Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for En2
Z-value: 1.01
Transcription factors associated with En2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
En2
|
ENSMUSG00000039095.9 | En2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| En2 | mm39_v1_chr5_+_28370687_28370720 | 0.31 | 7.4e-03 | Click! |
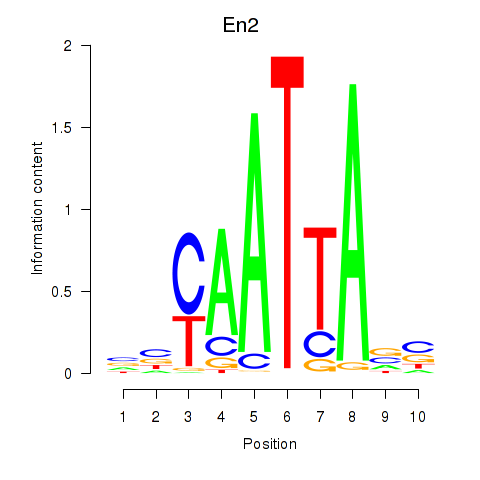
Activity profile of En2 motif
Sorted Z-values of En2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of En2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_135135735 | 9.17 |
ENSMUST00000201977.4
ENSMUST00000005507.10 |
Mlxipl
|
MLX interacting protein-like |
| chrM_+_10167 | 6.83 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr7_+_51528788 | 6.54 |
ENSMUST00000107591.9
|
Gas2
|
growth arrest specific 2 |
| chr1_+_127657142 | 6.44 |
ENSMUST00000038006.8
|
Acmsd
|
amino carboxymuconate semialdehyde decarboxylase |
| chr17_-_84154173 | 5.28 |
ENSMUST00000000687.9
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr17_-_84154196 | 4.89 |
ENSMUST00000234214.2
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chrM_+_9870 | 4.79 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chrM_+_3906 | 4.66 |
ENSMUST00000082396.1
|
mt-Nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr12_-_84497718 | 4.62 |
ENSMUST00000085192.7
ENSMUST00000220491.2 |
Aldh6a1
|
aldehyde dehydrogenase family 6, subfamily A1 |
| chr4_-_63072367 | 4.60 |
ENSMUST00000030041.5
|
Ambp
|
alpha 1 microglobulin/bikunin precursor |
| chr7_-_4909515 | 4.50 |
ENSMUST00000210663.2
|
Gm36210
|
predicted gene, 36210 |
| chr14_+_32043944 | 4.49 |
ENSMUST00000022480.8
ENSMUST00000228529.2 |
Ogdhl
|
oxoglutarate dehydrogenase-like |
| chr3_+_59989282 | 4.35 |
ENSMUST00000029326.6
|
Sucnr1
|
succinate receptor 1 |
| chr14_+_55797468 | 3.65 |
ENSMUST00000147981.2
ENSMUST00000133256.8 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr15_+_100251030 | 3.65 |
ENSMUST00000075675.7
ENSMUST00000088142.6 ENSMUST00000176287.2 |
Methig1
Mettl7a2
|
methyltransferase hypoxia inducible domain containing 1 methyltransferase like 7A2 |
| chr10_-_107321938 | 3.59 |
ENSMUST00000000445.2
|
Myf5
|
myogenic factor 5 |
| chrM_+_11735 | 3.40 |
ENSMUST00000082418.1
|
mt-Nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr15_+_21111428 | 3.33 |
ENSMUST00000075132.8
|
Cdh12
|
cadherin 12 |
| chr11_+_114741948 | 3.28 |
ENSMUST00000133245.2
ENSMUST00000122967.3 |
Gprc5c
|
G protein-coupled receptor, family C, group 5, member C |
| chr3_-_129518723 | 3.09 |
ENSMUST00000199615.5
ENSMUST00000197079.5 |
Egf
|
epidermal growth factor |
| chr15_+_39522905 | 3.04 |
ENSMUST00000226410.2
|
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chr17_+_7437500 | 3.00 |
ENSMUST00000024575.8
|
Rps6ka2
|
ribosomal protein S6 kinase, polypeptide 2 |
| chr8_+_84728123 | 2.99 |
ENSMUST00000060357.15
ENSMUST00000239176.2 |
1700067K01Rik
|
RIKEN cDNA 1700067K01 gene |
| chr9_+_32027335 | 2.94 |
ENSMUST00000174641.8
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr11_+_114742331 | 2.71 |
ENSMUST00000177952.8
|
Gprc5c
|
G protein-coupled receptor, family C, group 5, member C |
| chr2_+_118692435 | 2.68 |
ENSMUST00000028807.6
|
Ivd
|
isovaleryl coenzyme A dehydrogenase |
| chr14_+_33662976 | 2.67 |
ENSMUST00000100720.2
|
Gdf2
|
growth differentiation factor 2 |
| chr15_+_100262423 | 2.64 |
ENSMUST00000175683.8
ENSMUST00000177211.2 |
Higd1c
|
HIG1 domain family, member 1C |
| chr7_+_126550009 | 2.63 |
ENSMUST00000106332.3
|
Sez6l2
|
seizure related 6 homolog like 2 |
| chr7_+_126549692 | 2.61 |
ENSMUST00000106335.8
ENSMUST00000146017.3 |
Sez6l2
|
seizure related 6 homolog like 2 |
| chr7_+_44240310 | 2.54 |
ENSMUST00000107906.6
|
Kcnc3
|
potassium voltage gated channel, Shaw-related subfamily, member 3 |
| chr9_+_103940575 | 2.51 |
ENSMUST00000120854.8
|
Acad11
|
acyl-Coenzyme A dehydrogenase family, member 11 |
| chr13_+_23728222 | 2.50 |
ENSMUST00000075558.5
|
H3c7
|
H3 clustered histone 7 |
| chrM_+_7758 | 2.50 |
ENSMUST00000082407.1
|
mt-Atp8
|
mitochondrially encoded ATP synthase 8 |
| chr19_-_7943365 | 2.42 |
ENSMUST00000182102.8
ENSMUST00000075619.5 |
Slc22a27
|
solute carrier family 22, member 27 |
| chr14_+_55797934 | 2.41 |
ENSMUST00000121622.8
ENSMUST00000143431.2 ENSMUST00000150481.8 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chrM_+_7779 | 2.37 |
ENSMUST00000082408.1
|
mt-Atp6
|
mitochondrially encoded ATP synthase 6 |
| chr15_+_100232810 | 2.36 |
ENSMUST00000075420.6
|
Mettl7a3
|
methyltransferase like 7A3 |
| chr3_+_75464837 | 2.35 |
ENSMUST00000161776.8
ENSMUST00000029423.9 |
Serpini1
|
serine (or cysteine) peptidase inhibitor, clade I, member 1 |
| chr14_+_67953584 | 2.35 |
ENSMUST00000145542.8
ENSMUST00000125212.2 |
Kctd9
|
potassium channel tetramerisation domain containing 9 |
| chr8_+_94879235 | 2.30 |
ENSMUST00000034211.10
ENSMUST00000211930.2 ENSMUST00000211915.2 |
Mt3
|
metallothionein 3 |
| chr5_+_88731386 | 2.25 |
ENSMUST00000031229.11
|
Rufy3
|
RUN and FYVE domain containing 3 |
| chr14_+_58308004 | 2.23 |
ENSMUST00000165526.9
|
Fgf9
|
fibroblast growth factor 9 |
| chr13_-_113800172 | 2.22 |
ENSMUST00000054650.5
|
Hspb3
|
heat shock protein 3 |
| chrM_+_9459 | 2.21 |
ENSMUST00000082411.1
|
mt-Nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr5_+_88731366 | 2.18 |
ENSMUST00000199312.5
|
Rufy3
|
RUN and FYVE domain containing 3 |
| chr12_+_52746158 | 2.15 |
ENSMUST00000095737.5
|
Akap6
|
A kinase (PRKA) anchor protein 6 |
| chr14_+_67953547 | 2.14 |
ENSMUST00000078053.13
|
Kctd9
|
potassium channel tetramerisation domain containing 9 |
| chr19_-_56378309 | 2.14 |
ENSMUST00000166203.2
ENSMUST00000167239.8 |
Nrap
|
nebulin-related anchoring protein |
| chr12_-_85197985 | 2.14 |
ENSMUST00000019379.9
ENSMUST00000221972.2 |
Rps6kl1
|
ribosomal protein S6 kinase-like 1 |
| chr18_+_84869456 | 2.09 |
ENSMUST00000160180.9
|
Cyb5a
|
cytochrome b5 type A (microsomal) |
| chr12_-_108145469 | 2.00 |
ENSMUST00000125916.3
ENSMUST00000109879.8 |
Setd3
|
SET domain containing 3 |
| chr3_+_63203516 | 1.99 |
ENSMUST00000029400.7
|
Mme
|
membrane metallo endopeptidase |
| chr16_-_45544960 | 1.96 |
ENSMUST00000096057.5
|
Tagln3
|
transgelin 3 |
| chr14_+_67953687 | 1.95 |
ENSMUST00000150768.8
|
Kctd9
|
potassium channel tetramerisation domain containing 9 |
| chr12_-_108145498 | 1.93 |
ENSMUST00000071095.14
|
Setd3
|
SET domain containing 3 |
| chr12_+_84498196 | 1.93 |
ENSMUST00000137170.3
|
Lin52
|
lin-52 homolog (C. elegans) |
| chr2_+_9887427 | 1.90 |
ENSMUST00000114919.2
|
4930412O13Rik
|
RIKEN cDNA 4930412O13 gene |
| chr6_-_54969843 | 1.90 |
ENSMUST00000203208.2
|
Ggct
|
gamma-glutamyl cyclotransferase |
| chr6_-_54969928 | 1.89 |
ENSMUST00000131475.2
|
Ggct
|
gamma-glutamyl cyclotransferase |
| chr10_+_50770836 | 1.87 |
ENSMUST00000219436.2
|
Sim1
|
single-minded family bHLH transcription factor 1 |
| chr14_+_55798362 | 1.86 |
ENSMUST00000072530.11
ENSMUST00000128490.9 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr3_+_94320548 | 1.86 |
ENSMUST00000166032.8
ENSMUST00000200486.5 ENSMUST00000196386.5 ENSMUST00000045245.10 ENSMUST00000197901.5 ENSMUST00000198041.2 |
Tdrkh
Gm42463
|
tudor and KH domain containing protein predicted gene 42463 |
| chr7_+_141503411 | 1.82 |
ENSMUST00000078200.12
ENSMUST00000018971.15 |
Brsk2
|
BR serine/threonine kinase 2 |
| chr14_+_19801333 | 1.81 |
ENSMUST00000022340.5
|
Nid2
|
nidogen 2 |
| chr11_-_99213769 | 1.79 |
ENSMUST00000038004.3
|
Krt25
|
keratin 25 |
| chr12_+_108145802 | 1.79 |
ENSMUST00000221167.2
|
Ccnk
|
cyclin K |
| chr5_-_84565218 | 1.68 |
ENSMUST00000113401.4
|
Epha5
|
Eph receptor A5 |
| chr10_+_88721854 | 1.66 |
ENSMUST00000020255.8
|
Slc5a8
|
solute carrier family 5 (iodide transporter), member 8 |
| chr17_+_3447465 | 1.66 |
ENSMUST00000072156.7
|
Tiam2
|
T cell lymphoma invasion and metastasis 2 |
| chr9_+_107440445 | 1.64 |
ENSMUST00000010198.5
|
Tusc2
|
tumor suppressor 2, mitochondrial calcium regulator |
| chr19_+_25649767 | 1.63 |
ENSMUST00000053068.7
|
Dmrt2
|
doublesex and mab-3 related transcription factor 2 |
| chr5_-_131645437 | 1.62 |
ENSMUST00000161804.9
|
Auts2
|
autism susceptibility candidate 2 |
| chr6_+_92793440 | 1.60 |
ENSMUST00000057977.4
|
A730049H05Rik
|
RIKEN cDNA A730049H05 gene |
| chr14_+_55797443 | 1.56 |
ENSMUST00000117236.8
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr12_+_111780604 | 1.56 |
ENSMUST00000021714.9
ENSMUST00000223211.2 ENSMUST00000222843.2 ENSMUST00000221375.2 |
Zfyve21
|
zinc finger, FYVE domain containing 21 |
| chr12_+_38830812 | 1.52 |
ENSMUST00000160856.8
|
Etv1
|
ets variant 1 |
| chr12_+_38830283 | 1.51 |
ENSMUST00000162563.8
ENSMUST00000161164.8 ENSMUST00000160996.8 |
Etv1
|
ets variant 1 |
| chr4_+_150322151 | 1.50 |
ENSMUST00000141931.2
|
Eno1
|
enolase 1, alpha non-neuron |
| chr11_+_67689094 | 1.46 |
ENSMUST00000168612.8
|
Dhrs7c
|
dehydrogenase/reductase (SDR family) member 7C |
| chr2_+_30254239 | 1.46 |
ENSMUST00000077977.14
ENSMUST00000140075.9 ENSMUST00000142801.8 ENSMUST00000100214.10 |
Miga2
|
mitoguardin 2 |
| chr9_+_107440484 | 1.44 |
ENSMUST00000193418.2
|
Tusc2
|
tumor suppressor 2, mitochondrial calcium regulator |
| chrM_+_7006 | 1.43 |
ENSMUST00000082405.1
|
mt-Co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr14_+_60615128 | 1.42 |
ENSMUST00000022561.9
|
Amer2
|
APC membrane recruitment 2 |
| chr8_+_121842902 | 1.39 |
ENSMUST00000054691.8
|
Foxc2
|
forkhead box C2 |
| chr11_-_107228382 | 1.38 |
ENSMUST00000040380.13
|
Pitpnc1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr19_-_28945194 | 1.35 |
ENSMUST00000162110.8
|
Spata6l
|
spermatogenesis associated 6 like |
| chr11_+_75422925 | 1.31 |
ENSMUST00000169547.9
|
Slc43a2
|
solute carrier family 43, member 2 |
| chr8_+_94763826 | 1.31 |
ENSMUST00000109556.9
ENSMUST00000093301.9 ENSMUST00000060632.8 |
Ogfod1
|
2-oxoglutarate and iron-dependent oxygenase domain containing 1 |
| chr2_+_122016747 | 1.28 |
ENSMUST00000028661.6
|
Terb2
|
telomere repeat binding bouquet formation protein 2 |
| chr5_-_143846600 | 1.27 |
ENSMUST00000031613.11
ENSMUST00000100483.3 |
Aimp2
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 2 |
| chr12_+_108145997 | 1.27 |
ENSMUST00000101055.5
|
Ccnk
|
cyclin K |
| chr10_-_8632519 | 1.25 |
ENSMUST00000212869.2
|
Sash1
|
SAM and SH3 domain containing 1 |
| chr11_-_109502243 | 1.22 |
ENSMUST00000103060.10
ENSMUST00000047186.10 ENSMUST00000106689.2 |
Wipi1
|
WD repeat domain, phosphoinositide interacting 1 |
| chr3_-_54962899 | 1.21 |
ENSMUST00000199144.5
|
Ccna1
|
cyclin A1 |
| chr11_+_75422953 | 1.20 |
ENSMUST00000127226.3
|
Slc43a2
|
solute carrier family 43, member 2 |
| chr5_-_146158235 | 1.20 |
ENSMUST00000161859.8
|
Rnf6
|
ring finger protein (C3H2C3 type) 6 |
| chr16_+_37909363 | 1.17 |
ENSMUST00000023507.13
|
Gsk3b
|
glycogen synthase kinase 3 beta |
| chr12_+_76450941 | 1.16 |
ENSMUST00000080449.7
|
Hspa2
|
heat shock protein 2 |
| chr14_-_70666513 | 1.14 |
ENSMUST00000226426.2
ENSMUST00000048129.6 |
Piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr12_+_76451177 | 1.12 |
ENSMUST00000219555.2
|
Hspa2
|
heat shock protein 2 |
| chrM_+_8603 | 1.11 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr7_+_25380263 | 1.11 |
ENSMUST00000205325.2
ENSMUST00000206913.2 |
B9d2
|
B9 protein domain 2 |
| chr6_+_8948608 | 1.10 |
ENSMUST00000160300.2
|
Nxph1
|
neurexophilin 1 |
| chr11_+_75422516 | 1.10 |
ENSMUST00000149727.8
ENSMUST00000108433.8 ENSMUST00000042561.14 ENSMUST00000143035.8 |
Slc43a2
|
solute carrier family 43, member 2 |
| chr5_-_146158201 | 1.10 |
ENSMUST00000161574.8
|
Rnf6
|
ring finger protein (C3H2C3 type) 6 |
| chr2_-_84545504 | 1.10 |
ENSMUST00000035840.6
|
Zdhhc5
|
zinc finger, DHHC domain containing 5 |
| chr3_+_75982890 | 1.09 |
ENSMUST00000160261.8
|
Fstl5
|
follistatin-like 5 |
| chr2_+_22959452 | 1.09 |
ENSMUST00000155602.4
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr8_-_84321069 | 1.09 |
ENSMUST00000019382.17
ENSMUST00000212630.2 ENSMUST00000165740.9 |
Tecr
|
trans-2,3-enoyl-CoA reductase |
| chr12_-_56660054 | 1.08 |
ENSMUST00000072631.6
|
Nkx2-9
|
NK2 homeobox 9 |
| chr7_-_12096691 | 1.07 |
ENSMUST00000086228.3
|
Vmn1r84
|
vomeronasal 1 receptor 84 |
| chr4_+_150938376 | 1.06 |
ENSMUST00000073600.9
|
Errfi1
|
ERBB receptor feedback inhibitor 1 |
| chr18_-_43610829 | 1.06 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr1_+_104696235 | 1.04 |
ENSMUST00000062528.9
|
Cdh20
|
cadherin 20 |
| chr8_-_45835234 | 1.03 |
ENSMUST00000093526.13
|
Fam149a
|
family with sequence similarity 149, member A |
| chr9_+_56858162 | 1.01 |
ENSMUST00000068856.5
|
Snupn
|
snurportin 1 |
| chr10_+_73657689 | 1.00 |
ENSMUST00000064562.14
ENSMUST00000193174.6 ENSMUST00000105426.10 ENSMUST00000129404.9 ENSMUST00000131321.9 ENSMUST00000126920.9 ENSMUST00000147189.9 ENSMUST00000105424.10 ENSMUST00000092420.13 ENSMUST00000105429.10 ENSMUST00000193361.6 ENSMUST00000131724.9 ENSMUST00000152655.9 ENSMUST00000151116.9 ENSMUST00000155701.9 ENSMUST00000152819.9 ENSMUST00000125517.9 ENSMUST00000124046.8 ENSMUST00000146682.8 ENSMUST00000177107.8 ENSMUST00000149977.9 ENSMUST00000191854.6 |
Pcdh15
|
protocadherin 15 |
| chr6_+_37847721 | 0.99 |
ENSMUST00000031859.14
ENSMUST00000120428.8 |
Trim24
|
tripartite motif-containing 24 |
| chr10_+_128173603 | 0.98 |
ENSMUST00000005826.9
|
Cs
|
citrate synthase |
| chr2_+_27055245 | 0.98 |
ENSMUST00000000910.7
|
Dbh
|
dopamine beta hydroxylase |
| chr8_+_21382681 | 0.95 |
ENSMUST00000098908.4
|
Defb33
|
defensin beta 33 |
| chr5_-_143279378 | 0.94 |
ENSMUST00000212715.2
|
Zfp853
|
zinc finger protein 853 |
| chr6_-_131662707 | 0.93 |
ENSMUST00000072404.3
|
Tas2r104
|
taste receptor, type 2, member 104 |
| chr2_+_49956441 | 0.93 |
ENSMUST00000112712.10
ENSMUST00000128451.8 ENSMUST00000053208.14 |
Lypd6
|
LY6/PLAUR domain containing 6 |
| chr9_+_78355474 | 0.93 |
ENSMUST00000034896.13
|
Mto1
|
mitochondrial tRNA translation optimization 1 |
| chr10_-_40018243 | 0.92 |
ENSMUST00000092566.8
ENSMUST00000213488.2 |
Slc16a10
|
solute carrier family 16 (monocarboxylic acid transporters), member 10 |
| chr2_-_122016670 | 0.90 |
ENSMUST00000028665.5
|
Patl2
|
protein associated with topoisomerase II homolog 2 (yeast) |
| chr11_+_68986043 | 0.88 |
ENSMUST00000101004.9
|
Per1
|
period circadian clock 1 |
| chr17_+_69746321 | 0.87 |
ENSMUST00000169935.2
|
Akain1
|
A kinase (PRKA) anchor inhibitor 1 |
| chr18_+_23548192 | 0.86 |
ENSMUST00000222515.2
|
Dtna
|
dystrobrevin alpha |
| chr12_+_72488625 | 0.86 |
ENSMUST00000161284.3
ENSMUST00000162159.8 |
Lrrc9
|
leucine rich repeat containing 9 |
| chr11_-_99494134 | 0.85 |
ENSMUST00000072306.4
|
Gm11938
|
predicted gene 11938 |
| chr2_-_17465410 | 0.85 |
ENSMUST00000145492.2
|
Nebl
|
nebulette |
| chr11_+_102175985 | 0.84 |
ENSMUST00000156326.2
|
Tmub2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr11_-_69127848 | 0.84 |
ENSMUST00000021259.9
ENSMUST00000108665.8 ENSMUST00000108664.2 |
Gucy2e
|
guanylate cyclase 2e |
| chrX_+_20554193 | 0.84 |
ENSMUST00000115364.8
|
Cdk16
|
cyclin-dependent kinase 16 |
| chr2_+_177760959 | 0.83 |
ENSMUST00000108916.8
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr12_-_85327136 | 0.83 |
ENSMUST00000065913.8
ENSMUST00000008966.13 |
Acyp1
|
acylphosphatase 1, erythrocyte (common) type |
| chr2_+_109522781 | 0.83 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr5_-_146157711 | 0.82 |
ENSMUST00000169407.9
ENSMUST00000161331.8 ENSMUST00000159074.3 ENSMUST00000067837.10 |
Rnf6
|
ring finger protein (C3H2C3 type) 6 |
| chr18_+_39126325 | 0.82 |
ENSMUST00000137497.9
|
Arhgap26
|
Rho GTPase activating protein 26 |
| chr6_-_41752111 | 0.81 |
ENSMUST00000214976.3
|
Olfr459
|
olfactory receptor 459 |
| chr10_+_73657753 | 0.79 |
ENSMUST00000134009.9
ENSMUST00000177420.7 ENSMUST00000125006.9 |
Pcdh15
|
protocadherin 15 |
| chr2_-_88559941 | 0.76 |
ENSMUST00000099815.2
|
Olfr1197
|
olfactory receptor 1197 |
| chr10_-_25076008 | 0.76 |
ENSMUST00000100012.3
|
Akap7
|
A kinase (PRKA) anchor protein 7 |
| chrX_-_111315519 | 0.75 |
ENSMUST00000124335.8
|
Satl1
|
spermidine/spermine N1-acetyl transferase-like 1 |
| chr2_+_59442378 | 0.74 |
ENSMUST00000112568.8
ENSMUST00000037526.11 |
Tanc1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr9_+_20193647 | 0.74 |
ENSMUST00000071725.4
ENSMUST00000212983.3 |
Olfr39
|
olfactory receptor 39 |
| chr18_+_39126178 | 0.72 |
ENSMUST00000097593.9
|
Arhgap26
|
Rho GTPase activating protein 26 |
| chr8_-_35432783 | 0.72 |
ENSMUST00000033929.6
|
Tnks
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr7_-_119494669 | 0.71 |
ENSMUST00000098080.9
|
Dcun1d3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae) |
| chr1_-_92412835 | 0.70 |
ENSMUST00000214928.3
|
Olfr1416
|
olfactory receptor 1416 |
| chr4_+_109835224 | 0.69 |
ENSMUST00000061187.4
|
Dmrta2
|
doublesex and mab-3 related transcription factor like family A2 |
| chr11_+_68950533 | 0.69 |
ENSMUST00000051888.4
|
Borcs6
|
BLOC-1 related complex subunit 6 |
| chr12_-_91815855 | 0.68 |
ENSMUST00000167466.2
ENSMUST00000021347.12 ENSMUST00000178462.8 |
Sel1l
|
sel-1 suppressor of lin-12-like (C. elegans) |
| chr4_+_108576846 | 0.68 |
ENSMUST00000178992.2
|
3110021N24Rik
|
RIKEN cDNA 3110021N24 gene |
| chr3_+_103739877 | 0.67 |
ENSMUST00000062945.12
|
Bcl2l15
|
BCLl2-like 15 |
| chr8_-_84321032 | 0.66 |
ENSMUST00000163837.2
|
Tecr
|
trans-2,3-enoyl-CoA reductase |
| chr17_+_26342474 | 0.66 |
ENSMUST00000025014.10
ENSMUST00000236166.2 ENSMUST00000127647.3 |
Mrpl28
|
mitochondrial ribosomal protein L28 |
| chr16_-_89368059 | 0.65 |
ENSMUST00000171542.2
|
Krtap11-1
|
keratin associated protein 11-1 |
| chr8_+_107757847 | 0.64 |
ENSMUST00000034388.10
|
Vps4a
|
vacuolar protein sorting 4A |
| chr7_-_4815542 | 0.63 |
ENSMUST00000079496.9
|
Ube2s
|
ubiquitin-conjugating enzyme E2S |
| chr6_-_23650297 | 0.62 |
ENSMUST00000063548.4
|
Rnf133
|
ring finger protein 133 |
| chr2_+_177760768 | 0.60 |
ENSMUST00000108917.8
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr2_+_20742115 | 0.58 |
ENSMUST00000114606.8
ENSMUST00000114608.3 |
Etl4
|
enhancer trap locus 4 |
| chr4_+_116078830 | 0.56 |
ENSMUST00000030464.14
|
Pik3r3
|
phosphoinositide-3-kinase regulatory subunit 3 |
| chr2_-_111100733 | 0.56 |
ENSMUST00000099619.6
|
Olfr1277
|
olfactory receptor 1277 |
| chr18_+_37433852 | 0.55 |
ENSMUST00000051754.2
|
Pcdhb3
|
protocadherin beta 3 |
| chr19_-_6919755 | 0.54 |
ENSMUST00000099782.10
|
Gpr137
|
G protein-coupled receptor 137 |
| chr5_-_138185438 | 0.54 |
ENSMUST00000110937.8
ENSMUST00000139276.2 ENSMUST00000048698.14 ENSMUST00000123415.8 |
Taf6
|
TATA-box binding protein associated factor 6 |
| chr9_-_97252011 | 0.53 |
ENSMUST00000035026.5
|
Trim42
|
tripartite motif-containing 42 |
| chr9_+_38841385 | 0.53 |
ENSMUST00000058789.7
|
Olfr930
|
olfactory receptor 930 |
| chr3_-_92441809 | 0.53 |
ENSMUST00000193521.2
|
2310046K23Rik
|
RIKEN cDNA 2310046K23 gene |
| chr1_-_169796709 | 0.52 |
ENSMUST00000027989.13
ENSMUST00000111353.4 |
Hsd17b7
|
hydroxysteroid (17-beta) dehydrogenase 7 |
| chr10_+_85222677 | 0.52 |
ENSMUST00000105307.8
ENSMUST00000020231.10 |
Btbd11
|
BTB (POZ) domain containing 11 |
| chr7_-_25454177 | 0.52 |
ENSMUST00000206832.2
|
Hnrnpul1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr7_-_10488291 | 0.50 |
ENSMUST00000226874.2
ENSMUST00000227003.2 ENSMUST00000228561.2 ENSMUST00000228248.2 ENSMUST00000228526.2 ENSMUST00000228098.2 ENSMUST00000227940.2 ENSMUST00000228374.2 ENSMUST00000227702.2 |
Vmn1r71
|
vomeronasal 1 receptor 71 |
| chr9_-_96601574 | 0.49 |
ENSMUST00000128269.8
|
Zbtb38
|
zinc finger and BTB domain containing 38 |
| chr1_-_79417732 | 0.49 |
ENSMUST00000185234.2
ENSMUST00000049972.6 |
Scg2
|
secretogranin II |
| chr11_-_99241924 | 0.48 |
ENSMUST00000017732.3
|
Krt27
|
keratin 27 |
| chr6_-_87827993 | 0.48 |
ENSMUST00000204890.3
ENSMUST00000113617.3 ENSMUST00000113619.8 ENSMUST00000204653.3 ENSMUST00000032138.15 |
Cnbp
|
cellular nucleic acid binding protein |
| chr4_-_43823866 | 0.48 |
ENSMUST00000215406.2
ENSMUST00000079234.6 ENSMUST00000214843.2 |
Olfr156
|
olfactory receptor 156 |
| chr15_-_79658608 | 0.48 |
ENSMUST00000229644.2
ENSMUST00000023055.8 |
Dnal4
|
dynein, axonemal, light chain 4 |
| chr13_+_34918820 | 0.48 |
ENSMUST00000039605.8
|
Fam50b
|
family with sequence similarity 50, member B |
| chr7_+_25380195 | 0.47 |
ENSMUST00000205658.2
|
B9d2
|
B9 protein domain 2 |
| chr9_-_40039335 | 0.47 |
ENSMUST00000060345.6
|
Olfr985
|
olfactory receptor 985 |
| chr17_+_38104420 | 0.46 |
ENSMUST00000216051.3
|
Olfr123
|
olfactory receptor 123 |
| chr5_-_124387812 | 0.46 |
ENSMUST00000162812.8
|
Pitpnm2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chr2_-_86109346 | 0.44 |
ENSMUST00000217294.2
ENSMUST00000217245.2 ENSMUST00000216432.2 |
Olfr1051
|
olfactory receptor 1051 |
| chr16_-_22676264 | 0.44 |
ENSMUST00000232075.2
ENSMUST00000004576.8 |
Tbccd1
|
TBCC domain containing 1 |
| chr1_-_132318039 | 0.44 |
ENSMUST00000132435.2
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr5_-_74692327 | 0.43 |
ENSMUST00000072857.13
ENSMUST00000113542.9 ENSMUST00000151474.3 |
Scfd2
|
Sec1 family domain containing 2 |
| chr4_+_115747862 | 0.42 |
ENSMUST00000132221.3
ENSMUST00000165938.2 |
6430628N08Rik
|
RIKEN cDNA 6430628N08 gene |
| chr15_-_79658584 | 0.42 |
ENSMUST00000069877.12
|
Dnal4
|
dynein, axonemal, light chain 4 |
| chr18_+_23548534 | 0.42 |
ENSMUST00000221880.2
ENSMUST00000220904.2 ENSMUST00000047954.15 |
Dtna
|
dystrobrevin alpha |
| chr17_-_37409147 | 0.41 |
ENSMUST00000216376.2
ENSMUST00000217372.2 |
Olfr91
|
olfactory receptor 91 |
| chr7_-_25454126 | 0.41 |
ENSMUST00000108401.3
ENSMUST00000043765.14 |
Hnrnpul1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr17_-_36773221 | 0.41 |
ENSMUST00000169950.2
ENSMUST00000057502.14 |
H2-M10.4
|
histocompatibility 2, M region locus 10.4 |
| chr11_-_99228756 | 0.40 |
ENSMUST00000100482.3
|
Krt26
|
keratin 26 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 16.6 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 1.3 | 9.2 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 1.2 | 4.6 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 1.0 | 3.1 | GO:0052564 | interleukin-15 production(GO:0032618) response to immune response of other organism involved in symbiotic interaction(GO:0052564) response to host immune response(GO:0052572) |
| 0.8 | 2.3 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.6 | 12.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.6 | 2.3 | GO:0031662 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 0.5 | 3.8 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.5 | 3.6 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.5 | 2.9 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.5 | 1.8 | GO:0050973 | detection of mechanical stimulus involved in equilibrioception(GO:0050973) |
| 0.4 | 3.1 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.4 | 1.3 | GO:1903632 | positive regulation of aminoacyl-tRNA ligase activity(GO:1903632) |
| 0.4 | 1.6 | GO:0061055 | myotome development(GO:0061055) |
| 0.4 | 1.6 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.4 | 1.2 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.4 | 3.1 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.3 | 1.4 | GO:1902256 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.3 | 1.0 | GO:0006404 | RNA import into nucleus(GO:0006404) snRNA transport(GO:0051030) |
| 0.3 | 1.0 | GO:0006589 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 0.3 | 4.6 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.3 | 3.9 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.3 | 1.1 | GO:0000239 | pachytene(GO:0000239) |
| 0.3 | 2.0 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.3 | 1.3 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.2 | 0.7 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.2 | 3.0 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.2 | 0.9 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.2 | 3.0 | GO:0002035 | brain renin-angiotensin system(GO:0002035) |
| 0.2 | 2.7 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.2 | 0.6 | GO:0061738 | mitotic cytokinesis checkpoint(GO:0044878) late endosomal microautophagy(GO:0061738) |
| 0.2 | 9.2 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.2 | 1.1 | GO:0010616 | negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.2 | 2.4 | GO:0015747 | urate transport(GO:0015747) |
| 0.2 | 5.5 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.2 | 4.9 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.2 | 3.0 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 0.7 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 0.9 | GO:2000323 | circadian regulation of translation(GO:0097167) negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.1 | 1.3 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.1 | 1.3 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 5.2 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.1 | 4.4 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.1 | 2.2 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.1 | 1.0 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 | 2.7 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 | 1.7 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.1 | 2.5 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 1.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 1.9 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 0.9 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.3 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.1 | 1.2 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 0.3 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.1 | 0.4 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.1 | 1.8 | GO:0042761 | fatty acid elongation(GO:0030497) very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.1 | 0.9 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 1.8 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.1 | 3.8 | GO:0015807 | L-amino acid transport(GO:0015807) |
| 0.1 | 0.1 | GO:0002309 | T cell proliferation involved in immune response(GO:0002309) |
| 0.1 | 1.1 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.0 | 1.5 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 3.3 | GO:0001836 | release of cytochrome c from mitochondria(GO:0001836) |
| 0.0 | 2.2 | GO:0071385 | cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 | 0.3 | GO:0021815 | modulation of microtubule cytoskeleton involved in cerebral cortex radial glia guided migration(GO:0021815) |
| 0.0 | 1.5 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) |
| 0.0 | 0.7 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 1.1 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.2 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 | 0.5 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 5.8 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.2 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.7 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 2.5 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 1.6 | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.0 | 0.7 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.0 | 0.9 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 4.0 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.7 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.8 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.8 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.4 | GO:1903541 | regulation of exosomal secretion(GO:1903541) positive regulation of exosomal secretion(GO:1903543) |
| 0.0 | 1.9 | GO:0072163 | ureteric bud development(GO:0001657) mesonephric epithelium development(GO:0072163) mesonephric tubule development(GO:0072164) |
| 0.0 | 0.5 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 1.4 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0060205 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 1.0 | 3.1 | GO:0002945 | cyclin K-CDK12 complex(GO:0002944) cyclin K-CDK13 complex(GO:0002945) |
| 0.5 | 2.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.5 | 1.5 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 0.4 | 4.5 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.4 | 1.2 | GO:0016014 | dystrobrevin complex(GO:0016014) |
| 0.3 | 1.9 | GO:0071547 | piP-body(GO:0071547) |
| 0.3 | 4.9 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.3 | 2.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.2 | 9.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.2 | 22.9 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.2 | 1.1 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.2 | 0.9 | GO:0097123 | cyclin A1-CDK2 complex(GO:0097123) |
| 0.2 | 2.3 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.2 | 1.9 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 0.7 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.1 | 1.0 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 3.0 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 0.3 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 1.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 1.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 1.6 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.4 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.1 | 1.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 0.2 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 1.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 3.0 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 1.1 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 0.6 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 2.5 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.3 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.8 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.9 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 18.1 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 0.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 1.3 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.7 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 3.1 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.1 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 1.8 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 5.9 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 5.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 2.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.7 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 7.4 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 1.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 2.9 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.9 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.7 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 5.8 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 6.0 | GO:0005118 | sevenless binding(GO:0005118) |
| 1.1 | 4.6 | GO:0019862 | IgA binding(GO:0019862) |
| 0.8 | 2.5 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.7 | 3.0 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.7 | 21.9 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.6 | 4.5 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.6 | 3.8 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.5 | 3.9 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.4 | 10.2 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.3 | 1.3 | GO:0031544 | peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.3 | 1.0 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.3 | 0.8 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.3 | 7.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.2 | 2.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.2 | 1.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.2 | 0.8 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.2 | 0.7 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.2 | 0.9 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.2 | 0.5 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.2 | 1.7 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.1 | 3.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 2.4 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 1.0 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 1.0 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 2.7 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 3.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 3.1 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 2.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 1.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 6.4 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 1.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.9 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.1 | 2.3 | GO:0051861 | glycolipid binding(GO:0051861) |
| 0.1 | 0.9 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 2.1 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 4.6 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) thiolester hydrolase activity(GO:0016790) |
| 0.1 | 2.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 3.8 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 3.2 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 1.1 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 0.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 1.6 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 4.9 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.9 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.3 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 2.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 1.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 9.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.9 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 1.1 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 1.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0032142 | dinucleotide insertion or deletion binding(GO:0032139) single guanine insertion binding(GO:0032142) |
| 0.0 | 1.8 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.9 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 1.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.2 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 2.5 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 2.5 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.8 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 3.0 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.5 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 1.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 2.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.9 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 2.0 | GO:0008238 | exopeptidase activity(GO:0008238) |
| 0.0 | 0.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 1.4 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.9 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.0 | 1.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.4 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 1.2 | GO:0005550 | pheromone binding(GO:0005550) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 3.0 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 4.2 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 6.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 2.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 0.9 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.9 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 3.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 2.2 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 3.6 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 2.7 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.5 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 0.3 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 1.5 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 1.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 5.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.0 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 16.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.3 | 7.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 6.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 0.9 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 2.2 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 3.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 3.0 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 3.4 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 3.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 4.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 1.7 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 3.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 1.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 1.0 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 1.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.0 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 2.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 1.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 2.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 4.9 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 1.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.0 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.6 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.7 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 1.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.9 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 1.0 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.9 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |