |
ENSMUST00000161312.8
ENSMUST00000013458.9 |
Adh4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr15_+_4756684 | 1.84 |
ENSMUST00000161997.8
ENSMUST00000022788.15 |
C6
|
complement component 6 |
| chr6_+_29859685 | 1.84 |
ENSMUST00000134438.2
|
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr1_+_40554513 | 1.81 |
ENSMUST00000027237.12
|
Il18rap
|
interleukin 18 receptor accessory protein |
| chr17_-_35081456 | 1.80 |
ENSMUST00000025229.11
ENSMUST00000176203.9 ENSMUST00000128767.8 |
Cfb
|
complement factor B |
| chr7_+_100971034 | 1.78 |
ENSMUST00000173270.8
|
Stard10
|
START domain containing 10 |
| chr7_-_24997291 | 1.77 |
ENSMUST00000148150.8
ENSMUST00000155118.2 |
Pafah1b3
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 3 |
| chr5_+_31079177 | 1.77 |
ENSMUST00000031053.15
ENSMUST00000202752.2 |
Khk
|
ketohexokinase |
| chr2_-_93283024 | 1.77 |
ENSMUST00000111257.8
ENSMUST00000145553.8 |
Cd82
|
CD82 antigen |
| chr7_+_83281167 | 1.75 |
ENSMUST00000075418.15
|
Stard5
|
StAR-related lipid transfer (START) domain containing 5 |
| chr4_-_115980813 | 1.73 |
ENSMUST00000102704.4
ENSMUST00000102705.10 |
Rad54l
|
RAD54 like (S. cerevisiae) |
| chr5_+_53748323 | 1.72 |
ENSMUST00000201883.4
|
Rbpj
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr10_+_127157784 | 1.72 |
ENSMUST00000219511.2
|
Arhgap9
|
Rho GTPase activating protein 9 |
| chr1_+_135060431 | 1.72 |
ENSMUST00000187985.7
ENSMUST00000049449.11 |
Ptpn7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr3_-_121325887 | 1.72 |
ENSMUST00000039197.9
|
Slc44a3
|
solute carrier family 44, member 3 |
| chr9_+_107468146 | 1.72 |
ENSMUST00000195746.2
|
Ifrd2
|
interferon-related developmental regulator 2 |
| chr16_+_45044678 | 1.72 |
ENSMUST00000102802.10
ENSMUST00000063654.6 |
Btla
|
B and T lymphocyte associated |
| chrX_+_106193167 | 1.70 |
ENSMUST00000137107.2
ENSMUST00000067249.3 |
P2ry10b
|
purinergic receptor P2Y, G-protein coupled 10B |
| chr5_-_72893941 | 1.70 |
ENSMUST00000169534.6
|
Txk
|
TXK tyrosine kinase |
| chr7_+_83281193 | 1.69 |
ENSMUST00000117410.2
|
Stard5
|
StAR-related lipid transfer (START) domain containing 5 |
| chr9_-_35010357 | 1.66 |
ENSMUST00000214526.2
ENSMUST00000217149.2 |
St3gal4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr3_-_113325938 | 1.66 |
ENSMUST00000132353.2
|
Amy2a1
|
amylase 2a1 |
| chr9_-_119812829 | 1.65 |
ENSMUST00000216929.2
|
Csrnp1
|
cysteine-serine-rich nuclear protein 1 |
| chr9_+_65494429 | 1.64 |
ENSMUST00000134538.9
|
Pif1
|
PIF1 5'-to-3' DNA helicase |
| chrX_+_72760183 | 1.63 |
ENSMUST00000002084.14
|
Abcd1
|
ATP-binding cassette, sub-family D (ALD), member 1 |
| chr6_-_68784692 | 1.62 |
ENSMUST00000103334.4
|
Igkv4-90
|
immunoglobulin kappa chain variable 4-90 |
| chr3_-_30194559 | 1.62 |
ENSMUST00000108271.10
|
Mecom
|
MDS1 and EVI1 complex locus |
| chrX_+_47235313 | 1.59 |
ENSMUST00000033427.7
|
Sash3
|
SAM and SH3 domain containing 3 |
| chr5_+_13448647 | 1.58 |
ENSMUST00000125629.8
|
Sema3a
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr1_+_88093726 | 1.57 |
ENSMUST00000097659.5
|
Ugt1a5
|
UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr12_+_98234884 | 1.57 |
ENSMUST00000075072.6
|
Gpr65
|
G-protein coupled receptor 65 |
| chr11_-_86999481 | 1.57 |
ENSMUST00000051395.9
|
Prr11
|
proline rich 11 |
| chr12_-_114477427 | 1.56 |
ENSMUST00000191803.2
|
Ighv1-5
|
immunoglobulin heavy variable V1-5 |
| chr8_-_13250535 | 1.55 |
ENSMUST00000165605.4
ENSMUST00000209691.2 ENSMUST00000211128.2 ENSMUST00000210317.2 |
Grtp1
|
GH regulated TBC protein 1 |
| chr12_-_115459678 | 1.55 |
ENSMUST00000103534.2
|
Ighv1-63
|
immunoglobulin heavy variable V1-63 |
| chr6_-_70036183 | 1.53 |
ENSMUST00000197429.5
ENSMUST00000103376.3 |
Igkv7-33
|
immunoglobulin kappa chain variable 7-33 |
| chr10_-_18890281 | 1.52 |
ENSMUST00000146388.2
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr11_-_83540175 | 1.52 |
ENSMUST00000001008.6
|
Ccl3
|
chemokine (C-C motif) ligand 3 |
| chr16_-_23749544 | 1.51 |
ENSMUST00000061030.10
|
Rtp2
|
receptor transporter protein 2 |
| chrX_+_72760318 | 1.51 |
ENSMUST00000114461.3
|
Abcd1
|
ATP-binding cassette, sub-family D (ALD), member 1 |
| chr11_+_82006001 | 1.50 |
ENSMUST00000009329.3
|
Ccl8
|
chemokine (C-C motif) ligand 8 |
| chr2_+_101455079 | 1.50 |
ENSMUST00000111227.2
|
Rag2
|
recombination activating gene 2 |
| chr6_+_41118120 | 1.47 |
ENSMUST00000103273.3
|
Trbv15
|
T cell receptor beta, variable 15 |
| chr1_-_191307648 | 1.47 |
ENSMUST00000027933.11
|
Dtl
|
denticleless E3 ubiquitin protein ligase |
| chr6_-_87786736 | 1.46 |
ENSMUST00000032134.9
|
Rab43
|
RAB43, member RAS oncogene family |
| chr12_-_111947487 | 1.46 |
ENSMUST00000190536.2
|
Rd3l
|
retinal degeneration 3-like |
| chrX_+_135950334 | 1.44 |
ENSMUST00000047852.8
|
Fam199x
|
family with sequence similarity 199, X-linked |
| chr16_-_45830575 | 1.44 |
ENSMUST00000130481.2
|
Plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr2_+_101455022 | 1.43 |
ENSMUST00000044031.4
|
Rag2
|
recombination activating gene 2 |
| chr10_+_87926932 | 1.41 |
ENSMUST00000048621.8
|
Pmch
|
pro-melanin-concentrating hormone |
| chr11_-_99265721 | 1.39 |
ENSMUST00000006963.3
|
Krt28
|
keratin 28 |
| chr6_+_129374260 | 1.39 |
ENSMUST00000032262.14
|
Clec1b
|
C-type lectin domain family 1, member b |
| chr15_+_4756657 | 1.38 |
ENSMUST00000162585.8
|
C6
|
complement component 6 |
| chr8_+_23643279 | 1.38 |
ENSMUST00000071588.8
|
Nkx6-3
|
NK6 homeobox 3 |
| chr17_-_35304582 | 1.38 |
ENSMUST00000038507.7
|
Ly6g6f
|
lymphocyte antigen 6 complex, locus G6F |
| chr11_+_81948649 | 1.36 |
ENSMUST00000000342.3
|
Ccl11
|
chemokine (C-C motif) ligand 11 |
| chr10_-_120312374 | 1.35 |
ENSMUST00000072777.14
ENSMUST00000159699.2 |
Hmga2
|
high mobility group AT-hook 2 |
| chr4_-_44066960 | 1.35 |
ENSMUST00000173234.8
ENSMUST00000173274.2 |
Gne
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr6_+_29859660 | 1.34 |
ENSMUST00000128927.9
|
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr2_-_129213050 | 1.34 |
ENSMUST00000028881.14
|
Il1b
|
interleukin 1 beta |
| chr11_+_115044966 | 1.33 |
ENSMUST00000021076.6
|
Rab37
|
RAB37, member RAS oncogene family |
| chr7_+_19753097 | 1.32 |
ENSMUST00000117909.2
|
Nlrp9b
|
NLR family, pyrin domain containing 9B |
| chrX_+_106193060 | 1.32 |
ENSMUST00000125676.8
ENSMUST00000180182.2 |
P2ry10b
|
purinergic receptor P2Y, G-protein coupled 10B |
| chr4_-_56741398 | 1.31 |
ENSMUST00000095080.5
|
Actl7b
|
actin-like 7b |
| chr4_+_80828883 | 1.30 |
ENSMUST00000055922.4
|
Lurap1l
|
leucine rich adaptor protein 1-like |
| chr4_-_112632013 | 1.30 |
ENSMUST00000060327.4
|
Skint10
|
selection and upkeep of intraepithelial T cells 10 |
| chr5_-_113957318 | 1.30 |
ENSMUST00000201194.4
|
Selplg
|
selectin, platelet (p-selectin) ligand |
| chr3_-_75177378 | 1.29 |
ENSMUST00000039047.5
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr19_-_46314945 | 1.29 |
ENSMUST00000225781.2
ENSMUST00000223903.2 |
Psd
|
pleckstrin and Sec7 domain containing |
| chr6_+_124470053 | 1.29 |
ENSMUST00000049124.10
|
C1rl
|
complement component 1, r subcomponent-like |
| chr4_+_107111106 | 1.28 |
ENSMUST00000046558.8
|
Hspb11
|
heat shock protein family B (small), member 11 |
| chr11_+_87486472 | 1.28 |
ENSMUST00000134216.3
|
Mtmr4
|
myotubularin related protein 4 |
| chr13_-_63036096 | 1.27 |
ENSMUST00000092888.11
|
Fbp1
|
fructose bisphosphatase 1 |
| chr9_-_61934135 | 1.27 |
ENSMUST00000034817.11
|
Paqr5
|
progestin and adipoQ receptor family member V |
| chr3_+_40905066 | 1.26 |
ENSMUST00000191805.7
|
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr19_-_46315543 | 1.26 |
ENSMUST00000223917.2
ENSMUST00000224447.2 ENSMUST00000041391.5 ENSMUST00000096029.12 |
Psd
|
pleckstrin and Sec7 domain containing |
| chr19_+_5540591 | 1.25 |
ENSMUST00000237122.2
|
Cfl1
|
cofilin 1, non-muscle |
| chr11_+_105866030 | 1.23 |
ENSMUST00000001964.8
|
Ace
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 |
| chr6_+_70332836 | 1.23 |
ENSMUST00000103390.3
|
Igkv8-18
|
immunoglobulin kappa variable 8-18 |
| chr17_-_37511885 | 1.23 |
ENSMUST00000222190.2
|
Olfr94
|
olfactory receptor 94 |
| chr12_-_111947536 | 1.23 |
ENSMUST00000185354.2
|
Rd3l
|
retinal degeneration 3-like |
| chr6_+_41112064 | 1.21 |
ENSMUST00000103272.4
|
Trbv14
|
T cell receptor beta, variable 14 |
| chr16_+_35758836 | 1.21 |
ENSMUST00000114878.8
|
Parp9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr16_+_96081998 | 1.21 |
ENSMUST00000099497.4
|
B3galt5
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 5 |
| chr19_+_5540483 | 1.21 |
ENSMUST00000209469.2
ENSMUST00000116560.3 |
Cfl1
|
cofilin 1, non-muscle |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 9.3 | GO:0009608 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 1.2 | 3.7 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 1.2 | 7.3 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 1.0 | 7.2 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 1.0 | 3.0 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.9 | 2.7 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.9 | 2.6 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.9 | 3.5 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) |
| 0.8 | 2.5 | GO:2000769 | positive regulation of NMDA glutamate receptor activity(GO:1904783) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) |
| 0.8 | 3.2 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.8 | 2.4 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.8 | 3.1 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.7 | 2.2 | GO:0030887 | positive regulation of myeloid dendritic cell activation(GO:0030887) |
| 0.7 | 4.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.6 | 3.2 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.6 | 4.4 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.6 | 3.7 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.6 | 2.4 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.6 | 2.9 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) B cell homeostatic proliferation(GO:0002358) |
| 0.6 | 6.4 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.5 | 2.2 | GO:2000373 | regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.5 | 1.5 | GO:0070429 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) |
| 0.5 | 1.5 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.5 | 1.4 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.5 | 1.9 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.4 | 1.8 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.4 | 1.8 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.4 | 4.0 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.4 | 0.9 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.4 | 2.1 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.4 | 1.2 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.4 | 2.5 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.4 | 1.2 | GO:1904632 | negative regulation of interleukin-12 biosynthetic process(GO:0045083) response to diterpene(GO:1904629) cellular response to diterpene(GO:1904630) response to glucoside(GO:1904631) cellular response to glucoside(GO:1904632) |
| 0.4 | 5.8 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.4 | 3.8 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.4 | 0.8 | GO:0045404 | positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.4 | 1.1 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.4 | 4.8 | GO:0002295 | T-helper cell lineage commitment(GO:0002295) CD4-positive, alpha-beta T cell lineage commitment(GO:0043373) T-helper 17 cell lineage commitment(GO:0072540) |
| 0.3 | 1.7 | GO:1905068 | positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.3 | 2.4 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.3 | 1.0 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.3 | 1.0 | GO:0042128 | nitrate assimilation(GO:0042128) |
| 0.3 | 1.0 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.3 | 2.2 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.3 | 1.9 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.3 | 4.1 | GO:1903207 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.3 | 81.9 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.3 | 0.3 | GO:0039692 | single stranded viral RNA replication via double stranded DNA intermediate(GO:0039692) |
| 0.3 | 0.8 | GO:1904826 | regulation of hydrogen sulfide biosynthetic process(GO:1904826) positive regulation of hydrogen sulfide biosynthetic process(GO:1904828) |
| 0.3 | 0.8 | GO:0018900 | dichloromethane metabolic process(GO:0018900) |
| 0.3 | 4.3 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.3 | 2.1 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.3 | 3.9 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.3 | 1.8 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.3 | 2.0 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.2 | 1.0 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.2 | 0.5 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.2 | 0.9 | GO:0040030 | regulation of molecular function, epigenetic(GO:0040030) |
| 0.2 | 0.9 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.2 | 0.7 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.2 | 0.6 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.2 | 3.4 | GO:0035376 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.2 | 16.5 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.2 | 1.2 | GO:0045062 | interleukin-15-mediated signaling pathway(GO:0035723) extrathymic T cell selection(GO:0045062) cellular response to interleukin-15(GO:0071350) |
| 0.2 | 1.2 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.2 | 0.8 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) generation of ovulation cycle rhythm(GO:0060112) |
| 0.2 | 2.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.2 | 0.9 | GO:0019516 | lactate oxidation(GO:0019516) |
| 0.2 | 1.8 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.2 | 0.5 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.2 | 0.7 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.2 | 1.2 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.2 | 0.8 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.2 | 0.9 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.2 | 1.7 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.2 | 0.5 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) |
| 0.2 | 0.5 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.2 | 1.4 | GO:0060763 | mammary duct terminal end bud growth(GO:0060763) |
| 0.2 | 0.8 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.2 | 3.0 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 2.8 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.1 | 0.7 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.1 | 20.9 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 2.0 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.1 | 0.9 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) positive regulation of saliva secretion(GO:0046878) |
| 0.1 | 2.0 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.1 | 1.2 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 0.1 | 0.4 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) |
| 0.1 | 3.8 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 1.2 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 2.1 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.1 | 0.4 | GO:1902220 | positive regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902220) |
| 0.1 | 0.5 | GO:0000105 | histidine biosynthetic process(GO:0000105) 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.1 | 1.8 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 4.6 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.1 | 2.0 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.1 | 1.5 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.1 | 1.3 | GO:0060613 | fat pad development(GO:0060613) |
| 0.1 | 0.7 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.1 | 1.6 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.1 | 1.5 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.1 | 2.3 | GO:0045953 | negative regulation of natural killer cell mediated cytotoxicity(GO:0045953) |
| 0.1 | 3.2 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.1 | 2.0 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.1 | 0.9 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 | 0.6 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.1 | 0.2 | GO:0097195 | pilomotor reflex(GO:0097195) |
| 0.1 | 1.0 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.5 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.1 | 0.6 | GO:0002579 | positive regulation of antigen processing and presentation(GO:0002579) |
| 0.1 | 2.2 | GO:0018195 | peptidyl-arginine modification(GO:0018195) |
| 0.1 | 0.5 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 1.7 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.1 | 1.5 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 0.8 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 0.2 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.1 | 3.1 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 0.8 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.1 | 0.7 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.1 | 0.3 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.1 | 1.9 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 0.4 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.1 | 0.4 | GO:1902177 | positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902177) |
| 0.1 | 0.5 | GO:0002349 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.1 | 1.1 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 0.9 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.1 | 0.3 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 3.0 | GO:0002228 | natural killer cell mediated immunity(GO:0002228) |
| 0.1 | 0.5 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.1 | 3.0 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 0.7 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.1 | 0.6 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.1 | 1.1 | GO:0032674 | regulation of interleukin-5 production(GO:0032674) |
| 0.1 | 1.0 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.0 | 0.3 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 1.6 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.3 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 3.6 | GO:0090501 | RNA phosphodiester bond hydrolysis(GO:0090501) |
| 0.0 | 0.4 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 4.5 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 0.3 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.3 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.0 | 0.4 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.0 | 0.4 | GO:0046641 | positive regulation of alpha-beta T cell proliferation(GO:0046641) |
| 0.0 | 2.7 | GO:0070228 | regulation of lymphocyte apoptotic process(GO:0070228) |
| 0.0 | 0.5 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.8 | GO:0050849 | negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.0 | 2.5 | GO:0033077 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.0 | 0.9 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) |
| 0.0 | 2.8 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) |
| 0.0 | 0.4 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.6 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 1.4 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 0.7 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 1.0 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 1.1 | GO:0030903 | notochord development(GO:0030903) |
| 0.0 | 0.5 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.8 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 1.4 | GO:0030857 | negative regulation of epithelial cell differentiation(GO:0030857) |
| 0.0 | 2.1 | GO:0050710 | negative regulation of cytokine secretion(GO:0050710) |
| 0.0 | 1.4 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 2.7 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.2 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.1 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.0 | 0.9 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.0 | 1.0 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.3 | GO:0034116 | positive regulation of heterotypic cell-cell adhesion(GO:0034116) |
| 0.0 | 1.0 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.8 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 0.6 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.7 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.6 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.0 | 0.4 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.2 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.2 | GO:0071231 | neural crest cell migration involved in heart formation(GO:0003147) folic acid metabolic process(GO:0046655) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.0 | 0.2 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.3 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.2 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.5 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.1 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 1.9 | GO:0048525 | negative regulation of viral process(GO:0048525) |
| 0.0 | 0.2 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 0.1 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 1.0 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.4 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.0 | 0.9 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.1 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.0 | 0.1 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 0.7 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.3 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.4 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.1 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.0 | 0.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.7 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 2.6 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.3 | GO:1902572 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.9 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.0 | 0.1 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 1.0 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.8 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.0 | 0.6 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 1.1 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.2 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.5 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.1 | GO:2001199 | regulation of dendritic cell differentiation(GO:2001198) negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 0.0 | GO:0072125 | adrenal cortex development(GO:0035801) adrenal cortex formation(GO:0035802) negative regulation of glomerular mesangial cell proliferation(GO:0072125) posterior mesonephric tubule development(GO:0072166) metanephric comma-shaped body morphogenesis(GO:0072278) negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) negative regulation of glomerulus development(GO:0090194) negative regulation of cell proliferation involved in kidney development(GO:1901723) regulation of metanephric ureteric bud development(GO:2001074) positive regulation of metanephric ureteric bud development(GO:2001076) |
| 0.0 | 0.7 | GO:0006400 | tRNA modification(GO:0006400) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.8 | GO:0034359 | mature chylomicron(GO:0034359) |
| 0.7 | 4.5 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.5 | 12.4 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.4 | 3.5 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 0.4 | 6.6 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.4 | 4.7 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.4 | 3.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.3 | 5.2 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.3 | 2.0 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.2 | 3.7 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 1.7 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.2 | 1.5 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.2 | 2.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.2 | 20.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.2 | 2.6 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 2.7 | GO:0090543 | Flemming body(GO:0090543) |
| 0.1 | 1.0 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 3.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 1.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 1.8 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 1.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 5.2 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 0.7 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.1 | 2.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 2.2 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.1 | 0.5 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.1 | 0.2 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.1 | 4.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 1.2 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 0.8 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.3 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 3.8 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 1.6 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 1.2 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.6 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 2.2 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.1 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 0.5 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.8 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 1.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 11.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 69.3 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.8 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.7 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.4 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 3.4 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.3 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 2.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.3 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 1.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 4.3 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 1.0 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 2.6 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.1 | GO:0044299 | C-fiber(GO:0044299) |
| 0.0 | 0.1 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 1.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 3.3 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.2 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.1 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.6 | GO:0031985 | Golgi cisterna(GO:0031985) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 7.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 1.3 | 3.8 | GO:0033680 | ATP-dependent DNA/RNA helicase activity(GO:0033680) |
| 0.8 | 3.2 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.8 | 2.4 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.8 | 3.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.8 | 3.0 | GO:0023029 | MHC class Ib protein binding(GO:0023029) |
| 0.7 | 2.2 | GO:0032394 | MHC class Ib receptor activity(GO:0032394) |
| 0.6 | 1.9 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.6 | 1.9 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.6 | 1.8 | GO:0004454 | ketohexokinase activity(GO:0004454) |
| 0.6 | 5.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.6 | 4.0 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.6 | 4.0 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.5 | 4.8 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.5 | 16.7 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.5 | 1.4 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.4 | 5.8 | GO:0035473 | lipase binding(GO:0035473) |
| 0.4 | 2.6 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.4 | 4.0 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.4 | 15.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.3 | 3.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.3 | 3.4 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.3 | 2.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.3 | 1.0 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.3 | 10.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.3 | 1.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.3 | 1.3 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) |
| 0.2 | 1.2 | GO:0031711 | bradykinin receptor binding(GO:0031711) |
| 0.2 | 1.7 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.2 | 1.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.2 | 0.9 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.2 | 2.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.2 | 2.2 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.2 | 0.6 | GO:0052692 | raffinose alpha-galactosidase activity(GO:0052692) |
| 0.2 | 1.0 | GO:0043546 | molybdenum ion binding(GO:0030151) molybdopterin cofactor binding(GO:0043546) |
| 0.2 | 1.5 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.2 | 1.9 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 2.2 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.2 | 0.7 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.2 | 2.0 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.2 | 0.5 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.2 | 1.5 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.2 | 4.2 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.2 | 6.6 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.2 | 3.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.2 | 1.4 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.2 | 0.6 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.2 | 1.7 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.2 | 20.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.2 | 2.6 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.2 | 2.0 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.2 | 0.5 | GO:0000401 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.2 | 0.9 | GO:0031781 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.2 | 2.3 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 3.8 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 1.0 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 2.0 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 1.8 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.1 | 0.9 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 1.0 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 2.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.5 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 0.5 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 1.8 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.7 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.1 | 0.6 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.1 | 0.3 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.1 | 3.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.8 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.1 | 0.4 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.1 | 0.7 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.1 | 1.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 2.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.9 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 3.1 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 2.5 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 0.8 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 2.5 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 0.4 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.1 | 1.3 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.1 | 0.4 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 1.1 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.1 | 5.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 6.4 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 4.7 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 0.5 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 1.2 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 0.9 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.1 | 3.5 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 1.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 0.4 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.1 | 2.2 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 1.0 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 0.3 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 1.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 3.0 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 0.3 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 1.0 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 1.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 2.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.6 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.8 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.9 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.3 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.4 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.3 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.0 | 0.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.1 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 2.0 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.8 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 1.9 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.8 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.6 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) aromatase activity(GO:0070330) |
| 0.0 | 0.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 1.0 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 4.9 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 0.4 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 2.4 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 3.0 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.8 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.2 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.0 | 0.4 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.2 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.2 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 6.8 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.3 | GO:0001164 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 3.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.3 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 1.3 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 3.8 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 1.1 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 2.7 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 12.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 2.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.2 | 2.5 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 8.6 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 0.5 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 4.9 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 2.9 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 4.0 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 1.7 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 1.8 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 3.8 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 5.1 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 2.8 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 3.9 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.0 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 2.6 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.5 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 1.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.8 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 3.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.7 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 2.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.2 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.2 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.9 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 1.6 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.4 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.5 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 5.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.2 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 2.0 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.4 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.5 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.4 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 5.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.3 | 7.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.2 | 2.7 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.2 | 7.8 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.2 | 0.6 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.2 | 1.9 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 4.7 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.2 | 5.5 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.2 | 0.8 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.1 | 3.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 3.8 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 1.0 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 1.0 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 2.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 5.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 3.9 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 5.2 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 0.4 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 2.7 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 0.9 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 0.9 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 3.8 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 1.0 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 1.3 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 1.7 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.1 | 1.1 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 1.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 3.1 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 1.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 3.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.0 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.5 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.9 | REACTOME SIGNALING BY ERBB4 | Genes involved in Signaling by ERBB4 |
| 0.0 | 0.7 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 1.6 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 1.5 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.5 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 1.3 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.7 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 3.7 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.2 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 1.0 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.4 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 1.6 | REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | Genes involved in Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer |
| 0.0 | 0.4 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.5 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.7 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 1.0 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.5 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 3.3 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.7 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 3.2 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.0 | 2.2 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |