Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
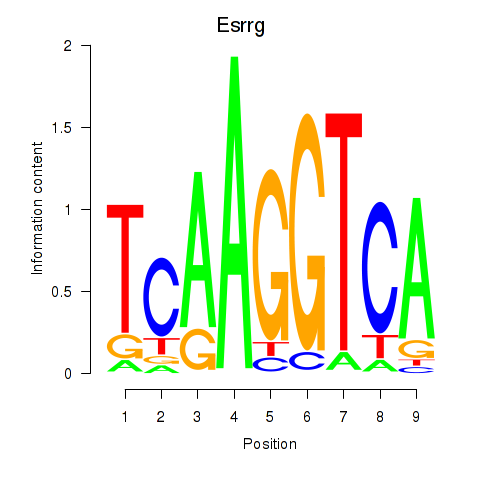
Results for Esrrg
Z-value: 1.17
Transcription factors associated with Esrrg
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Esrrg
|
ENSMUSG00000026610.14 | Esrrg |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Esrrg | mm39_v1_chr1_+_187340952_187340988 | 0.15 | 2.2e-01 | Click! |
Activity profile of Esrrg motif
Sorted Z-values of Esrrg motif
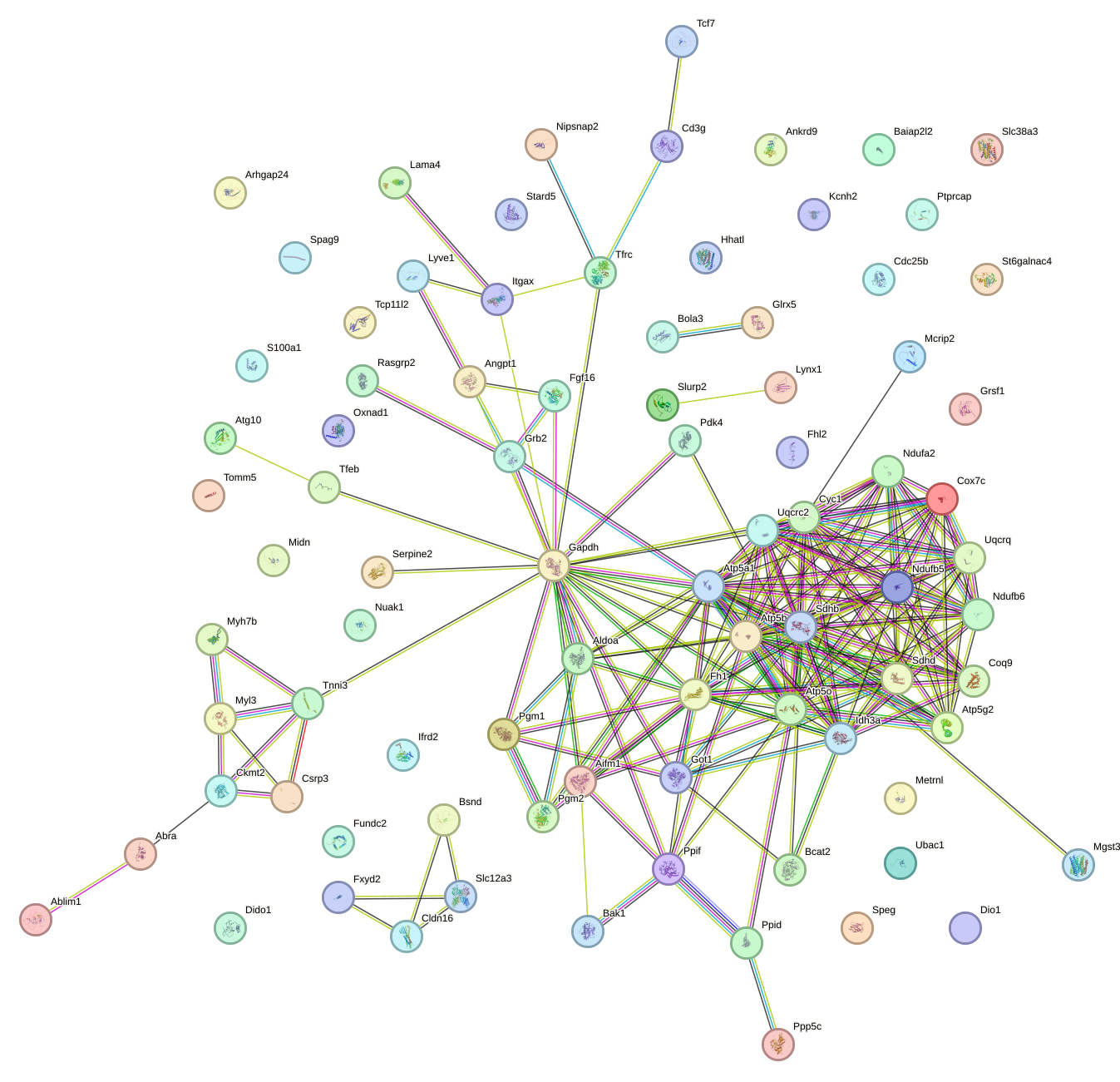
Network of associatons between targets according to the STRING database.
First level regulatory network of Esrrg
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_110595224 | 13.26 |
ENSMUST00000136695.3
|
Myl3
|
myosin, light polypeptide 3 |
| chr5_+_122239007 | 9.21 |
ENSMUST00000014080.13
ENSMUST00000111750.8 |
Myl2
|
myosin, light polypeptide 2, regulatory, cardiac, slow |
| chr15_-_41733099 | 8.33 |
ENSMUST00000054742.7
|
Abra
|
actin-binding Rho activating protein |
| chr5_+_122239030 | 8.20 |
ENSMUST00000139213.8
ENSMUST00000111751.8 ENSMUST00000155612.8 |
Myl2
|
myosin, light polypeptide 2, regulatory, cardiac, slow |
| chr13_-_92024994 | 8.20 |
ENSMUST00000022122.4
|
Ckmt2
|
creatine kinase, mitochondrial 2 |
| chr7_-_48494959 | 7.38 |
ENSMUST00000208050.2
|
Csrp3
|
cysteine and glycine-rich protein 3 |
| chr9_-_44891626 | 6.60 |
ENSMUST00000002101.12
ENSMUST00000160886.2 |
Cd3g
|
CD3 antigen, gamma polypeptide |
| chr6_+_70703409 | 6.21 |
ENSMUST00000103410.3
|
Igkc
|
immunoglobulin kappa constant |
| chr9_-_121621544 | 5.81 |
ENSMUST00000035110.11
|
Hhatl
|
hedgehog acyltransferase-like |
| chr2_-_73741664 | 5.68 |
ENSMUST00000111996.8
ENSMUST00000018914.3 |
Atp5g3
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C3 (subunit 9) |
| chr1_-_167221344 | 5.64 |
ENSMUST00000028005.3
|
Mgst3
|
microsomal glutathione S-transferase 3 |
| chr6_-_5496261 | 5.51 |
ENSMUST00000203347.3
ENSMUST00000019721.7 |
Pdk4
|
pyruvate dehydrogenase kinase, isoenzyme 4 |
| chr15_-_74624811 | 5.40 |
ENSMUST00000189128.2
ENSMUST00000023259.15 |
Lynx1
|
Ly6/neurotoxin 1 |
| chr19_-_43512929 | 5.23 |
ENSMUST00000026196.14
|
Got1
|
glutamic-oxaloacetic transaminase 1, soluble |
| chr14_+_25694594 | 5.19 |
ENSMUST00000022419.7
|
Ppif
|
peptidylprolyl isomerase F (cyclophilin F) |
| chr6_-_125142539 | 5.02 |
ENSMUST00000183272.2
ENSMUST00000182052.8 ENSMUST00000182277.2 |
Gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr7_-_4525426 | 4.94 |
ENSMUST00000209148.2
ENSMUST00000098859.10 |
Tnni3
|
troponin I, cardiac 3 |
| chr19_+_6450553 | 4.94 |
ENSMUST00000146831.8
ENSMUST00000035716.15 ENSMUST00000138555.8 ENSMUST00000167240.8 |
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr9_+_54493618 | 4.93 |
ENSMUST00000217484.2
|
Idh3a
|
isocitrate dehydrogenase 3 (NAD+) alpha |
| chr9_+_54493784 | 4.89 |
ENSMUST00000167866.2
|
Idh3a
|
isocitrate dehydrogenase 3 (NAD+) alpha |
| chr5_+_129802127 | 4.89 |
ENSMUST00000086046.10
ENSMUST00000186265.6 |
Nipsnap2
|
nipsnap homolog 2 |
| chr19_+_6449887 | 4.66 |
ENSMUST00000146601.8
ENSMUST00000150713.8 |
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr10_+_127919142 | 4.61 |
ENSMUST00000026459.6
|
Atp5b
|
ATP synthase, H+ transporting mitochondrial F1 complex, beta subunit |
| chr15_-_42540363 | 4.49 |
ENSMUST00000022921.7
|
Angpt1
|
angiopoietin 1 |
| chr7_-_4525793 | 4.38 |
ENSMUST00000140424.8
|
Tnni3
|
troponin I, cardiac 3 |
| chr4_+_99786611 | 4.24 |
ENSMUST00000058351.16
|
Pgm1
|
phosphoglucomutase 1 |
| chr9_+_45311000 | 4.23 |
ENSMUST00000216289.2
|
Fxyd2
|
FXYD domain-containing ion transport regulator 2 |
| chr2_+_155453103 | 3.99 |
ENSMUST00000092995.6
|
Myh7b
|
myosin, heavy chain 7B, cardiac muscle, beta |
| chr7_-_16761790 | 3.91 |
ENSMUST00000003183.12
|
Ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr7_-_16761732 | 3.78 |
ENSMUST00000142597.2
|
Ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr2_+_32477069 | 3.69 |
ENSMUST00000102818.11
|
St6galnac4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr4_-_131802606 | 3.61 |
ENSMUST00000146021.8
|
Epb41
|
erythrocyte membrane protein band 4.1 |
| chrX_+_156482116 | 3.40 |
ENSMUST00000112521.8
|
Smpx
|
small muscle protein, X-linked |
| chr19_+_6450641 | 3.30 |
ENSMUST00000113467.2
|
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chrX_+_74425990 | 3.29 |
ENSMUST00000033541.5
|
Fundc2
|
FUN14 domain containing 2 |
| chr9_+_37313287 | 3.28 |
ENSMUST00000115048.10
ENSMUST00000115046.9 ENSMUST00000102895.7 ENSMUST00000239486.2 |
Robo4
|
roundabout guidance receptor 4 |
| chr3_-_90421557 | 3.26 |
ENSMUST00000107340.2
ENSMUST00000060738.9 |
S100a1
|
S100 calcium binding protein A1 |
| chr7_-_126398165 | 3.21 |
ENSMUST00000205890.2
ENSMUST00000205336.2 ENSMUST00000087566.11 |
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr16_+_32427738 | 3.13 |
ENSMUST00000023486.15
|
Tfrc
|
transferrin receptor |
| chr19_+_6449776 | 3.10 |
ENSMUST00000113468.8
|
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr9_-_50515089 | 3.06 |
ENSMUST00000000175.6
|
Sdhd
|
succinate dehydrogenase complex, subunit D, integral membrane protein |
| chrX_+_104807868 | 3.01 |
ENSMUST00000033581.4
|
Fgf16
|
fibroblast growth factor 16 |
| chr8_+_95564949 | 2.97 |
ENSMUST00000034234.15
ENSMUST00000159871.4 |
Coq9
|
coenzyme Q9 |
| chr7_+_120234399 | 2.94 |
ENSMUST00000033176.7
ENSMUST00000208400.2 |
Uqcrc2
|
ubiquinol cytochrome c reductase core protein 2 |
| chr12_+_104998895 | 2.90 |
ENSMUST00000223244.2
ENSMUST00000021522.5 |
Glrx5
|
glutaredoxin 5 |
| chr18_+_77861656 | 2.85 |
ENSMUST00000114748.2
|
Atp5a1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1 |
| chr19_-_47079052 | 2.69 |
ENSMUST00000235771.2
ENSMUST00000096014.5 ENSMUST00000236170.2 |
Atp5md
|
ATP synthase membrane subunit DAPIT |
| chr6_-_124698805 | 2.68 |
ENSMUST00000173315.8
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr9_+_37313193 | 2.66 |
ENSMUST00000214185.3
|
Robo4
|
roundabout guidance receptor 4 |
| chr4_+_99812912 | 2.62 |
ENSMUST00000102783.5
|
Pgm1
|
phosphoglucomutase 1 |
| chr15_-_79169671 | 2.56 |
ENSMUST00000170955.2
ENSMUST00000165408.8 |
Baiap2l2
|
BAI1-associated protein 2-like 2 |
| chr3_+_32791139 | 2.55 |
ENSMUST00000127477.8
ENSMUST00000121778.8 ENSMUST00000154257.8 |
Ndufb5
|
NADH:ubiquinone oxidoreductase subunit B5 |
| chr4_-_106349480 | 2.46 |
ENSMUST00000054472.4
|
Bsnd
|
barttin CLCNK type accessory beta subunit |
| chr16_-_91728531 | 2.40 |
ENSMUST00000023677.10
|
Atp5o
|
ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
| chr4_+_140688514 | 2.39 |
ENSMUST00000010007.9
|
Sdhb
|
succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
| chr16_+_32427789 | 2.37 |
ENSMUST00000120680.2
|
Tfrc
|
transferrin receptor |
| chr1_-_175453117 | 2.34 |
ENSMUST00000027810.14
|
Fh1
|
fumarate hydratase 1 |
| chr15_+_76227695 | 2.32 |
ENSMUST00000023210.8
ENSMUST00000231045.2 |
Cyc1
|
cytochrome c-1 |
| chr6_+_83326424 | 2.32 |
ENSMUST00000136501.2
|
Bola3
|
bolA-like 3 (E. coli) |
| chr2_+_131028861 | 2.26 |
ENSMUST00000028804.15
ENSMUST00000079857.9 |
Cdc25b
|
cell division cycle 25B |
| chr19_-_57303328 | 2.25 |
ENSMUST00000111524.8
|
Ablim1
|
actin-binding LIM protein 1 |
| chr16_-_91728162 | 2.25 |
ENSMUST00000139277.8
ENSMUST00000154661.8 |
Atp5o
|
ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
| chr1_-_43235914 | 2.24 |
ENSMUST00000187357.2
|
Fhl2
|
four and a half LIM domains 2 |
| chr17_+_48047955 | 2.13 |
ENSMUST00000086932.10
|
Tfeb
|
transcription factor EB |
| chr13_-_21685588 | 2.13 |
ENSMUST00000044043.3
|
Cox5b-ps
|
cytochrome c oxidase subunit 5B, pseudogene |
| chr12_-_110945415 | 2.12 |
ENSMUST00000135131.2
ENSMUST00000043459.13 ENSMUST00000128353.8 |
Ankrd9
|
ankyrin repeat domain 9 |
| chr10_+_84412490 | 2.11 |
ENSMUST00000020223.8
|
Tcp11l2
|
t-complex 11 (mouse) like 2 |
| chr17_-_26087696 | 2.11 |
ENSMUST00000236479.2
ENSMUST00000235806.2 ENSMUST00000026828.7 |
Mcrip2
|
MAPK regulated corepressor interacting protein 2 |
| chr2_-_25911691 | 2.08 |
ENSMUST00000036509.14
|
Ubac1
|
ubiquitin associated domain containing 1 |
| chr8_+_95055829 | 2.00 |
ENSMUST00000034218.5
ENSMUST00000212134.2 |
Slc12a3
|
solute carrier family 12, member 3 |
| chr14_+_31807760 | 1.99 |
ENSMUST00000170600.8
ENSMUST00000168986.7 ENSMUST00000169649.2 |
Oxnad1
|
oxidoreductase NAD-binding domain containing 1 |
| chr9_+_107468146 | 1.98 |
ENSMUST00000195746.2
|
Ifrd2
|
interferon-related developmental regulator 2 |
| chr10_-_84276454 | 1.98 |
ENSMUST00000020220.15
|
Nuak1
|
NUAK family, SNF1-like kinase, 1 |
| chr9_-_107546166 | 1.97 |
ENSMUST00000177567.8
|
Slc38a3
|
solute carrier family 38, member 3 |
| chr18_-_36877571 | 1.97 |
ENSMUST00000014438.5
|
Ndufa2
|
NADH:ubiquinone oxidoreductase subunit A2 |
| chr4_-_40279382 | 1.96 |
ENSMUST00000108108.3
ENSMUST00000095128.10 |
Ndufb6
|
NADH:ubiquinone oxidoreductase subunit B6 |
| chr15_-_35938155 | 1.96 |
ENSMUST00000156915.3
|
Cox6c
|
cytochrome c oxidase subunit 6C |
| chr15_-_102579463 | 1.95 |
ENSMUST00000185641.7
|
Atp5g2
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C2 (subunit 9) |
| chr12_-_110945376 | 1.93 |
ENSMUST00000142012.2
|
Ankrd9
|
ankyrin repeat domain 9 |
| chr1_-_79838897 | 1.93 |
ENSMUST00000190724.2
|
Serpine2
|
serine (or cysteine) peptidase inhibitor, clade E, member 2 |
| chr11_+_93886906 | 1.91 |
ENSMUST00000041956.14
|
Spag9
|
sperm associated antigen 9 |
| chr11_-_53321606 | 1.91 |
ENSMUST00000061326.5
ENSMUST00000109021.4 |
Uqcrq
|
ubiquinol-cytochrome c reductase, complex III subunit VII |
| chr7_+_83281193 | 1.89 |
ENSMUST00000117410.2
|
Stard5
|
StAR-related lipid transfer (START) domain containing 5 |
| chr17_+_6926452 | 1.88 |
ENSMUST00000097430.10
|
Sytl3
|
synaptotagmin-like 3 |
| chr13_-_86194889 | 1.82 |
ENSMUST00000131011.2
|
Cox7c
|
cytochrome c oxidase subunit 7C |
| chr10_+_38841511 | 1.80 |
ENSMUST00000019992.6
|
Lama4
|
laminin, alpha 4 |
| chr8_+_121395047 | 1.78 |
ENSMUST00000181795.2
|
Cox4i1
|
cytochrome c oxidase subunit 4I1 |
| chr10_-_43416941 | 1.75 |
ENSMUST00000147196.3
|
Mtres1
|
mitochondrial transcription rescue factor 1 |
| chr11_-_52165682 | 1.72 |
ENSMUST00000238914.2
|
Tcf7
|
transcription factor 7, T cell specific |
| chr2_-_25911544 | 1.70 |
ENSMUST00000136750.3
|
Ubac1
|
ubiquitin associated domain containing 1 |
| chr4_-_107164315 | 1.70 |
ENSMUST00000126291.2
ENSMUST00000106748.2 ENSMUST00000129138.2 |
Dio1
|
deiodinase, iodothyronine, type I |
| chr16_-_95260104 | 1.69 |
ENSMUST00000176345.10
ENSMUST00000121809.11 ENSMUST00000233664.2 ENSMUST00000122199.10 |
Erg
|
ETS transcription factor |
| chr9_-_107546195 | 1.64 |
ENSMUST00000192990.6
|
Slc38a3
|
solute carrier family 38, member 3 |
| chr11_+_121593582 | 1.61 |
ENSMUST00000125580.2
|
Metrnl
|
meteorin, glial cell differentiation regulator-like |
| chr5_+_102916637 | 1.58 |
ENSMUST00000112852.8
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr1_+_75377616 | 1.53 |
ENSMUST00000122266.3
|
Speg
|
SPEG complex locus |
| chr17_-_71575584 | 1.52 |
ENSMUST00000233148.2
|
Emilin2
|
elastin microfibril interfacer 2 |
| chr7_+_127728712 | 1.52 |
ENSMUST00000033053.8
ENSMUST00000205460.2 |
Itgax
|
integrin alpha X |
| chr7_-_110462446 | 1.50 |
ENSMUST00000033050.5
|
Lyve1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr19_-_37184692 | 1.50 |
ENSMUST00000132580.8
ENSMUST00000079754.11 ENSMUST00000136286.8 ENSMUST00000126188.8 ENSMUST00000126781.2 |
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr1_+_36730530 | 1.46 |
ENSMUST00000081180.7
ENSMUST00000193210.6 ENSMUST00000195151.6 |
Cox5b
|
cytochrome c oxidase subunit 5B |
| chr5_-_88823472 | 1.44 |
ENSMUST00000113234.8
ENSMUST00000153565.8 |
Grsf1
|
G-rich RNA sequence binding factor 1 |
| chr17_-_27247581 | 1.42 |
ENSMUST00000143158.3
|
Bak1
|
BCL2-antagonist/killer 1 |
| chr12_-_113896002 | 1.42 |
ENSMUST00000103463.3
|
Ighv14-1
|
immunoglobulin heavy variable 14-1 |
| chr10_+_79984097 | 1.39 |
ENSMUST00000099492.10
ENSMUST00000042057.12 |
Midn
|
midnolin |
| chrX_-_47602395 | 1.38 |
ENSMUST00000114945.9
ENSMUST00000037349.8 |
Aifm1
|
apoptosis-inducing factor, mitochondrion-associated 1 |
| chr4_-_45108038 | 1.38 |
ENSMUST00000107809.9
ENSMUST00000107808.3 ENSMUST00000107807.2 ENSMUST00000107810.3 |
Tomm5
|
translocase of outer mitochondrial membrane 5 |
| chr19_+_4203603 | 1.38 |
ENSMUST00000236632.2
|
Ptprcap
|
protein tyrosine phosphatase, receptor type, C polypeptide-associated protein |
| chr16_+_26281885 | 1.37 |
ENSMUST00000161053.8
ENSMUST00000115302.2 |
Cldn16
|
claudin 16 |
| chr11_-_115590133 | 1.34 |
ENSMUST00000106499.8
|
Grb2
|
growth factor receptor bound protein 2 |
| chr11_-_115590318 | 1.32 |
ENSMUST00000106497.8
|
Grb2
|
growth factor receptor bound protein 2 |
| chr7_+_45224524 | 1.31 |
ENSMUST00000210811.2
|
Bcat2
|
branched chain aminotransferase 2, mitochondrial |
| chr8_+_85696396 | 1.30 |
ENSMUST00000109733.8
|
Prdx2
|
peroxiredoxin 2 |
| chr14_-_36690726 | 1.29 |
ENSMUST00000090024.11
|
Ccser2
|
coiled-coil serine rich 2 |
| chr13_-_91372072 | 1.28 |
ENSMUST00000022119.6
|
Atg10
|
autophagy related 10 |
| chr8_+_85696453 | 1.26 |
ENSMUST00000125893.8
|
Prdx2
|
peroxiredoxin 2 |
| chr12_+_85645801 | 1.25 |
ENSMUST00000177587.9
|
Jdp2
|
Jun dimerization protein 2 |
| chr12_+_85646162 | 1.24 |
ENSMUST00000050687.14
|
Jdp2
|
Jun dimerization protein 2 |
| chr18_-_67582191 | 1.23 |
ENSMUST00000025408.10
|
Afg3l2
|
AFG3-like AAA ATPase 2 |
| chr2_-_155771938 | 1.22 |
ENSMUST00000152766.8
ENSMUST00000139232.8 ENSMUST00000109632.8 ENSMUST00000006036.13 ENSMUST00000142655.2 ENSMUST00000159238.2 |
Uqcc1
|
ubiquinol-cytochrome c reductase complex assembly factor 1 |
| chr11_+_76297969 | 1.20 |
ENSMUST00000021203.7
ENSMUST00000152183.2 |
Timm22
|
translocase of inner mitochondrial membrane 22 |
| chr17_+_85265420 | 1.19 |
ENSMUST00000080217.14
ENSMUST00000112304.10 |
Ppm1b
|
protein phosphatase 1B, magnesium dependent, beta isoform |
| chr11_+_4186391 | 1.18 |
ENSMUST00000075221.3
|
Osm
|
oncostatin M |
| chr7_-_105131407 | 1.18 |
ENSMUST00000047040.4
|
Cavin3
|
caveolae associated 3 |
| chr10_-_4338032 | 1.16 |
ENSMUST00000100078.10
|
Zbtb2
|
zinc finger and BTB domain containing 2 |
| chr2_+_140012560 | 1.16 |
ENSMUST00000044825.5
|
Ndufaf5
|
NADH:ubiquinone oxidoreductase complex assembly factor 5 |
| chr16_+_4501934 | 1.15 |
ENSMUST00000060067.12
ENSMUST00000115854.4 ENSMUST00000229529.2 |
Dnaja3
|
DnaJ heat shock protein family (Hsp40) member A3 |
| chr14_-_36690636 | 1.15 |
ENSMUST00000067700.13
|
Ccser2
|
coiled-coil serine rich 2 |
| chr9_+_65536892 | 1.14 |
ENSMUST00000169003.8
|
Rbpms2
|
RNA binding protein with multiple splicing 2 |
| chr19_+_6973511 | 1.12 |
ENSMUST00000088223.7
|
Trpt1
|
tRNA phosphotransferase 1 |
| chr16_-_20245071 | 1.12 |
ENSMUST00000115547.9
ENSMUST00000096199.5 |
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr7_-_4448180 | 1.11 |
ENSMUST00000138798.2
|
Rdh13
|
retinol dehydrogenase 13 (all-trans and 9-cis) |
| chr7_+_110368037 | 1.10 |
ENSMUST00000213373.2
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chrX_-_70536449 | 1.08 |
ENSMUST00000037391.12
ENSMUST00000114586.9 ENSMUST00000114587.3 |
Cd99l2
|
CD99 antigen-like 2 |
| chr4_+_34893772 | 1.06 |
ENSMUST00000029975.10
ENSMUST00000135871.8 ENSMUST00000108130.2 |
Cga
|
glycoprotein hormones, alpha subunit |
| chrX_-_70536198 | 1.06 |
ENSMUST00000080035.11
|
Cd99l2
|
CD99 antigen-like 2 |
| chr5_-_24650164 | 1.05 |
ENSMUST00000115043.8
ENSMUST00000115041.2 ENSMUST00000030800.13 |
Fastk
|
Fas-activated serine/threonine kinase |
| chr8_-_70975734 | 1.04 |
ENSMUST00000137610.3
|
Kxd1
|
KxDL motif containing 1 |
| chr14_+_58313964 | 0.97 |
ENSMUST00000166770.2
|
Fgf9
|
fibroblast growth factor 9 |
| chrX_+_56008685 | 0.97 |
ENSMUST00000096431.10
|
Adgrg4
|
adhesion G protein-coupled receptor G4 |
| chr1_+_93096316 | 0.96 |
ENSMUST00000138595.3
|
Crocc2
|
ciliary rootlet coiled-coil, rootletin family member 2 |
| chr14_+_54429757 | 0.94 |
ENSMUST00000103714.2
|
Traj27
|
T cell receptor alpha joining 27 |
| chr10_+_78410803 | 0.93 |
ENSMUST00000218763.2
ENSMUST00000220430.2 ENSMUST00000218885.2 ENSMUST00000218215.2 ENSMUST00000218271.2 |
Ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr17_-_32639936 | 0.92 |
ENSMUST00000170392.9
ENSMUST00000237165.2 ENSMUST00000235892.2 ENSMUST00000114455.3 |
Pglyrp2
|
peptidoglycan recognition protein 2 |
| chr7_-_4448631 | 0.88 |
ENSMUST00000008579.14
|
Rdh13
|
retinol dehydrogenase 13 (all-trans and 9-cis) |
| chr14_+_58310143 | 0.88 |
ENSMUST00000022545.14
|
Fgf9
|
fibroblast growth factor 9 |
| chr4_+_57821050 | 0.87 |
ENSMUST00000238994.2
|
Pakap
|
paralemmin A kinase anchor protein |
| chr16_-_20245138 | 0.86 |
ENSMUST00000079158.13
|
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr3_+_146205864 | 0.86 |
ENSMUST00000119130.2
|
Gng5
|
guanine nucleotide binding protein (G protein), gamma 5 |
| chr11_-_69451012 | 0.84 |
ENSMUST00000004036.6
|
Efnb3
|
ephrin B3 |
| chr19_-_7218363 | 0.84 |
ENSMUST00000236769.2
|
Naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chr8_-_70975813 | 0.83 |
ENSMUST00000121623.8
ENSMUST00000093456.12 ENSMUST00000118850.8 |
Kxd1
|
KxDL motif containing 1 |
| chr19_-_6973393 | 0.82 |
ENSMUST00000041686.10
ENSMUST00000180765.2 |
Nudt22
|
nudix (nucleoside diphosphate linked moiety X)-type motif 22 |
| chr16_-_8455525 | 0.82 |
ENSMUST00000052505.10
|
Tmem186
|
transmembrane protein 186 |
| chr8_+_85696695 | 0.82 |
ENSMUST00000164807.2
|
Prdx2
|
peroxiredoxin 2 |
| chr14_+_79086665 | 0.81 |
ENSMUST00000227255.2
|
Vwa8
|
von Willebrand factor A domain containing 8 |
| chr4_+_138161958 | 0.81 |
ENSMUST00000044058.11
ENSMUST00000105813.8 ENSMUST00000105815.2 |
Mul1
|
mitochondrial ubiquitin ligase activator of NFKB 1 |
| chr17_+_47696329 | 0.81 |
ENSMUST00000145462.2
|
Guca1b
|
guanylate cyclase activator 1B |
| chr17_+_47696312 | 0.80 |
ENSMUST00000024774.14
|
Guca1b
|
guanylate cyclase activator 1B |
| chr7_+_75879603 | 0.79 |
ENSMUST00000156166.8
|
Agbl1
|
ATP/GTP binding protein-like 1 |
| chr2_-_102903680 | 0.76 |
ENSMUST00000132449.8
ENSMUST00000111183.2 ENSMUST00000011058.9 |
Pdhx
|
pyruvate dehydrogenase complex, component X |
| chr9_+_108367801 | 0.75 |
ENSMUST00000006854.13
|
Usp19
|
ubiquitin specific peptidase 19 |
| chr15_-_76083575 | 0.74 |
ENSMUST00000169438.8
|
Plec
|
plectin |
| chr19_+_37184927 | 0.74 |
ENSMUST00000024078.15
ENSMUST00000112391.8 |
Marchf5
|
membrane associated ring-CH-type finger 5 |
| chr7_-_101899294 | 0.73 |
ENSMUST00000106923.2
ENSMUST00000098230.11 |
Rhog
|
ras homolog family member G |
| chr4_-_107164347 | 0.73 |
ENSMUST00000082426.11
|
Dio1
|
deiodinase, iodothyronine, type I |
| chr1_+_135727571 | 0.73 |
ENSMUST00000148201.8
|
Tnni1
|
troponin I, skeletal, slow 1 |
| chr8_-_84771610 | 0.72 |
ENSMUST00000061923.5
|
Rln3
|
relaxin 3 |
| chr8_-_71315902 | 0.70 |
ENSMUST00000212611.2
|
Kcnn1
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1 |
| chr17_-_33879224 | 0.69 |
ENSMUST00000130946.8
|
Hnrnpm
|
heterogeneous nuclear ribonucleoprotein M |
| chrX_-_7884688 | 0.69 |
ENSMUST00000033503.3
|
Glod5
|
glyoxalase domain containing 5 |
| chr4_-_82423511 | 0.67 |
ENSMUST00000050872.15
ENSMUST00000064770.9 |
Nfib
|
nuclear factor I/B |
| chr8_-_13544478 | 0.66 |
ENSMUST00000033828.7
|
Gas6
|
growth arrest specific 6 |
| chr14_-_31807552 | 0.63 |
ENSMUST00000022461.11
ENSMUST00000067955.12 ENSMUST00000124303.9 |
Dph3
|
diphthamine biosynthesis 3 |
| chr2_+_158636727 | 0.63 |
ENSMUST00000029186.14
ENSMUST00000109478.9 ENSMUST00000156893.2 |
Dhx35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr14_+_79086492 | 0.62 |
ENSMUST00000040990.7
|
Vwa8
|
von Willebrand factor A domain containing 8 |
| chr10_-_127016448 | 0.61 |
ENSMUST00000222911.3
ENSMUST00000095270.3 |
Slc26a10
|
solute carrier family 26, member 10 |
| chr1_+_36510670 | 0.61 |
ENSMUST00000153128.2
|
Cnnm4
|
cyclin M4 |
| chr11_+_93935156 | 0.61 |
ENSMUST00000024979.15
|
Spag9
|
sperm associated antigen 9 |
| chr1_+_6805048 | 0.58 |
ENSMUST00000139838.8
|
St18
|
suppression of tumorigenicity 18 |
| chr11_+_70655035 | 0.55 |
ENSMUST00000060444.6
|
Zfp3
|
zinc finger protein 3 |
| chrX_+_159551171 | 0.55 |
ENSMUST00000112368.3
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr11_-_94568228 | 0.53 |
ENSMUST00000116349.9
|
Xylt2
|
xylosyltransferase II |
| chr13_-_8994336 | 0.51 |
ENSMUST00000021573.6
|
Gm9745
|
predicted gene 9745 |
| chr1_-_16589425 | 0.51 |
ENSMUST00000159558.8
ENSMUST00000054668.13 ENSMUST00000162627.8 ENSMUST00000162007.8 ENSMUST00000128957.9 ENSMUST00000115359.10 ENSMUST00000151888.8 |
Stau2
|
staufen double-stranded RNA binding protein 2 |
| chr11_-_79418500 | 0.48 |
ENSMUST00000154415.2
|
Evi2a
|
ecotropic viral integration site 2a |
| chrX_-_104918911 | 0.47 |
ENSMUST00000200471.2
|
Atrx
|
ATRX, chromatin remodeler |
| chr2_-_77000878 | 0.47 |
ENSMUST00000111833.3
|
Ccdc141
|
coiled-coil domain containing 141 |
| chr4_-_19708910 | 0.45 |
ENSMUST00000108246.9
|
Wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr7_-_100232276 | 0.45 |
ENSMUST00000152876.3
ENSMUST00000150042.8 ENSMUST00000132888.9 |
Mrpl48
|
mitochondrial ribosomal protein L48 |
| chr6_+_15727798 | 0.45 |
ENSMUST00000128849.3
|
Mdfic
|
MyoD family inhibitor domain containing |
| chr3_+_68776884 | 0.44 |
ENSMUST00000054551.3
|
1110032F04Rik
|
RIKEN cDNA 1110032F04 gene |
| chr6_-_71800788 | 0.44 |
ENSMUST00000065103.4
|
Mrpl35
|
mitochondrial ribosomal protein L35 |
| chr9_-_121686601 | 0.43 |
ENSMUST00000213124.2
ENSMUST00000215300.2 ENSMUST00000213147.2 |
Higd1a
|
HIG1 domain family, member 1A |
| chr14_-_36641470 | 0.43 |
ENSMUST00000182042.2
|
Ccser2
|
coiled-coil serine rich 2 |
| chr2_+_74656145 | 0.43 |
ENSMUST00000028511.8
|
Mtx2
|
metaxin 2 |
| chrX_-_104919201 | 0.43 |
ENSMUST00000198209.2
|
Atrx
|
ATRX, chromatin remodeler |
| chr7_+_28050077 | 0.42 |
ENSMUST00000082134.6
|
Rps16
|
ribosomal protein S16 |
| chr17_-_35978438 | 0.41 |
ENSMUST00000043674.15
|
Vars2
|
valyl-tRNA synthetase 2, mitochondrial |
| chr3_-_32791296 | 0.40 |
ENSMUST00000043966.8
|
Mrpl47
|
mitochondrial ribosomal protein L47 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 17.4 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 2.5 | 7.6 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 2.5 | 7.4 | GO:1903920 | positive regulation of actin filament severing(GO:1903920) |
| 1.9 | 5.8 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 1.7 | 5.0 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 1.4 | 8.4 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 1.4 | 5.5 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 1.3 | 7.8 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 1.3 | 5.0 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 1.2 | 4.9 | GO:2000983 | regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 1.2 | 3.6 | GO:2000487 | asparagine transport(GO:0006867) positive regulation of glutamine transport(GO:2000487) |
| 1.1 | 9.8 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 1.0 | 5.2 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.7 | 1.4 | GO:0002352 | B cell negative selection(GO:0002352) |
| 0.7 | 4.6 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.6 | 1.9 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.6 | 3.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.6 | 3.0 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) |
| 0.6 | 2.2 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.5 | 2.1 | GO:0009816 | defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.5 | 7.7 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.4 | 1.3 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.4 | 2.9 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.4 | 10.5 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.4 | 1.2 | GO:0030961 | peptidyl-arginine hydroxylation(GO:0030961) |
| 0.3 | 5.5 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.3 | 2.7 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.3 | 2.9 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.3 | 2.5 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.3 | 1.2 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.3 | 1.2 | GO:1901003 | negative regulation of fermentation(GO:1901003) |
| 0.3 | 1.7 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.3 | 3.4 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 0.3 | 1.9 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.3 | 2.3 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.2 | 2.0 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.2 | 1.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.2 | 3.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 3.3 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.2 | 11.6 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.2 | 3.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.2 | 3.6 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.2 | 1.1 | GO:0060331 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.2 | 2.0 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.2 | 1.6 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
| 0.2 | 7.9 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.2 | 16.0 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.2 | 1.4 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.2 | 1.7 | GO:0033153 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.2 | 1.2 | GO:0006498 | N-terminal protein lipidation(GO:0006498) N-terminal protein myristoylation(GO:0006499) |
| 0.2 | 1.2 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.2 | 0.7 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.2 | 8.3 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.2 | 0.8 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.2 | 0.8 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.2 | 1.6 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.2 | 0.9 | GO:0032827 | negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.2 | 0.9 | GO:0097394 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.1 | 2.4 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.1 | 2.5 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) |
| 0.1 | 0.9 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 | 2.7 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.1 | 3.0 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 0.5 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.1 | 0.5 | GO:0098963 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.1 | 1.3 | GO:0006983 | ER overload response(GO:0006983) |
| 0.1 | 1.2 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.1 | 1.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 1.9 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.1 | 1.6 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.1 | 1.5 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.1 | 0.2 | GO:0032278 | positive regulation of gonadotropin secretion(GO:0032278) positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.1 | 0.4 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.1 | 0.8 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.1 | 0.3 | GO:0060450 | positive regulation of hindgut contraction(GO:0060450) |
| 0.1 | 0.8 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.1 | 5.4 | GO:0099601 | regulation of neurotransmitter receptor activity(GO:0099601) |
| 0.1 | 2.6 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 1.1 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.1 | 1.1 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 1.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 6.6 | GO:0070228 | regulation of lymphocyte apoptotic process(GO:0070228) |
| 0.1 | 0.2 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.1 | 0.4 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.1 | 2.5 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.1 | 1.1 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 | 1.4 | GO:0070633 | transepithelial transport(GO:0070633) |
| 0.1 | 0.4 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.1 | 0.3 | GO:0019046 | release from viral latency(GO:0019046) regulation of DNA strand elongation(GO:0060382) |
| 0.1 | 3.7 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 0.3 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.1 | 0.5 | GO:0009240 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.1 | 0.2 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.1 | 1.1 | GO:0070508 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.1 | 7.3 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 4.2 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.6 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 | 2.5 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.2 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.7 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.0 | 0.6 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 1.8 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.6 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.8 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 1.5 | GO:0050798 | activated T cell proliferation(GO:0050798) |
| 0.0 | 0.1 | GO:1900247 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.0 | 0.3 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 2.8 | GO:0061726 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.7 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.0 | 2.4 | GO:0042446 | hormone biosynthetic process(GO:0042446) |
| 0.0 | 0.3 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.4 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.0 | 0.3 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.3 | GO:0090201 | negative regulation of release of cytochrome c from mitochondria(GO:0090201) |
| 0.0 | 4.3 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.0 | 2.2 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.2 | GO:0097066 | response to thyroid hormone(GO:0097066) |
| 0.0 | 1.5 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 4.6 | GO:0006575 | cellular modified amino acid metabolic process(GO:0006575) |
| 0.0 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.2 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.0 | 1.6 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 4.0 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 0.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.4 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.9 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 1.4 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.6 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.6 | GO:0021799 | cerebral cortex radially oriented cell migration(GO:0021799) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 30.7 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 1.4 | 5.4 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 1.3 | 5.2 | GO:0005757 | mitochondrial permeability transition pore complex(GO:0005757) |
| 0.8 | 5.0 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.8 | 7.5 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.8 | 9.3 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.8 | 12.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.7 | 7.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.5 | 7.7 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.4 | 3.4 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.4 | 5.5 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.4 | 1.2 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.3 | 1.4 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.3 | 1.2 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.3 | 5.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.3 | 2.7 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.3 | 18.8 | GO:0031672 | A band(GO:0031672) |
| 0.2 | 1.9 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.2 | 0.9 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.2 | 2.6 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.2 | 1.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.2 | 5.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.2 | 0.9 | GO:1990421 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 0.1 | 2.3 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.1 | 6.5 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 0.4 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.1 | 3.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 0.8 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 0.3 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.1 | 1.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 1.9 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 0.2 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.1 | 0.7 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 7.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 1.8 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 0.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 3.6 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 1.8 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 1.5 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 8.8 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 7.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 14.0 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 2.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 4.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 5.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 4.4 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 6.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 2.3 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 2.1 | GO:0001650 | fibrillar center(GO:0001650) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 9.8 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 1.9 | 7.5 | GO:0043532 | angiostatin binding(GO:0043532) |
| 1.8 | 5.4 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 1.7 | 5.2 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 1.4 | 5.5 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 1.3 | 9.3 | GO:0030172 | troponin C binding(GO:0030172) |
| 1.3 | 5.0 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 1.1 | 6.9 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 1.1 | 5.5 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 1.1 | 7.4 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.9 | 3.6 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.8 | 8.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.7 | 5.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.7 | 12.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.6 | 2.4 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.6 | 29.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.5 | 5.4 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.5 | 1.6 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.4 | 1.2 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.4 | 3.7 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.3 | 3.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.3 | 1.4 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.3 | 4.5 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.2 | 7.0 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 1.9 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.2 | 3.4 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.2 | 2.7 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.2 | 0.5 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.2 | 2.7 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.2 | 5.6 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.2 | 1.3 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.2 | 0.9 | GO:0016019 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.1 | 3.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 2.0 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.1 | 5.2 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.1 | 2.5 | GO:0048273 | MAP-kinase scaffold activity(GO:0005078) mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.1 | 7.9 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 0.9 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 4.9 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.8 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 1.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 1.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 0.5 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.1 | 2.5 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.1 | 1.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 2.8 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.8 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.1 | 0.9 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 2.0 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.2 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.1 | 0.4 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.1 | 7.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.5 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 1.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 4.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 1.4 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.3 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.2 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.7 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.7 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.6 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 1.7 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 4.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 2.3 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.4 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 1.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 9.6 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 2.3 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.8 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.3 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 2.3 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 1.2 | GO:0030145 | protein serine/threonine phosphatase activity(GO:0004722) manganese ion binding(GO:0030145) |
| 0.0 | 10.3 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.4 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 1.0 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 2.7 | GO:0001047 | core promoter binding(GO:0001047) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 17.4 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.3 | 18.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 2.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 6.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 15.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 3.6 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 4.5 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 8.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 2.7 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.1 | 2.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 1.8 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 1.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 2.5 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 1.6 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.2 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 5.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 2.5 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.8 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 1.4 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.9 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.7 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.4 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.4 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.8 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 3.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 2.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 40.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.6 | 15.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.6 | 6.6 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.6 | 12.1 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.5 | 6.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.4 | 16.0 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.3 | 6.9 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.3 | 5.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.3 | 20.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 7.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.2 | 2.7 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.2 | 3.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 3.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 5.5 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.1 | 5.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 2.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 5.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.8 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 1.9 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 2.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 2.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 3.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 1.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.1 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 1.4 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 1.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 5.6 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.7 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.9 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.5 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 2.2 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.9 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |