Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Ep300
Z-value: 0.99
Transcription factors associated with Ep300
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ep300
|
ENSMUSG00000055024.13 | Ep300 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ep300 | mm39_v1_chr15_+_81511486_81511626 | 0.24 | 4.1e-02 | Click! |
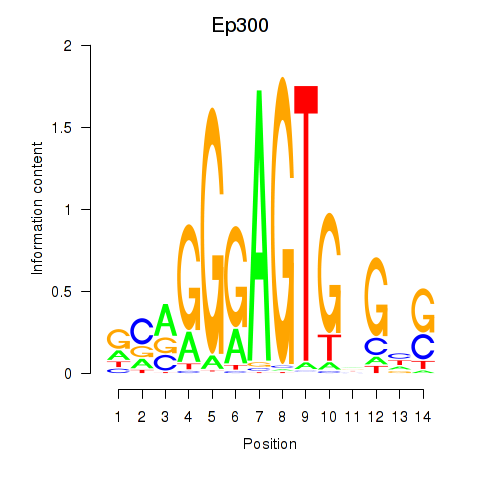
Activity profile of Ep300 motif
Sorted Z-values of Ep300 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Ep300
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_108610032 | 8.16 |
ENSMUST00000033341.12
|
Tub
|
tubby bipartite transcription factor |
| chr1_-_87501548 | 7.70 |
ENSMUST00000068681.12
|
Ngef
|
neuronal guanine nucleotide exchange factor |
| chr2_+_102488985 | 7.69 |
ENSMUST00000080210.10
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr15_-_79389442 | 7.24 |
ENSMUST00000057801.8
|
Kcnj4
|
potassium inwardly-rectifying channel, subfamily J, member 4 |
| chr2_-_155356716 | 7.13 |
ENSMUST00000029131.11
|
Ggt7
|
gamma-glutamyltransferase 7 |
| chr9_-_29323032 | 6.99 |
ENSMUST00000115236.2
|
Ntm
|
neurotrimin |
| chr6_+_110622533 | 6.91 |
ENSMUST00000071076.13
ENSMUST00000172951.2 |
Grm7
|
glutamate receptor, metabotropic 7 |
| chr9_-_29323500 | 6.42 |
ENSMUST00000115237.8
|
Ntm
|
neurotrimin |
| chr9_-_57513510 | 6.06 |
ENSMUST00000215487.2
ENSMUST00000045068.10 |
Cplx3
|
complexin 3 |
| chr11_+_98239230 | 5.80 |
ENSMUST00000078694.13
|
Ppp1r1b
|
protein phosphatase 1, regulatory inhibitor subunit 1B |
| chr2_+_91480513 | 5.47 |
ENSMUST00000090614.11
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chr15_+_74388044 | 5.39 |
ENSMUST00000042035.16
|
Adgrb1
|
adhesion G protein-coupled receptor B1 |
| chrX_-_151820545 | 5.23 |
ENSMUST00000051484.5
|
Mageh1
|
MAGE family member H1 |
| chr5_+_14075281 | 5.16 |
ENSMUST00000073957.8
|
Sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr1_-_83385911 | 5.08 |
ENSMUST00000160953.8
|
Sphkap
|
SPHK1 interactor, AKAP domain containing |
| chr2_+_25293140 | 4.92 |
ENSMUST00000154809.8
ENSMUST00000055921.14 ENSMUST00000141567.8 |
Npdc1
|
neural proliferation, differentiation and control 1 |
| chr2_-_26012751 | 4.87 |
ENSMUST00000140993.2
ENSMUST00000028300.6 |
Nacc2
|
nucleus accumbens associated 2, BEN and BTB (POZ) domain containing |
| chr8_+_23247760 | 4.80 |
ENSMUST00000033941.7
|
Plat
|
plasminogen activator, tissue |
| chr8_+_39472981 | 4.77 |
ENSMUST00000239508.1
ENSMUST00000239509.1 |
TUSC3
|
tumor suppressor candidate 3 |
| chr2_+_102489558 | 4.69 |
ENSMUST00000111213.8
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr19_-_7083094 | 4.57 |
ENSMUST00000113383.4
|
Flrt1
|
fibronectin leucine rich transmembrane protein 1 |
| chr2_-_31735937 | 4.50 |
ENSMUST00000028188.8
|
Fibcd1
|
fibrinogen C domain containing 1 |
| chr14_-_20027279 | 4.44 |
ENSMUST00000160013.8
|
Gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr4_-_148123223 | 4.27 |
ENSMUST00000030879.12
ENSMUST00000137724.8 |
Clcn6
|
chloride channel, voltage-sensitive 6 |
| chr8_-_70686746 | 4.25 |
ENSMUST00000130319.2
|
Armc6
|
armadillo repeat containing 6 |
| chrX_+_119199956 | 4.22 |
ENSMUST00000113364.10
ENSMUST00000050239.16 ENSMUST00000113358.10 |
Pcdh11x
|
protocadherin 11 X-linked |
| chrX_+_60753074 | 4.17 |
ENSMUST00000075983.6
|
Ldoc1
|
regulator of NFKB signaling |
| chr15_-_34679321 | 4.07 |
ENSMUST00000040791.9
|
Nipal2
|
NIPA-like domain containing 2 |
| chr6_-_136150491 | 3.97 |
ENSMUST00000111905.8
ENSMUST00000152012.8 ENSMUST00000143943.8 ENSMUST00000125905.2 |
Grin2b
|
glutamate receptor, ionotropic, NMDA2B (epsilon 2) |
| chr10_-_57408512 | 3.96 |
ENSMUST00000169122.8
|
Serinc1
|
serine incorporator 1 |
| chr3_-_54823287 | 3.95 |
ENSMUST00000070342.4
|
Sertm1
|
serine rich and transmembrane domain containing 1 |
| chr4_-_152533265 | 3.90 |
ENSMUST00000159840.8
ENSMUST00000105648.10 |
Kcnab2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
| chr10_-_81318489 | 3.87 |
ENSMUST00000120508.8
ENSMUST00000238823.2 ENSMUST00000118763.8 |
Celf5
|
CUGBP, Elav-like family member 5 |
| chr1_+_55445033 | 3.85 |
ENSMUST00000042986.10
|
Plcl1
|
phospholipase C-like 1 |
| chr6_+_99669640 | 3.85 |
ENSMUST00000101122.3
|
Gpr27
|
G protein-coupled receptor 27 |
| chr2_+_91480460 | 3.85 |
ENSMUST00000111331.9
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chr14_-_20027219 | 3.79 |
ENSMUST00000055100.14
ENSMUST00000162425.8 |
Gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr14_-_20026761 | 3.70 |
ENSMUST00000161247.2
|
Gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr1_+_183078573 | 3.59 |
ENSMUST00000109166.8
|
Aida
|
axin interactor, dorsalization associated |
| chr16_-_9812787 | 3.56 |
ENSMUST00000199708.5
|
Grin2a
|
glutamate receptor, ionotropic, NMDA2A (epsilon 1) |
| chr14_-_20027112 | 3.46 |
ENSMUST00000159028.8
|
Gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr3_-_108062172 | 3.46 |
ENSMUST00000062028.8
|
Gpr61
|
G protein-coupled receptor 61 |
| chr1_-_183078488 | 3.40 |
ENSMUST00000057062.12
|
Brox
|
BRO1 domain and CAAX motif containing |
| chr13_-_19803928 | 3.40 |
ENSMUST00000221014.2
ENSMUST00000002885.8 ENSMUST00000220944.2 |
Epdr1
|
ependymin related protein 1 (zebrafish) |
| chr1_+_75427080 | 3.37 |
ENSMUST00000113577.8
|
Asic4
|
acid-sensing (proton-gated) ion channel family member 4 |
| chr10_-_57408585 | 3.29 |
ENSMUST00000020027.11
|
Serinc1
|
serine incorporator 1 |
| chr1_+_75427343 | 3.25 |
ENSMUST00000037708.10
|
Asic4
|
acid-sensing (proton-gated) ion channel family member 4 |
| chr2_-_102230602 | 3.00 |
ENSMUST00000152929.2
|
Trim44
|
tripartite motif-containing 44 |
| chr8_-_86567506 | 3.00 |
ENSMUST00000034140.9
|
Itfg1
|
integrin alpha FG-GAP repeat containing 1 |
| chr15_+_89383799 | 2.96 |
ENSMUST00000109309.9
|
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr17_-_24863907 | 2.88 |
ENSMUST00000234505.2
|
Slc9a3r2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 |
| chr19_+_23736205 | 2.87 |
ENSMUST00000025830.9
|
Apba1
|
amyloid beta (A4) precursor protein binding, family A, member 1 |
| chr9_+_74944994 | 2.82 |
ENSMUST00000007800.8
|
Arpp19
|
cAMP-regulated phosphoprotein 19 |
| chr9_+_6168638 | 2.77 |
ENSMUST00000214892.2
|
Pdgfd
|
platelet-derived growth factor, D polypeptide |
| chr10_-_30531832 | 2.74 |
ENSMUST00000217138.2
ENSMUST00000217644.2 ENSMUST00000216172.2 |
Ncoa7
|
nuclear receptor coactivator 7 |
| chr4_+_86493905 | 2.69 |
ENSMUST00000091064.8
|
Rraga
|
Ras-related GTP binding A |
| chr2_-_91480096 | 2.67 |
ENSMUST00000099714.10
ENSMUST00000111333.2 |
Zfp408
|
zinc finger protein 408 |
| chr15_-_13173736 | 2.66 |
ENSMUST00000036439.6
|
Cdh6
|
cadherin 6 |
| chr10_-_30531768 | 2.66 |
ENSMUST00000092610.12
|
Ncoa7
|
nuclear receptor coactivator 7 |
| chr17_-_24863956 | 2.62 |
ENSMUST00000019684.13
|
Slc9a3r2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 |
| chr16_-_13804699 | 2.60 |
ENSMUST00000117803.2
|
Ifitm7
|
interferon induced transmembrane protein 7 |
| chr14_+_77394173 | 2.58 |
ENSMUST00000022589.9
|
Enox1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr10_-_127370408 | 2.57 |
ENSMUST00000095266.3
|
Nxph4
|
neurexophilin 4 |
| chrX_-_106859842 | 2.57 |
ENSMUST00000120722.2
|
2610002M06Rik
|
RIKEN cDNA 2610002M06 gene |
| chr10_-_83484467 | 2.51 |
ENSMUST00000146876.9
ENSMUST00000176294.2 |
Appl2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr4_+_150321142 | 2.47 |
ENSMUST00000150175.8
|
Eno1
|
enolase 1, alpha non-neuron |
| chr4_+_103476777 | 2.42 |
ENSMUST00000106827.8
|
Dab1
|
disabled 1 |
| chr1_-_91386976 | 2.38 |
ENSMUST00000069620.10
|
Per2
|
period circadian clock 2 |
| chr7_-_79570342 | 2.38 |
ENSMUST00000075657.8
|
Ap3s2
|
adaptor-related protein complex 3, sigma 2 subunit |
| chr2_-_52566583 | 2.31 |
ENSMUST00000178799.8
|
Cacnb4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr7_-_140462187 | 2.28 |
ENSMUST00000211179.2
|
Sirt3
|
sirtuin 3 |
| chr16_-_52272828 | 2.27 |
ENSMUST00000170035.8
ENSMUST00000164728.8 ENSMUST00000168071.2 |
Alcam
|
activated leukocyte cell adhesion molecule |
| chr19_+_47167259 | 2.26 |
ENSMUST00000111808.11
|
Neurl1a
|
neuralized E3 ubiquitin protein ligase 1A |
| chr1_+_133291302 | 2.24 |
ENSMUST00000135222.9
|
Etnk2
|
ethanolamine kinase 2 |
| chr2_+_163444214 | 2.20 |
ENSMUST00000171696.8
ENSMUST00000109408.10 |
Ttpal
|
tocopherol (alpha) transfer protein-like |
| chr11_-_120042019 | 2.15 |
ENSMUST00000179094.8
ENSMUST00000103018.11 ENSMUST00000045402.14 ENSMUST00000076697.13 ENSMUST00000053692.9 |
Slc38a10
|
solute carrier family 38, member 10 |
| chr9_+_74945021 | 2.09 |
ENSMUST00000168301.2
|
Arpp19
|
cAMP-regulated phosphoprotein 19 |
| chrX_+_149830166 | 2.05 |
ENSMUST00000026296.8
|
Fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr11_+_76070483 | 2.05 |
ENSMUST00000129853.8
|
Tlcd3a
|
TLC domain containing 3A |
| chr18_+_24737009 | 2.03 |
ENSMUST00000234266.2
ENSMUST00000025120.8 |
Elp2
|
elongator acetyltransferase complex subunit 2 |
| chr16_-_94327689 | 2.03 |
ENSMUST00000023615.7
|
Vps26c
|
VPS26 endosomal protein sorting factor C |
| chr1_+_75483721 | 2.03 |
ENSMUST00000037330.5
|
Inha
|
inhibin alpha |
| chr7_+_96171549 | 2.02 |
ENSMUST00000129737.2
|
Tenm4
|
teneurin transmembrane protein 4 |
| chr13_+_120151982 | 1.98 |
ENSMUST00000179869.3
ENSMUST00000224188.2 |
Hmgcs1
|
3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 |
| chr3_+_90444537 | 1.97 |
ENSMUST00000098911.10
|
S100a16
|
S100 calcium binding protein A16 |
| chr5_+_145217272 | 1.96 |
ENSMUST00000200246.2
|
Zscan25
|
zinc finger and SCAN domain containing 25 |
| chr5_-_39801940 | 1.90 |
ENSMUST00000152057.2
ENSMUST00000053116.7 |
Hs3st1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
| chr4_-_118266416 | 1.89 |
ENSMUST00000075406.12
|
Szt2
|
SZT2 subunit of KICSTOR complex |
| chr17_+_87061117 | 1.88 |
ENSMUST00000024954.11
|
Epas1
|
endothelial PAS domain protein 1 |
| chr3_-_63872079 | 1.88 |
ENSMUST00000161659.8
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr8_-_65302573 | 1.88 |
ENSMUST00000210166.2
|
Klhl2
|
kelch-like 2, Mayven |
| chr2_-_102231208 | 1.87 |
ENSMUST00000102573.8
|
Trim44
|
tripartite motif-containing 44 |
| chr7_-_140462221 | 1.83 |
ENSMUST00000026559.14
|
Sirt3
|
sirtuin 3 |
| chr3_-_90340910 | 1.81 |
ENSMUST00000196530.2
|
Ints3
|
integrator complex subunit 3 |
| chr10_-_116309764 | 1.79 |
ENSMUST00000068233.11
|
Kcnmb4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4 |
| chr9_-_79700789 | 1.77 |
ENSMUST00000120690.2
|
Tmem30a
|
transmembrane protein 30A |
| chr3_+_88461059 | 1.76 |
ENSMUST00000008748.8
|
Ubqln4
|
ubiquilin 4 |
| chr6_-_149003171 | 1.73 |
ENSMUST00000111557.8
|
Dennd5b
|
DENN/MADD domain containing 5B |
| chr4_+_148123490 | 1.72 |
ENSMUST00000097788.11
|
Mthfr
|
methylenetetrahydrofolate reductase |
| chr8_-_85526972 | 1.70 |
ENSMUST00000099070.10
|
Nfix
|
nuclear factor I/X |
| chr8_-_71060911 | 1.67 |
ENSMUST00000210580.2
ENSMUST00000211608.2 ENSMUST00000049908.11 |
Ssbp4
|
single stranded DNA binding protein 4 |
| chr10_+_79752797 | 1.62 |
ENSMUST00000045529.3
|
Kiss1r
|
KISS1 receptor |
| chr3_+_90444613 | 1.61 |
ENSMUST00000107335.2
|
S100a16
|
S100 calcium binding protein A16 |
| chr7_-_62069887 | 1.58 |
ENSMUST00000094340.4
|
Mkrn3
|
makorin, ring finger protein, 3 |
| chrX_-_47123719 | 1.57 |
ENSMUST00000039026.8
|
Apln
|
apelin |
| chr10_-_127047396 | 1.55 |
ENSMUST00000013970.9
|
Pip4k2c
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma |
| chr3_-_90340830 | 1.54 |
ENSMUST00000029542.12
|
Ints3
|
integrator complex subunit 3 |
| chr3_+_116306719 | 1.54 |
ENSMUST00000000349.11
ENSMUST00000197201.5 |
Dbt
|
dihydrolipoamide branched chain transacylase E2 |
| chr9_-_79700660 | 1.51 |
ENSMUST00000034878.12
|
Tmem30a
|
transmembrane protein 30A |
| chr6_-_39534765 | 1.51 |
ENSMUST00000036877.10
ENSMUST00000154149.2 |
Dennd2a
|
DENN/MADD domain containing 2A |
| chr7_+_126295114 | 1.49 |
ENSMUST00000106369.2
|
Bola2
|
bolA-like 2 (E. coli) |
| chr12_+_4284009 | 1.49 |
ENSMUST00000179139.3
|
Ptrhd1
|
peptidyl-tRNA hydrolase domain containing 1 |
| chr11_-_76070325 | 1.48 |
ENSMUST00000167114.8
ENSMUST00000094015.11 ENSMUST00000108419.9 ENSMUST00000170730.3 ENSMUST00000129256.2 ENSMUST00000056601.11 |
Vps53
|
VPS53 GARP complex subunit |
| chr5_-_122752508 | 1.47 |
ENSMUST00000127220.8
ENSMUST00000031426.14 |
Ift81
|
intraflagellar transport 81 |
| chr9_+_57847387 | 1.46 |
ENSMUST00000043059.9
|
Sema7a
|
sema domain, immunoglobulin domain (Ig), and GPI membrane anchor, (semaphorin) 7A |
| chr4_+_150321659 | 1.45 |
ENSMUST00000133839.8
|
Eno1
|
enolase 1, alpha non-neuron |
| chr19_-_46327071 | 1.45 |
ENSMUST00000235977.2
ENSMUST00000167861.8 ENSMUST00000051234.9 ENSMUST00000236066.2 |
Cuedc2
|
CUE domain containing 2 |
| chr19_-_46327024 | 1.45 |
ENSMUST00000236046.2
ENSMUST00000236980.2 ENSMUST00000235485.2 ENSMUST00000236061.2 ENSMUST00000236236.2 ENSMUST00000236768.2 ENSMUST00000236651.2 |
Cuedc2
|
CUE domain containing 2 |
| chr11_-_76969230 | 1.34 |
ENSMUST00000102494.8
|
Nsrp1
|
nuclear speckle regulatory protein 1 |
| chr7_+_142623241 | 1.33 |
ENSMUST00000137856.2
|
Tssc4
|
tumor-suppressing subchromosomal transferable fragment 4 |
| chr8_+_61446221 | 1.32 |
ENSMUST00000120689.8
ENSMUST00000034065.14 ENSMUST00000211256.2 ENSMUST00000211672.2 |
Nek1
|
NIMA (never in mitosis gene a)-related expressed kinase 1 |
| chr15_-_83054698 | 1.31 |
ENSMUST00000162178.8
|
Cyb5r3
|
cytochrome b5 reductase 3 |
| chr1_-_60605867 | 1.31 |
ENSMUST00000027168.12
ENSMUST00000090293.11 ENSMUST00000140485.8 |
Raph1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr4_+_150321272 | 1.31 |
ENSMUST00000080926.13
|
Eno1
|
enolase 1, alpha non-neuron |
| chr7_-_139616234 | 1.31 |
ENSMUST00000209574.2
|
Tubgcp2
|
tubulin, gamma complex associated protein 2 |
| chr11_-_116197994 | 1.29 |
ENSMUST00000124281.2
|
Exoc7
|
exocyst complex component 7 |
| chr12_+_78273356 | 1.27 |
ENSMUST00000110388.10
|
Gphn
|
gephyrin |
| chr7_-_127475949 | 1.25 |
ENSMUST00000106262.2
ENSMUST00000106263.8 ENSMUST00000054415.12 |
Zfp668
|
zinc finger protein 668 |
| chr2_+_163444248 | 1.23 |
ENSMUST00000152135.8
|
Ttpal
|
tocopherol (alpha) transfer protein-like |
| chr10_-_79752746 | 1.21 |
ENSMUST00000218750.2
ENSMUST00000045628.15 ENSMUST00000171416.2 |
R3hdm4
|
R3H domain containing 4 |
| chr18_-_24736521 | 1.20 |
ENSMUST00000154205.2
|
Slc39a6
|
solute carrier family 39 (metal ion transporter), member 6 |
| chr2_+_179684288 | 1.19 |
ENSMUST00000041126.9
|
Ss18l1
|
SS18, nBAF chromatin remodeling complex subunit like 1 |
| chr15_+_6416229 | 1.18 |
ENSMUST00000110664.9
ENSMUST00000110663.9 ENSMUST00000161812.8 ENSMUST00000160134.8 |
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr7_-_139616167 | 1.17 |
ENSMUST00000026547.9
|
Tubgcp2
|
tubulin, gamma complex associated protein 2 |
| chr8_+_70686836 | 1.16 |
ENSMUST00000164403.8
ENSMUST00000093458.11 |
Sugp2
|
SURP and G patch domain containing 2 |
| chr4_+_118266582 | 1.15 |
ENSMUST00000144577.2
|
Med8
|
mediator complex subunit 8 |
| chr17_+_78507669 | 1.15 |
ENSMUST00000112498.3
|
Crim1
|
cysteine rich transmembrane BMP regulator 1 (chordin like) |
| chr7_+_65343156 | 1.14 |
ENSMUST00000032726.14
ENSMUST00000107495.5 ENSMUST00000143508.3 ENSMUST00000129166.3 ENSMUST00000206517.2 ENSMUST00000206837.2 ENSMUST00000206628.2 ENSMUST00000206361.2 |
Tm2d3
|
TM2 domain containing 3 |
| chr12_+_78273144 | 1.12 |
ENSMUST00000052472.6
|
Gphn
|
gephyrin |
| chr11_-_116226175 | 1.11 |
ENSMUST00000036215.8
|
Foxj1
|
forkhead box J1 |
| chr6_+_146789978 | 1.10 |
ENSMUST00000016631.14
ENSMUST00000203730.3 ENSMUST00000111623.9 |
Ppfibp1
|
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
| chr2_+_126850613 | 1.09 |
ENSMUST00000110394.8
ENSMUST00000002063.15 |
Ap4e1
|
adaptor-related protein complex AP-4, epsilon 1 |
| chr10_+_62088104 | 1.07 |
ENSMUST00000020278.6
|
Tacr2
|
tachykinin receptor 2 |
| chr7_+_142622986 | 1.06 |
ENSMUST00000060433.10
ENSMUST00000133410.3 ENSMUST00000105920.8 ENSMUST00000177841.8 ENSMUST00000147995.2 |
Tssc4
|
tumor-suppressing subchromosomal transferable fragment 4 |
| chr6_-_113508536 | 1.06 |
ENSMUST00000032425.7
|
Emc3
|
ER membrane protein complex subunit 3 |
| chr7_-_126294902 | 1.04 |
ENSMUST00000144897.2
|
Slx1b
|
SLX1 structure-specific endonuclease subunit homolog B (S. cerevisiae) |
| chr11_-_116197523 | 1.02 |
ENSMUST00000133468.2
ENSMUST00000106411.10 ENSMUST00000106413.10 ENSMUST00000021147.14 |
Exoc7
|
exocyst complex component 7 |
| chr13_-_55510595 | 1.02 |
ENSMUST00000021940.8
|
Lman2
|
lectin, mannose-binding 2 |
| chr9_+_20914211 | 1.01 |
ENSMUST00000214124.2
ENSMUST00000216818.2 |
Mrpl4
|
mitochondrial ribosomal protein L4 |
| chr3_-_107992662 | 1.01 |
ENSMUST00000078912.7
|
Ampd2
|
adenosine monophosphate deaminase 2 |
| chrX_-_48877080 | 1.00 |
ENSMUST00000114893.8
|
Igsf1
|
immunoglobulin superfamily, member 1 |
| chr4_-_116413092 | 1.00 |
ENSMUST00000069674.6
ENSMUST00000106478.9 |
Tmem69
|
transmembrane protein 69 |
| chr2_-_90900525 | 0.99 |
ENSMUST00000153367.2
ENSMUST00000079976.10 |
Slc39a13
|
solute carrier family 39 (metal ion transporter), member 13 |
| chr16_+_18066730 | 0.99 |
ENSMUST00000115640.8
ENSMUST00000140206.8 |
Trmt2a
|
TRM2 tRNA methyltransferase 2A |
| chr4_+_148123554 | 0.98 |
ENSMUST00000141283.8
|
Mthfr
|
methylenetetrahydrofolate reductase |
| chr19_+_47167444 | 0.98 |
ENSMUST00000235326.2
|
Neurl1a
|
neuralized E3 ubiquitin protein ligase 1A |
| chr9_-_110237276 | 0.96 |
ENSMUST00000040021.12
|
Ptpn23
|
protein tyrosine phosphatase, non-receptor type 23 |
| chr3_-_146518706 | 0.96 |
ENSMUST00000102515.10
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr11_-_116197478 | 0.95 |
ENSMUST00000126731.8
|
Exoc7
|
exocyst complex component 7 |
| chr6_+_65648574 | 0.93 |
ENSMUST00000054351.6
|
Ndnf
|
neuron-derived neurotrophic factor |
| chr1_+_37469220 | 0.93 |
ENSMUST00000114925.10
ENSMUST00000027285.13 ENSMUST00000144617.8 ENSMUST00000193979.6 ENSMUST00000118059.3 ENSMUST00000193713.2 |
Unc50
|
unc-50 homolog |
| chr16_+_18066825 | 0.92 |
ENSMUST00000100099.10
|
Trmt2a
|
TRM2 tRNA methyltransferase 2A |
| chr8_+_106877025 | 0.92 |
ENSMUST00000212963.2
ENSMUST00000034377.8 |
Pla2g15
|
phospholipase A2, group XV |
| chr18_-_24736848 | 0.92 |
ENSMUST00000070726.10
|
Slc39a6
|
solute carrier family 39 (metal ion transporter), member 6 |
| chr9_+_55116209 | 0.92 |
ENSMUST00000034859.15
|
Fbxo22
|
F-box protein 22 |
| chr10_+_122514669 | 0.90 |
ENSMUST00000161487.8
ENSMUST00000067918.12 |
Ppm1h
|
protein phosphatase 1H (PP2C domain containing) |
| chr4_+_118266526 | 0.90 |
ENSMUST00000084319.11
ENSMUST00000106384.10 ENSMUST00000126089.8 ENSMUST00000073881.8 ENSMUST00000019229.15 |
Med8
|
mediator complex subunit 8 |
| chr12_-_44257109 | 0.89 |
ENSMUST00000015049.5
|
Dnajb9
|
DnaJ heat shock protein family (Hsp40) member B9 |
| chr9_+_21914513 | 0.88 |
ENSMUST00000215795.2
|
Prkcsh
|
protein kinase C substrate 80K-H |
| chr2_+_166857113 | 0.87 |
ENSMUST00000018143.16
ENSMUST00000176066.8 ENSMUST00000150571.2 |
Ddx27
|
DEAD box helicase 27 |
| chr3_-_63872189 | 0.87 |
ENSMUST00000029402.15
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr10_+_59057767 | 0.87 |
ENSMUST00000182161.2
|
Sowahc
|
sosondowah ankyrin repeat domain family member C |
| chr15_-_78088423 | 0.86 |
ENSMUST00000005860.16
|
Pvalb
|
parvalbumin |
| chr7_-_109351399 | 0.85 |
ENSMUST00000128043.2
ENSMUST00000033333.13 |
Tmem9b
|
TMEM9 domain family, member B |
| chr18_-_73887528 | 0.83 |
ENSMUST00000041138.3
|
Elac1
|
elaC ribonuclease Z 1 |
| chr9_+_55116474 | 0.81 |
ENSMUST00000146201.8
|
Fbxo22
|
F-box protein 22 |
| chr9_+_74945101 | 0.80 |
ENSMUST00000167885.8
ENSMUST00000169188.2 |
Arpp19
|
cAMP-regulated phosphoprotein 19 |
| chr15_+_12117899 | 0.79 |
ENSMUST00000122941.8
|
Zfr
|
zinc finger RNA binding protein |
| chr5_+_122529941 | 0.79 |
ENSMUST00000102525.11
|
Arpc3
|
actin related protein 2/3 complex, subunit 3 |
| chr18_-_68433398 | 0.78 |
ENSMUST00000042852.7
|
Fam210a
|
family with sequence similarity 210, member A |
| chr5_-_36987917 | 0.76 |
ENSMUST00000031002.10
|
Man2b2
|
mannosidase 2, alpha B2 |
| chr2_+_29779227 | 0.75 |
ENSMUST00000123335.8
|
Odf2
|
outer dense fiber of sperm tails 2 |
| chr9_+_20914012 | 0.74 |
ENSMUST00000003386.7
|
Mrpl4
|
mitochondrial ribosomal protein L4 |
| chr9_-_121106209 | 0.74 |
ENSMUST00000051479.13
ENSMUST00000171923.8 |
Ulk4
|
unc-51-like kinase 4 |
| chr9_+_105520154 | 0.72 |
ENSMUST00000190358.2
ENSMUST00000191268.7 ENSMUST00000065778.13 ENSMUST00000188784.2 |
Pik3r4
|
phosphoinositide-3-kinase regulatory subunit 4 |
| chr11_-_113456568 | 0.69 |
ENSMUST00000071539.10
ENSMUST00000106633.10 ENSMUST00000042657.16 ENSMUST00000149034.8 |
Slc39a11
|
solute carrier family 39 (metal ion transporter), member 11 |
| chr15_+_6416079 | 0.69 |
ENSMUST00000080880.12
|
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr3_+_126390951 | 0.68 |
ENSMUST00000171289.8
|
Camk2d
|
calcium/calmodulin-dependent protein kinase II, delta |
| chr2_-_120439826 | 0.68 |
ENSMUST00000102497.10
|
Lrrc57
|
leucine rich repeat containing 57 |
| chr3_+_108444837 | 0.67 |
ENSMUST00000029485.6
|
1700013F07Rik
|
RIKEN cDNA 1700013F07 gene |
| chr15_-_83054369 | 0.67 |
ENSMUST00000162834.3
|
Cyb5r3
|
cytochrome b5 reductase 3 |
| chr7_-_131918926 | 0.66 |
ENSMUST00000080215.6
|
Chst15
|
carbohydrate sulfotransferase 15 |
| chr13_-_100867398 | 0.66 |
ENSMUST00000225990.2
ENSMUST00000091299.8 |
Cdk7
|
cyclin-dependent kinase 7 |
| chr1_-_60606237 | 0.65 |
ENSMUST00000142258.3
|
Raph1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr1_+_87501721 | 0.63 |
ENSMUST00000166259.8
ENSMUST00000172222.8 ENSMUST00000163606.8 |
Neu2
|
neuraminidase 2 |
| chr14_+_56905698 | 0.62 |
ENSMUST00000116468.2
|
Mphosph8
|
M-phase phosphoprotein 8 |
| chr11_+_76092833 | 0.62 |
ENSMUST00000094014.10
|
Tlcd3a
|
TLC domain containing 3A |
| chr2_-_120439981 | 0.62 |
ENSMUST00000133612.2
ENSMUST00000102498.8 ENSMUST00000102499.8 |
Lrrc57
|
leucine rich repeat containing 57 |
| chr19_+_8713156 | 0.61 |
ENSMUST00000210512.2
ENSMUST00000049424.11 |
Wdr74
|
WD repeat domain 74 |
| chr16_+_48104098 | 0.60 |
ENSMUST00000096045.9
ENSMUST00000050705.4 |
Dppa4
|
developmental pluripotency associated 4 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 12.4 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 1.6 | 4.9 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 1.6 | 9.3 | GO:1903336 | endosome localization(GO:0032439) negative regulation of vacuolar transport(GO:1903336) |
| 1.2 | 6.9 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 1.0 | 7.3 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 1.0 | 5.8 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 1.0 | 4.8 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.9 | 3.6 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.8 | 2.4 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.8 | 4.0 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.8 | 2.4 | GO:0031554 | regulation of DNA-templated transcription, termination(GO:0031554) |
| 0.8 | 3.8 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.6 | 1.9 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.6 | 2.4 | GO:0007529 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) |
| 0.6 | 2.8 | GO:2000439 | positive regulation of monocyte extravasation(GO:2000439) |
| 0.5 | 2.7 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.5 | 1.6 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.5 | 8.2 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.5 | 7.1 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.5 | 2.0 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.5 | 2.0 | GO:0060912 | cardiac cell fate specification(GO:0060912) |
| 0.5 | 3.0 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.5 | 7.7 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.5 | 1.0 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.5 | 3.8 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.4 | 3.6 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.4 | 3.8 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.4 | 4.2 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.4 | 1.9 | GO:1903181 | regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) |
| 0.4 | 1.1 | GO:0002663 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.4 | 2.9 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.3 | 1.0 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.3 | 4.1 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.3 | 1.9 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.3 | 0.9 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.3 | 2.7 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.3 | 5.4 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.3 | 1.1 | GO:0033685 | negative regulation of luteinizing hormone secretion(GO:0033685) |
| 0.3 | 3.3 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.2 | 1.4 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.2 | 2.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.2 | 4.1 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.2 | 1.5 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.2 | 3.9 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.2 | 3.3 | GO:2000535 | entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.2 | 5.2 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.2 | 1.8 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.2 | 2.3 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.2 | 6.3 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.2 | 1.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 4.5 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.1 | 1.3 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 7.2 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 0.8 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.1 | 3.5 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 3.4 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 1.6 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 | 5.1 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 1.7 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.1 | 0.5 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) |
| 0.1 | 1.5 | GO:0010935 | regulation of macrophage cytokine production(GO:0010935) |
| 0.1 | 2.4 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 1.0 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 5.2 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.1 | 1.0 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 | 10.0 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.9 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.1 | 0.4 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 1.7 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 0.6 | GO:0006689 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 2.0 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.1 | 0.8 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.7 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 5.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 0.4 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.1 | 4.3 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 0.2 | GO:0090309 | C-5 methylation of cytosine(GO:0090116) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 0.4 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 1.6 | GO:1903963 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 2.3 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.3 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.7 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 4.2 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 3.5 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.5 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 2.8 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 6.7 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.0 | 0.7 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 8.8 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 0.6 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 2.0 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.0 | 1.2 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.0 | 1.0 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.3 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.8 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.4 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 2.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:1900110 | negative regulation of histone H3-K9 dimethylation(GO:1900110) |
| 0.0 | 2.0 | GO:0048675 | axon extension(GO:0048675) |
| 0.0 | 3.9 | GO:0030073 | insulin secretion(GO:0030073) |
| 0.0 | 1.0 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.0 | 3.0 | GO:0071229 | cellular response to acid chemical(GO:0071229) |
| 0.0 | 1.7 | GO:0051592 | response to calcium ion(GO:0051592) |
| 0.0 | 0.2 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.7 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.1 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.7 | GO:0019319 | hexose biosynthetic process(GO:0019319) |
| 0.0 | 0.8 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 2.6 | GO:0007034 | vacuolar transport(GO:0007034) |
| 0.0 | 0.2 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 9.3 | GO:0097443 | sorting endosome(GO:0097443) |
| 1.3 | 3.9 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.9 | 4.6 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.7 | 2.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) inhibin A complex(GO:0043512) |
| 0.6 | 2.5 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.6 | 3.4 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.4 | 6.9 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.4 | 5.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.4 | 1.9 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.3 | 7.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.3 | 1.5 | GO:0000938 | GARP complex(GO:0000938) EARP complex(GO:1990745) |
| 0.3 | 2.0 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.3 | 7.4 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.3 | 3.3 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.2 | 0.7 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.2 | 15.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.2 | 12.4 | GO:0030673 | axolemma(GO:0030673) |
| 0.2 | 1.8 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 2.4 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.1 | 0.7 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 2.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 4.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 1.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 0.5 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.1 | 2.3 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 5.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 1.0 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 2.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 3.0 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 1.5 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 11.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.3 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.6 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 2.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.2 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.8 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 1.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 11.1 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 1.1 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 1.6 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.4 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.9 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 1.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 5.3 | GO:0099572 | postsynaptic specialization(GO:0099572) |
| 0.0 | 0.1 | GO:0097574 | lateral part of cell(GO:0097574) basolateral part of cell(GO:1990794) rod bipolar cell terminal bouton(GO:1990795) |
| 0.0 | 1.6 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 8.8 | GO:0000323 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 1.0 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 9.8 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.9 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 6.1 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 1.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 5.9 | GO:0005929 | cilium(GO:0005929) |
| 0.0 | 1.7 | GO:0030027 | lamellipodium(GO:0030027) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 12.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 1.9 | 5.8 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 1.7 | 6.9 | GO:0070905 | serine binding(GO:0070905) |
| 1.0 | 3.8 | GO:0045183 | translation factor activity, non-nucleic acid binding(GO:0045183) |
| 0.9 | 2.7 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.7 | 4.0 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.7 | 2.0 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.6 | 7.1 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.6 | 2.2 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.6 | 7.3 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.6 | 2.8 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.6 | 15.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.5 | 1.6 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.5 | 2.4 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.4 | 3.0 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.4 | 4.5 | GO:0008061 | chitin binding(GO:0008061) |
| 0.4 | 5.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.3 | 6.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.3 | 3.0 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.3 | 3.6 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.3 | 1.9 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.3 | 1.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.2 | 4.1 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.2 | 1.0 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.2 | 3.3 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.2 | 4.1 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.2 | 4.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 7.2 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.2 | 6.1 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.2 | 2.0 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.2 | 0.5 | GO:0016900 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.1 | 3.9 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.1 | 1.9 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 2.4 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 0.6 | GO:0016997 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 4.9 | GO:0004407 | histone deacetylase activity(GO:0004407) protein deacetylase activity(GO:0033558) |
| 0.1 | 6.6 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.1 | 6.3 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 2.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 0.7 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.1 | 2.8 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 7.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 5.5 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 4.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 4.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.0 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.9 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 0.5 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 1.0 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 1.4 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 5.1 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 2.0 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 1.0 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 1.6 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.1 | 1.8 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.3 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.0 | 1.6 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 2.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.8 | GO:0015923 | alpha-mannosidase activity(GO:0004559) mannosidase activity(GO:0015923) |
| 0.0 | 1.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 1.0 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 8.2 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 2.4 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 2.9 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 6.0 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.7 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.4 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.4 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 1.3 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 1.9 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.8 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.0 | 1.9 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.0 | 2.0 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.1 | GO:0047874 | dolichyldiphosphatase activity(GO:0047874) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 2.1 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 1.0 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 2.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 1.0 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 15.4 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.3 | 19.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.2 | 2.0 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 2.0 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 10.0 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 4.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 4.1 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 2.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 1.9 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 5.5 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 2.8 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 3.3 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 3.0 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 5.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.9 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 1.0 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 1.9 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 4.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.1 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 4.0 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.8 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.9 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.4 | 22.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.3 | 9.0 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.2 | 6.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 2.0 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.2 | 6.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 2.5 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 7.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 5.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 2.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 9.9 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 9.8 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.9 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 2.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.9 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 5.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 2.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 2.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 3.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 2.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 7.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.9 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 7.0 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.7 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 4.0 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.0 | 0.7 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 1.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 2.3 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 2.1 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.1 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 2.3 | REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 0.0 | 0.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |