Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
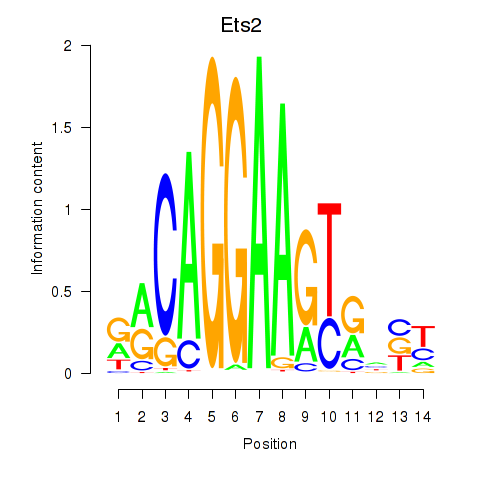
Results for Ets2
Z-value: 1.15
Transcription factors associated with Ets2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ets2
|
ENSMUSG00000022895.17 | Ets2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ets2 | mm39_v1_chr16_+_95502911_95502960 | -0.43 | 1.7e-04 | Click! |
Activity profile of Ets2 motif
Sorted Z-values of Ets2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Ets2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_69983459 | 11.49 |
ENSMUST00000102572.8
|
Asgr2
|
asialoglycoprotein receptor 2 |
| chr11_-_48707763 | 10.85 |
ENSMUST00000140800.2
|
Trim41
|
tripartite motif-containing 41 |
| chr11_+_69983479 | 10.20 |
ENSMUST00000143772.8
|
Asgr2
|
asialoglycoprotein receptor 2 |
| chr11_+_69983531 | 9.98 |
ENSMUST00000124721.2
|
Asgr2
|
asialoglycoprotein receptor 2 |
| chr11_-_48708159 | 9.86 |
ENSMUST00000047145.14
|
Trim41
|
tripartite motif-containing 41 |
| chrX_+_169106356 | 9.33 |
ENSMUST00000178693.4
|
Asmt
|
acetylserotonin O-methyltransferase |
| chr12_-_28673311 | 8.05 |
ENSMUST00000036136.9
|
Colec11
|
collectin sub-family member 11 |
| chr10_+_128104525 | 7.91 |
ENSMUST00000050901.5
|
Apof
|
apolipoprotein F |
| chr12_-_103925197 | 7.66 |
ENSMUST00000122229.8
|
Serpina1e
|
serine (or cysteine) peptidase inhibitor, clade A, member 1E |
| chr12_-_28673259 | 7.18 |
ENSMUST00000220836.2
|
Colec11
|
collectin sub-family member 11 |
| chr12_-_103706774 | 6.93 |
ENSMUST00000186166.7
|
Serpina1b
|
serine (or cysteine) preptidase inhibitor, clade A, member 1B |
| chr8_+_46080746 | 6.28 |
ENSMUST00000145458.9
ENSMUST00000134321.8 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr12_+_87204374 | 5.74 |
ENSMUST00000222222.2
|
Gstz1
|
glutathione transferase zeta 1 (maleylacetoacetate isomerase) |
| chr17_+_32904629 | 5.57 |
ENSMUST00000008801.7
|
Cyp4f15
|
cytochrome P450, family 4, subfamily f, polypeptide 15 |
| chr17_+_32904601 | 5.49 |
ENSMUST00000168171.8
|
Cyp4f15
|
cytochrome P450, family 4, subfamily f, polypeptide 15 |
| chr9_-_103097022 | 5.22 |
ENSMUST00000168142.8
|
Trf
|
transferrin |
| chr10_+_87697155 | 4.96 |
ENSMUST00000122100.3
|
Igf1
|
insulin-like growth factor 1 |
| chr10_+_87696339 | 4.72 |
ENSMUST00000121161.8
|
Igf1
|
insulin-like growth factor 1 |
| chr10_-_95159933 | 4.70 |
ENSMUST00000053594.7
|
Cradd
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr2_-_25390625 | 4.61 |
ENSMUST00000040042.11
|
C8g
|
complement component 8, gamma polypeptide |
| chr1_-_121255448 | 4.54 |
ENSMUST00000186915.2
ENSMUST00000160968.8 ENSMUST00000162582.2 |
Insig2
|
insulin induced gene 2 |
| chr11_-_109613040 | 4.46 |
ENSMUST00000020938.8
|
Fam20a
|
FAM20A, golgi associated secretory pathway pseudokinase |
| chr10_+_21253190 | 4.40 |
ENSMUST00000042699.14
|
Aldh8a1
|
aldehyde dehydrogenase 8 family, member A1 |
| chr10_-_59057570 | 4.38 |
ENSMUST00000220156.2
ENSMUST00000165971.3 |
Septin10
|
septin 10 |
| chr1_-_121255400 | 4.33 |
ENSMUST00000159085.8
ENSMUST00000159125.2 ENSMUST00000161818.2 |
Insig2
|
insulin induced gene 2 |
| chr1_-_121255503 | 4.30 |
ENSMUST00000160688.2
|
Insig2
|
insulin induced gene 2 |
| chr5_+_111881790 | 4.23 |
ENSMUST00000180627.2
|
Gm26897
|
predicted gene, 26897 |
| chr16_-_38253507 | 4.11 |
ENSMUST00000002926.8
|
Pla1a
|
phospholipase A1 member A |
| chr19_+_18609291 | 4.10 |
ENSMUST00000042392.14
ENSMUST00000237347.2 |
Nmrk1
|
nicotinamide riboside kinase 1 |
| chr1_+_63312420 | 3.94 |
ENSMUST00000239483.2
ENSMUST00000114132.8 ENSMUST00000126932.2 |
Zdbf2
|
zinc finger, DBF-type containing 2 |
| chr1_-_121255753 | 3.83 |
ENSMUST00000003818.14
|
Insig2
|
insulin induced gene 2 |
| chr6_+_54249817 | 3.64 |
ENSMUST00000204921.3
ENSMUST00000203091.3 ENSMUST00000204115.3 ENSMUST00000203941.3 ENSMUST00000204746.2 |
Chn2
|
chimerin 2 |
| chr4_+_43562706 | 3.63 |
ENSMUST00000167751.2
ENSMUST00000132631.2 |
Creb3
|
cAMP responsive element binding protein 3 |
| chr3_+_94600863 | 3.52 |
ENSMUST00000090848.10
ENSMUST00000173981.8 ENSMUST00000173849.8 ENSMUST00000174223.2 |
Selenbp2
|
selenium binding protein 2 |
| chr12_+_37292029 | 3.51 |
ENSMUST00000160390.2
|
Agmo
|
alkylglycerol monooxygenase |
| chr5_-_38637624 | 3.51 |
ENSMUST00000067886.12
|
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr8_+_22966736 | 3.40 |
ENSMUST00000067786.9
|
Slc20a2
|
solute carrier family 20, member 2 |
| chr8_+_22966889 | 3.27 |
ENSMUST00000209305.2
|
Slc20a2
|
solute carrier family 20, member 2 |
| chr2_+_65499097 | 3.19 |
ENSMUST00000200829.4
|
Scn2a
|
sodium channel, voltage-gated, type II, alpha |
| chr19_+_25384024 | 3.19 |
ENSMUST00000146647.3
|
Kank1
|
KN motif and ankyrin repeat domains 1 |
| chr9_-_29323032 | 3.16 |
ENSMUST00000115236.2
|
Ntm
|
neurotrimin |
| chr9_+_110306020 | 3.05 |
ENSMUST00000198858.5
|
Kif9
|
kinesin family member 9 |
| chr8_+_110806390 | 3.04 |
ENSMUST00000212754.2
ENSMUST00000058804.10 |
Zfp612
|
zinc finger protein 612 |
| chr4_-_107780716 | 3.00 |
ENSMUST00000106719.8
ENSMUST00000106720.9 ENSMUST00000131644.2 ENSMUST00000030345.15 |
Cpt2
|
carnitine palmitoyltransferase 2 |
| chr7_-_30672747 | 2.98 |
ENSMUST00000205961.2
|
Lsr
|
lipolysis stimulated lipoprotein receptor |
| chr1_+_185187000 | 2.94 |
ENSMUST00000061093.7
|
Slc30a10
|
solute carrier family 30, member 10 |
| chr16_-_38533597 | 2.92 |
ENSMUST00000023487.5
|
Arhgap31
|
Rho GTPase activating protein 31 |
| chr19_+_4036562 | 2.85 |
ENSMUST00000236224.2
ENSMUST00000236510.2 ENSMUST00000237910.2 ENSMUST00000235612.2 ENSMUST00000054030.8 |
Acy3
|
aspartoacylase (aminoacylase) 3 |
| chr3_+_129326004 | 2.83 |
ENSMUST00000199910.5
ENSMUST00000197070.5 ENSMUST00000071402.7 |
Elovl6
|
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
| chr9_+_110306052 | 2.79 |
ENSMUST00000197248.5
ENSMUST00000061155.12 ENSMUST00000198043.5 ENSMUST00000084952.8 |
Kif9
|
kinesin family member 9 |
| chrX_+_156601431 | 2.78 |
ENSMUST00000087157.5
|
Klhl34
|
kelch-like 34 |
| chr5_-_135518098 | 2.77 |
ENSMUST00000201998.2
|
Hip1
|
huntingtin interacting protein 1 |
| chr11_-_4110286 | 2.74 |
ENSMUST00000093381.11
ENSMUST00000101626.9 |
Ccdc157
|
coiled-coil domain containing 157 |
| chr9_-_29323500 | 2.73 |
ENSMUST00000115237.8
|
Ntm
|
neurotrimin |
| chr4_-_118477960 | 2.72 |
ENSMUST00000071972.11
|
Cfap57
|
cilia and flagella associated protein 57 |
| chrX_-_103024847 | 2.69 |
ENSMUST00000121153.8
ENSMUST00000070705.6 |
Rlim
|
ring finger protein, LIM domain interacting |
| chr3_-_51184730 | 2.68 |
ENSMUST00000195432.2
ENSMUST00000091144.11 ENSMUST00000156983.3 |
Elf2
|
E74-like factor 2 |
| chr16_+_3690232 | 2.68 |
ENSMUST00000151988.8
|
Naa60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit |
| chr15_-_37458768 | 2.66 |
ENSMUST00000116445.9
|
Ncald
|
neurocalcin delta |
| chr7_+_143027473 | 2.59 |
ENSMUST00000052348.12
|
Slc22a18
|
solute carrier family 22 (organic cation transporter), member 18 |
| chr9_-_54568950 | 2.59 |
ENSMUST00000128624.2
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr6_+_5725811 | 2.52 |
ENSMUST00000115554.4
ENSMUST00000153942.2 |
Dync1i1
|
dynein cytoplasmic 1 intermediate chain 1 |
| chr18_-_7004717 | 2.51 |
ENSMUST00000079788.7
|
Mkx
|
mohawk homeobox |
| chr5_-_38637474 | 2.51 |
ENSMUST00000143758.8
ENSMUST00000156272.8 |
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr15_-_78687216 | 2.50 |
ENSMUST00000164826.8
|
Card10
|
caspase recruitment domain family, member 10 |
| chr8_-_26275182 | 2.49 |
ENSMUST00000038498.10
|
Bag4
|
BCL2-associated athanogene 4 |
| chr9_+_32027335 | 2.48 |
ENSMUST00000174641.8
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr17_-_23805187 | 2.47 |
ENSMUST00000227952.2
ENSMUST00000115516.11 |
Zfp13
|
zinc finger protein 13 |
| chr11_-_82655132 | 2.46 |
ENSMUST00000021040.10
ENSMUST00000100722.5 |
Cct6b
|
chaperonin containing Tcp1, subunit 6b (zeta) |
| chr1_-_24139263 | 2.46 |
ENSMUST00000187369.7
ENSMUST00000187752.7 ENSMUST00000186999.7 |
Fam135a
|
family with sequence similarity 135, member A |
| chr7_-_126062272 | 2.43 |
ENSMUST00000032974.13
|
Atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr3_+_98289854 | 2.43 |
ENSMUST00000178372.2
|
Zfp697
|
zinc finger protein 697 |
| chr3_+_98289755 | 2.41 |
ENSMUST00000056096.15
|
Zfp697
|
zinc finger protein 697 |
| chr2_+_32536594 | 2.40 |
ENSMUST00000113272.8
ENSMUST00000009705.14 ENSMUST00000167841.8 |
Eng
|
endoglin |
| chr17_-_56312555 | 2.40 |
ENSMUST00000043785.8
|
Stap2
|
signal transducing adaptor family member 2 |
| chr3_+_129326285 | 2.40 |
ENSMUST00000197235.5
|
Elovl6
|
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
| chr11_-_55310724 | 2.39 |
ENSMUST00000108858.8
ENSMUST00000141530.2 |
Sparc
|
secreted acidic cysteine rich glycoprotein |
| chr6_+_5725639 | 2.35 |
ENSMUST00000115556.8
ENSMUST00000115555.8 ENSMUST00000115559.10 |
Dync1i1
|
dynein cytoplasmic 1 intermediate chain 1 |
| chr15_-_54141816 | 2.35 |
ENSMUST00000079772.4
|
Tnfrsf11b
|
tumor necrosis factor receptor superfamily, member 11b (osteoprotegerin) |
| chr9_-_54569128 | 2.35 |
ENSMUST00000034822.12
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr14_+_79086665 | 2.33 |
ENSMUST00000227255.2
|
Vwa8
|
von Willebrand factor A domain containing 8 |
| chr11_-_53321242 | 2.30 |
ENSMUST00000109019.8
|
Uqcrq
|
ubiquinol-cytochrome c reductase, complex III subunit VII |
| chr9_+_21504018 | 2.29 |
ENSMUST00000062125.11
|
Timm29
|
translocase of inner mitochondrial membrane 29 |
| chr8_+_87350672 | 2.28 |
ENSMUST00000034141.18
ENSMUST00000122188.10 |
Lonp2
|
lon peptidase 2, peroxisomal |
| chr11_-_96932220 | 2.25 |
ENSMUST00000021251.7
|
Lrrc46
|
leucine rich repeat containing 46 |
| chr13_-_59970885 | 2.25 |
ENSMUST00000225179.2
ENSMUST00000225576.2 ENSMUST00000071703.6 |
Tut7
|
terminal uridylyl transferase 7 |
| chr11_+_49554430 | 2.23 |
ENSMUST00000043873.10
ENSMUST00000076006.5 |
Scgb3a1
|
secretoglobin, family 3A, member 1 |
| chr6_-_122317156 | 2.23 |
ENSMUST00000159384.8
|
Phc1
|
polyhomeotic 1 |
| chr18_+_37822865 | 2.23 |
ENSMUST00000195112.2
|
Pcdhgb2
|
protocadherin gamma subfamily B, 2 |
| chr3_-_27950491 | 2.19 |
ENSMUST00000058077.4
|
Tmem212
|
transmembrane protein 212 |
| chr1_-_4567577 | 2.19 |
ENSMUST00000192650.6
|
Sox17
|
SRY (sex determining region Y)-box 17 |
| chr16_-_17906886 | 2.17 |
ENSMUST00000132241.2
ENSMUST00000139861.2 ENSMUST00000003620.13 |
Prodh
|
proline dehydrogenase |
| chr1_+_16758731 | 2.16 |
ENSMUST00000190366.2
|
Ly96
|
lymphocyte antigen 96 |
| chr16_+_48104098 | 2.15 |
ENSMUST00000096045.9
ENSMUST00000050705.4 |
Dppa4
|
developmental pluripotency associated 4 |
| chr9_-_60595401 | 2.15 |
ENSMUST00000114034.9
ENSMUST00000065603.12 |
Lrrc49
|
leucine rich repeat containing 49 |
| chr1_-_24139387 | 2.13 |
ENSMUST00000027337.15
|
Fam135a
|
family with sequence similarity 135, member A |
| chr2_+_27567246 | 2.12 |
ENSMUST00000166775.8
|
Rxra
|
retinoid X receptor alpha |
| chr6_-_42669963 | 2.12 |
ENSMUST00000045140.5
|
Tcaf1
|
TRPM8 channel-associated factor 1 |
| chr12_-_58315949 | 2.09 |
ENSMUST00000062254.4
|
Clec14a
|
C-type lectin domain family 14, member a |
| chr14_+_79086492 | 2.07 |
ENSMUST00000040990.7
|
Vwa8
|
von Willebrand factor A domain containing 8 |
| chr1_+_183078573 | 2.06 |
ENSMUST00000109166.8
|
Aida
|
axin interactor, dorsalization associated |
| chr7_+_130375799 | 2.05 |
ENSMUST00000048453.7
ENSMUST00000208593.2 |
Btbd16
|
BTB (POZ) domain containing 16 |
| chr16_-_64591509 | 2.03 |
ENSMUST00000076991.7
|
4930453N24Rik
|
RIKEN cDNA 4930453N24 gene |
| chr1_-_183078488 | 2.02 |
ENSMUST00000057062.12
|
Brox
|
BRO1 domain and CAAX motif containing |
| chr9_+_32135781 | 2.02 |
ENSMUST00000183121.2
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chrX_+_106860083 | 2.02 |
ENSMUST00000143975.8
ENSMUST00000144695.8 ENSMUST00000167154.2 |
Tent5d
|
terminal nucleotidyltransferase 5D |
| chrX_-_151820545 | 2.01 |
ENSMUST00000051484.5
|
Mageh1
|
MAGE family member H1 |
| chr5_+_143166759 | 2.00 |
ENSMUST00000031574.10
|
Spdye4b
|
speedy/RINGO cell cycle regulator family, member E4B |
| chr17_+_43700327 | 1.98 |
ENSMUST00000113599.2
ENSMUST00000224278.2 ENSMUST00000225466.2 |
Adgrf5
|
adhesion G protein-coupled receptor F5 |
| chr9_+_32135540 | 1.97 |
ENSMUST00000168954.9
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr2_-_26012751 | 1.95 |
ENSMUST00000140993.2
ENSMUST00000028300.6 |
Nacc2
|
nucleus accumbens associated 2, BEN and BTB (POZ) domain containing |
| chr7_+_44499374 | 1.95 |
ENSMUST00000141311.8
ENSMUST00000107880.9 ENSMUST00000208384.2 |
Akt1s1
|
AKT1 substrate 1 (proline-rich) |
| chr10_+_70276604 | 1.94 |
ENSMUST00000173042.9
ENSMUST00000062883.7 |
Fam13c
|
family with sequence similarity 13, member C |
| chr8_+_120163857 | 1.94 |
ENSMUST00000152420.8
ENSMUST00000212112.2 ENSMUST00000098365.4 |
Osgin1
|
oxidative stress induced growth inhibitor 1 |
| chr2_+_27567213 | 1.93 |
ENSMUST00000077257.12
|
Rxra
|
retinoid X receptor alpha |
| chr11_-_5492175 | 1.92 |
ENSMUST00000020776.5
|
Ccdc117
|
coiled-coil domain containing 117 |
| chr15_+_92495007 | 1.91 |
ENSMUST00000035399.10
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chr3_+_10077608 | 1.91 |
ENSMUST00000029046.9
|
Fabp5
|
fatty acid binding protein 5, epidermal |
| chr7_-_67294943 | 1.91 |
ENSMUST00000190276.7
ENSMUST00000032775.12 ENSMUST00000053950.10 ENSMUST00000189836.2 |
Lrrc28
|
leucine rich repeat containing 28 |
| chr7_-_4527228 | 1.91 |
ENSMUST00000154913.2
|
Tnni3
|
troponin I, cardiac 3 |
| chr1_+_16758629 | 1.90 |
ENSMUST00000026881.11
|
Ly96
|
lymphocyte antigen 96 |
| chr3_-_33898405 | 1.90 |
ENSMUST00000029222.8
|
Ccdc39
|
coiled-coil domain containing 39 |
| chr11_+_87482971 | 1.87 |
ENSMUST00000103179.10
ENSMUST00000092802.12 ENSMUST00000146871.8 |
Mtmr4
|
myotubularin related protein 4 |
| chr5_-_139799780 | 1.84 |
ENSMUST00000146780.3
|
Tmem184a
|
transmembrane protein 184a |
| chr7_+_44499818 | 1.84 |
ENSMUST00000136232.2
ENSMUST00000207223.2 |
Akt1s1
|
AKT1 substrate 1 (proline-rich) |
| chr10_+_4561974 | 1.84 |
ENSMUST00000105590.8
ENSMUST00000067086.14 |
Esr1
|
estrogen receptor 1 (alpha) |
| chr11_-_68762664 | 1.83 |
ENSMUST00000101017.9
|
Ndel1
|
nudE neurodevelopment protein 1 like 1 |
| chr6_-_42670021 | 1.82 |
ENSMUST00000121083.8
|
Tcaf1
|
TRPM8 channel-associated factor 1 |
| chr8_+_105858432 | 1.80 |
ENSMUST00000161289.2
|
Ces4a
|
carboxylesterase 4A |
| chr17_+_27248233 | 1.80 |
ENSMUST00000053683.7
ENSMUST00000236222.2 |
Ggnbp1
|
gametogenetin binding protein 1 |
| chr2_+_32489710 | 1.79 |
ENSMUST00000131229.8
ENSMUST00000140983.8 |
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr11_-_70537878 | 1.78 |
ENSMUST00000014750.15
|
Slc25a11
|
solute carrier family 25 (mitochondrial carrier oxoglutarate carrier), member 11 |
| chr6_+_21986445 | 1.78 |
ENSMUST00000115382.8
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr2_-_130126339 | 1.77 |
ENSMUST00000239288.2
ENSMUST00000028892.11 |
Idh3b
|
isocitrate dehydrogenase 3 (NAD+) beta |
| chr19_+_6111204 | 1.77 |
ENSMUST00000162726.5
|
Znhit2
|
zinc finger, HIT domain containing 2 |
| chr18_-_39062201 | 1.76 |
ENSMUST00000134864.2
|
Fgf1
|
fibroblast growth factor 1 |
| chr17_+_29487881 | 1.76 |
ENSMUST00000234845.2
ENSMUST00000235038.2 ENSMUST00000235050.2 ENSMUST00000120346.9 ENSMUST00000234377.2 ENSMUST00000235074.2 ENSMUST00000235040.2 ENSMUST00000234256.2 ENSMUST00000234459.2 |
BC004004
|
cDNA sequence BC004004 |
| chr9_-_63509699 | 1.75 |
ENSMUST00000171243.2
ENSMUST00000163982.8 ENSMUST00000163624.8 |
Iqch
|
IQ motif containing H |
| chr4_+_155171034 | 1.74 |
ENSMUST00000030915.11
ENSMUST00000155775.8 ENSMUST00000127457.2 |
Morn1
|
MORN repeat containing 1 |
| chr7_-_139616234 | 1.74 |
ENSMUST00000209574.2
|
Tubgcp2
|
tubulin, gamma complex associated protein 2 |
| chr12_+_102915102 | 1.74 |
ENSMUST00000101099.12
|
Unc79
|
unc-79 homolog |
| chr5_-_71705532 | 1.73 |
ENSMUST00000050129.6
|
Cox7b2
|
cytochrome c oxidase subunit 7B2 |
| chr9_+_70114623 | 1.73 |
ENSMUST00000034745.9
|
Myo1e
|
myosin IE |
| chr3_-_116601700 | 1.72 |
ENSMUST00000159742.8
|
Agl
|
amylo-1,6-glucosidase, 4-alpha-glucanotransferase |
| chr3_+_32583602 | 1.72 |
ENSMUST00000091257.11
|
Mfn1
|
mitofusin 1 |
| chr6_-_119365632 | 1.72 |
ENSMUST00000169744.8
|
Adipor2
|
adiponectin receptor 2 |
| chr3_-_127202663 | 1.70 |
ENSMUST00000182008.8
ENSMUST00000182547.8 |
Ank2
|
ankyrin 2, brain |
| chr5_-_36987917 | 1.70 |
ENSMUST00000031002.10
|
Man2b2
|
mannosidase 2, alpha B2 |
| chr9_-_7184440 | 1.70 |
ENSMUST00000140466.8
|
Dync2h1
|
dynein cytoplasmic 2 heavy chain 1 |
| chr7_+_109617456 | 1.69 |
ENSMUST00000084731.5
|
Ipo7
|
importin 7 |
| chr12_-_4088905 | 1.69 |
ENSMUST00000111178.2
|
Efr3b
|
EFR3 homolog B |
| chrX_+_60753074 | 1.68 |
ENSMUST00000075983.6
|
Ldoc1
|
regulator of NFKB signaling |
| chr4_-_140344373 | 1.68 |
ENSMUST00000154979.2
|
Arhgef10l
|
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr9_+_57913694 | 1.68 |
ENSMUST00000188116.7
|
Cyp11a1
|
cytochrome P450, family 11, subfamily a, polypeptide 1 |
| chr17_+_3447465 | 1.66 |
ENSMUST00000072156.7
|
Tiam2
|
T cell lymphoma invasion and metastasis 2 |
| chr3_+_32583681 | 1.65 |
ENSMUST00000147350.8
|
Mfn1
|
mitofusin 1 |
| chr10_-_83484576 | 1.65 |
ENSMUST00000020500.14
|
Appl2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr3_-_101831729 | 1.62 |
ENSMUST00000190824.7
|
Slc22a15
|
solute carrier family 22 (organic anion/cation transporter), member 15 |
| chr9_+_108708939 | 1.61 |
ENSMUST00000192235.2
|
Celsr3
|
cadherin, EGF LAG seven-pass G-type receptor 3 |
| chr19_+_8828132 | 1.61 |
ENSMUST00000235683.2
ENSMUST00000096257.3 |
Lrrn4cl
|
LRRN4 C-terminal like |
| chr4_-_140309113 | 1.60 |
ENSMUST00000147426.2
ENSMUST00000105797.9 |
Arhgef10l
|
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr3_-_127202635 | 1.59 |
ENSMUST00000182959.8
|
Ank2
|
ankyrin 2, brain |
| chr3_-_127202693 | 1.59 |
ENSMUST00000182078.9
|
Ank2
|
ankyrin 2, brain |
| chr5_+_29940935 | 1.58 |
ENSMUST00000114839.8
ENSMUST00000198694.5 ENSMUST00000012734.10 ENSMUST00000196528.5 |
Dnajb6
|
DnaJ heat shock protein family (Hsp40) member B6 |
| chr8_-_123768984 | 1.57 |
ENSMUST00000212937.2
|
Ankrd11
|
ankyrin repeat domain 11 |
| chr9_-_79700660 | 1.57 |
ENSMUST00000034878.12
|
Tmem30a
|
transmembrane protein 30A |
| chr15_-_10714653 | 1.57 |
ENSMUST00000169385.3
|
Rai14
|
retinoic acid induced 14 |
| chr12_-_31549538 | 1.56 |
ENSMUST00000064240.14
ENSMUST00000185739.8 ENSMUST00000188326.3 ENSMUST00000101499.10 ENSMUST00000085487.12 |
Cbll1
|
Casitas B-lineage lymphoma-like 1 |
| chr12_+_65012564 | 1.56 |
ENSMUST00000066296.9
ENSMUST00000223166.2 |
Togaram1
|
TOG array regulator of axonemal microtubules 1 |
| chr7_+_127475968 | 1.55 |
ENSMUST00000131000.2
|
Zfp646
|
zinc finger protein 646 |
| chr4_-_149569614 | 1.54 |
ENSMUST00000126896.2
ENSMUST00000105693.2 ENSMUST00000030845.13 |
Nmnat1
|
nicotinamide nucleotide adenylyltransferase 1 |
| chr16_-_38342949 | 1.54 |
ENSMUST00000002925.6
|
Timmdc1
|
translocase of inner mitochondrial membrane domain containing 1 |
| chr19_+_6413703 | 1.54 |
ENSMUST00000131252.8
ENSMUST00000113489.8 |
Sf1
|
splicing factor 1 |
| chr4_+_129823042 | 1.54 |
ENSMUST00000084263.6
|
Spocd1
|
SPOC domain containing 1 |
| chr2_-_163261439 | 1.53 |
ENSMUST00000046908.10
|
Oser1
|
oxidative stress responsive serine rich 1 |
| chr9_-_21504101 | 1.52 |
ENSMUST00000180365.9
ENSMUST00000213809.2 ENSMUST00000034700.15 |
Yipf2
|
Yip1 domain family, member 2 |
| chr9_-_21504032 | 1.52 |
ENSMUST00000078572.9
|
Yipf2
|
Yip1 domain family, member 2 |
| chr5_-_139799953 | 1.51 |
ENSMUST00000044002.10
|
Tmem184a
|
transmembrane protein 184a |
| chr8_-_37419567 | 1.51 |
ENSMUST00000163663.3
|
Dlc1
|
deleted in liver cancer 1 |
| chr7_-_30292351 | 1.50 |
ENSMUST00000108151.3
|
Zbtb32
|
zinc finger and BTB domain containing 32 |
| chrX_-_149224054 | 1.50 |
ENSMUST00000059256.8
|
Tmem29
|
transmembrane protein 29 |
| chr1_+_74640590 | 1.50 |
ENSMUST00000087183.11
ENSMUST00000148456.8 ENSMUST00000113694.8 |
Stk36
|
serine/threonine kinase 36 |
| chr8_-_22966831 | 1.49 |
ENSMUST00000163774.3
ENSMUST00000033935.16 |
Smim19
|
small integral membrane protein 19 |
| chr17_-_24863956 | 1.49 |
ENSMUST00000019684.13
|
Slc9a3r2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 |
| chr5_+_67418137 | 1.48 |
ENSMUST00000161369.3
|
Tmem33
|
transmembrane protein 33 |
| chr2_+_31462659 | 1.48 |
ENSMUST00000113482.8
|
Fubp3
|
far upstream element (FUSE) binding protein 3 |
| chr10_+_77458109 | 1.48 |
ENSMUST00000174510.8
ENSMUST00000172813.2 |
Ube2g2
|
ubiquitin-conjugating enzyme E2G 2 |
| chr9_-_79700789 | 1.47 |
ENSMUST00000120690.2
|
Tmem30a
|
transmembrane protein 30A |
| chr2_+_91033230 | 1.47 |
ENSMUST00000150403.8
ENSMUST00000002172.14 ENSMUST00000238832.2 ENSMUST00000239169.2 ENSMUST00000155418.2 |
Acp2
|
acid phosphatase 2, lysosomal |
| chr11_-_93846453 | 1.47 |
ENSMUST00000072566.5
|
Nme2
|
NME/NM23 nucleoside diphosphate kinase 2 |
| chr2_+_130248398 | 1.46 |
ENSMUST00000055421.6
|
Tmem239
|
transmembrane 239 |
| chr12_+_75355082 | 1.46 |
ENSMUST00000118602.8
ENSMUST00000118966.8 ENSMUST00000055390.6 |
Rhoj
|
ras homolog family member J |
| chr19_+_11747721 | 1.42 |
ENSMUST00000167199.3
|
Mrpl16
|
mitochondrial ribosomal protein L16 |
| chr3_+_115801869 | 1.42 |
ENSMUST00000106502.2
|
Extl2
|
exostosin-like glycosyltransferase 2 |
| chr11_-_50216426 | 1.42 |
ENSMUST00000179865.8
ENSMUST00000020637.9 |
Canx
|
calnexin |
| chr16_-_4698148 | 1.40 |
ENSMUST00000037843.7
|
Ubald1
|
UBA-like domain containing 1 |
| chr9_-_119019428 | 1.38 |
ENSMUST00000127794.2
|
Slc22a14
|
solute carrier family 22 (organic cation transporter), member 14 |
| chr7_-_139616167 | 1.37 |
ENSMUST00000026547.9
|
Tubgcp2
|
tubulin, gamma complex associated protein 2 |
| chr17_-_3608056 | 1.36 |
ENSMUST00000041003.8
|
Tfb1m
|
transcription factor B1, mitochondrial |
| chr3_-_116601815 | 1.35 |
ENSMUST00000040603.14
|
Agl
|
amylo-1,6-glucosidase, 4-alpha-glucanotransferase |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 9.3 | GO:2000019 | negative regulation of male gonad development(GO:2000019) |
| 2.4 | 2.4 | GO:1905072 | cardiac jelly development(GO:1905072) |
| 1.4 | 9.7 | GO:1904075 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 1.2 | 3.6 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 1.1 | 3.2 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 1.0 | 3.0 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 1.0 | 2.9 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.9 | 17.0 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.9 | 1.8 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.8 | 2.5 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.8 | 2.5 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 0.8 | 2.5 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.8 | 2.4 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) relaxation of skeletal muscle(GO:0090076) |
| 0.8 | 4.1 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 0.7 | 2.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.7 | 2.8 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.7 | 2.8 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.7 | 3.4 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.7 | 5.9 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.7 | 2.0 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 0.6 | 5.7 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.6 | 3.2 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.6 | 2.5 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.6 | 1.8 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.6 | 4.1 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.6 | 4.1 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.6 | 3.3 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.5 | 2.2 | GO:0003142 | cardiogenic plate morphogenesis(GO:0003142) regulation of transcription from RNA polymerase II promoter involved in definitive endodermal cell fate specification(GO:0060807) |
| 0.5 | 2.1 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.5 | 1.5 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.5 | 3.9 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.5 | 2.9 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.5 | 4.4 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.4 | 4.9 | GO:2000580 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.4 | 5.2 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.4 | 1.2 | GO:2000328 | positive regulation of memory T cell differentiation(GO:0043382) regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.4 | 1.2 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.4 | 1.4 | GO:2000395 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.3 | 1.4 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.3 | 2.3 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.3 | 1.6 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.3 | 1.9 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.3 | 2.7 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.3 | 2.9 | GO:2000535 | entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.3 | 7.9 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.3 | 3.2 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.3 | 1.2 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.3 | 1.7 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.3 | 2.0 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.3 | 0.8 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.3 | 6.0 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.3 | 1.9 | GO:0060287 | determination of pancreatic left/right asymmetry(GO:0035469) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.3 | 4.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.3 | 3.5 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.3 | 2.2 | GO:1904378 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.3 | 4.5 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.3 | 3.1 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.3 | 1.0 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.3 | 1.8 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.2 | 2.7 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.2 | 1.5 | GO:1990839 | response to endothelin(GO:1990839) |
| 0.2 | 6.9 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.2 | 1.0 | GO:0019064 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.2 | 2.9 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.2 | 0.7 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.2 | 0.7 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.2 | 1.4 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.2 | 2.4 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.2 | 3.0 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.2 | 0.9 | GO:0000105 | histidine biosynthetic process(GO:0000105) 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.2 | 2.5 | GO:0007549 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.2 | 1.8 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.2 | 0.7 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.2 | 0.9 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.2 | 1.8 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.2 | 0.7 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.2 | 0.6 | GO:0046038 | GMP catabolic process(GO:0046038) |
| 0.2 | 5.1 | GO:0019363 | pyridine nucleotide biosynthetic process(GO:0019363) |
| 0.2 | 1.0 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.2 | 0.6 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.2 | 0.8 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.2 | 3.5 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.2 | 1.7 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.2 | 33.1 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.2 | 1.7 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.2 | 0.9 | GO:0060684 | epithelial-mesenchymal cell signaling(GO:0060684) |
| 0.2 | 3.3 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.2 | 1.3 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.2 | 1.2 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.2 | 0.5 | GO:2000554 | regulation of T-helper 1 cell cytokine production(GO:2000554) positive regulation of T-helper 1 cell cytokine production(GO:2000556) |
| 0.2 | 0.7 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.2 | 3.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.2 | 0.9 | GO:0072737 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.2 | 1.2 | GO:1901300 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) |
| 0.2 | 0.3 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 0.2 | 5.8 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 0.2 | 1.7 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.2 | 2.1 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.2 | 0.8 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.1 | 1.8 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.5 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.1 | 4.9 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.1 | 1.5 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 0.4 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 1.7 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 0.4 | GO:1904959 | regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.1 | 3.1 | GO:0009251 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 0.5 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.1 | 2.8 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 3.2 | GO:0072662 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.1 | 1.0 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 2.8 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.1 | 2.5 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 0.7 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.1 | 1.0 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.1 | 0.3 | GO:0001546 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 0.9 | GO:0002553 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.1 | 1.5 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 1.5 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 1.3 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.1 | 1.5 | GO:0030238 | male sex determination(GO:0030238) |
| 0.1 | 0.3 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.1 | 1.6 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.1 | 1.5 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.1 | 1.0 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.1 | 1.0 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 2.0 | GO:0030157 | pancreatic juice secretion(GO:0030157) |
| 0.1 | 0.9 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 0.4 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.1 | 0.2 | GO:0001844 | protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:0001844) |
| 0.1 | 5.8 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.9 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.1 | 2.7 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.1 | 0.7 | GO:0072092 | ureteric bud invasion(GO:0072092) metanephric renal vesicle formation(GO:0072093) |
| 0.1 | 1.1 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 5.7 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 0.9 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.1 | 0.6 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.3 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.1 | 1.5 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 1.2 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.1 | 0.5 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.1 | 0.4 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.1 | 1.9 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.1 | 1.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 0.6 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.1 | 1.0 | GO:1903350 | response to dopamine(GO:1903350) cellular response to dopamine(GO:1903351) |
| 0.1 | 4.0 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 0.4 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.1 | 0.4 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 0.5 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 1.3 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.1 | 2.6 | GO:0015893 | drug transport(GO:0015893) |
| 0.1 | 0.5 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 0.4 | GO:0021886 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.1 | 0.7 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.1 | 2.7 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 0.8 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.1 | 1.5 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 0.9 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.4 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 0.2 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.1 | 0.4 | GO:0015867 | ATP transport(GO:0015867) |
| 0.1 | 0.4 | GO:1990009 | retinal cell apoptotic process(GO:1990009) |
| 0.1 | 1.7 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.1 | 0.2 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 1.1 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.1 | 0.5 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.1 | 1.6 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.1 | 2.4 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.1 | 2.4 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 0.9 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.1 | GO:0030167 | proteoglycan catabolic process(GO:0030167) heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 1.3 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.7 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 1.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.3 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 0.5 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.4 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.2 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.5 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.2 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.4 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 0.0 | 0.7 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.7 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.0 | 2.6 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.0 | 3.5 | GO:1903749 | positive regulation of establishment of protein localization to mitochondrion(GO:1903749) positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 1.1 | GO:0040020 | regulation of meiotic nuclear division(GO:0040020) |
| 0.0 | 1.4 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 1.9 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 2.1 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 2.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.3 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.4 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.6 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) |
| 0.0 | 2.0 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.9 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.9 | GO:0006110 | regulation of glycolytic process(GO:0006110) |
| 0.0 | 2.2 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.0 | 0.7 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.8 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 0.2 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.1 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.3 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.2 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.6 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 1.0 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 1.2 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.5 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.5 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 2.2 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.2 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 0.0 | 2.6 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 0.7 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.9 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.6 | GO:0050802 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) circadian sleep/wake cycle, sleep(GO:0050802) |
| 0.0 | 0.3 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.9 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.5 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.9 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.4 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.4 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.7 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.1 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 | 0.7 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.0 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 3.8 | GO:0043434 | response to peptide hormone(GO:0043434) |
| 0.0 | 0.7 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.1 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.2 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 1.0 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 0.1 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.2 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 0.5 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.2 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.5 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 | 0.1 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 1.0 | GO:0008637 | apoptotic mitochondrial changes(GO:0008637) |
| 0.0 | 0.4 | GO:0002042 | cell migration involved in sprouting angiogenesis(GO:0002042) |
| 0.0 | 0.7 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.0 | 0.1 | GO:0070234 | positive regulation of T cell apoptotic process(GO:0070234) |
| 0.0 | 0.5 | GO:0001779 | natural killer cell differentiation(GO:0001779) |
| 0.0 | 0.3 | GO:0001569 | patterning of blood vessels(GO:0001569) |
| 0.0 | 2.5 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.0 | 0.4 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.8 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.9 | GO:0031497 | chromatin assembly(GO:0031497) |
| 0.0 | 0.3 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 1.7 | GO:0098656 | anion transmembrane transport(GO:0098656) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 17.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 1.6 | 4.9 | GO:1990257 | piccolo-bassoon transport vesicle(GO:1990257) |
| 1.0 | 9.7 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.8 | 3.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.8 | 2.3 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.6 | 2.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.6 | 2.8 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.5 | 4.6 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.5 | 4.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.5 | 1.5 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.5 | 5.2 | GO:0097433 | dense body(GO:0097433) |
| 0.5 | 1.8 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.4 | 11.2 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.4 | 1.2 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.4 | 1.5 | GO:0071920 | cleavage body(GO:0071920) |
| 0.4 | 2.2 | GO:0070449 | elongin complex(GO:0070449) |
| 0.3 | 3.8 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.3 | 2.4 | GO:0031673 | H zone(GO:0031673) |
| 0.3 | 1.1 | GO:0008623 | CHRAC(GO:0008623) |
| 0.3 | 1.9 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.3 | 3.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.3 | 1.8 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.2 | 1.7 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.2 | 2.2 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.2 | 0.9 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.2 | 2.9 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.2 | 0.9 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.2 | 0.6 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.2 | 2.3 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.2 | 2.0 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.2 | 0.8 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.2 | 1.3 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.2 | 2.8 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.2 | 1.8 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 2.0 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 2.5 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.5 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 3.6 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 1.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 3.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 4.1 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 5.9 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.4 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.1 | 1.4 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 0.6 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 12.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 0.5 | GO:0005879 | axonemal microtubule(GO:0005879) symmetric synapse(GO:0032280) |
| 0.1 | 0.5 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 2.5 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 1.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 1.5 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 1.0 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.3 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 0.7 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 0.5 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 0.4 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 5.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.7 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 0.6 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 1.8 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 1.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 0.8 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 2.2 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 1.8 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.4 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.4 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 1.6 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.4 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 1.0 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.1 | 0.9 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.8 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.2 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.6 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 3.3 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 1.0 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.4 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 0.3 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 1.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 4.4 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.9 | GO:0098984 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.0 | 0.1 | GO:0036501 | UFD1-NPL4 complex(GO:0036501) |
| 0.0 | 1.0 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 1.0 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 6.4 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.4 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.3 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.4 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.9 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 3.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 6.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 2.8 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.5 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 2.0 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 2.0 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.4 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.8 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.9 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.4 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 2.6 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 0.6 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 4.8 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 1.0 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.4 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 5.0 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 0.0 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 1.6 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.9 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 1.5 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 7.8 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 2.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.9 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 3.1 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.3 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 2.2 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 7.6 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 0.9 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.3 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.2 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 1.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.5 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.2 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.7 | GO:0016034 | maleylacetoacetate isomerase activity(GO:0016034) |
| 1.7 | 5.1 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 1.3 | 6.7 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 1.2 | 3.6 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 1.2 | 3.5 | GO:0050479 | glyceryl-ether monooxygenase activity(GO:0050479) |
| 1.0 | 3.1 | GO:0004133 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.8 | 2.4 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.7 | 5.2 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.7 | 4.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.7 | 4.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.6 | 15.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.6 | 1.8 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.6 | 1.8 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.6 | 2.3 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.5 | 4.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.5 | 3.0 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.5 | 2.9 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.5 | 4.9 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.5 | 1.4 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.4 | 5.2 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.4 | 1.7 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 0.4 | 1.5 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.4 | 1.8 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.4 | 2.9 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.4 | 1.8 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.4 | 6.0 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.3 | 2.4 | GO:0005534 | galactose binding(GO:0005534) |
| 0.3 | 1.7 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.3 | 1.3 | GO:0031544 | peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.3 | 4.9 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.3 | 4.4 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.3 | 9.3 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.3 | 0.9 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.3 | 1.5 | GO:0033677 | DNA/RNA helicase activity(GO:0033677) |
| 0.3 | 2.3 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.3 | 1.9 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.3 | 4.8 | GO:0019841 | retinol binding(GO:0019841) |
| 0.3 | 11.9 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.3 | 1.3 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.3 | 4.7 | GO:0008430 | selenium binding(GO:0008430) |
| 0.3 | 1.3 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.3 | 0.5 | GO:0019970 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.2 | 1.0 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.2 | 0.9 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.2 | 2.8 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.2 | 1.3 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.2 | 0.6 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.2 | 2.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.2 | 1.5 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 0.2 | 0.8 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.2 | 2.0 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.2 | 3.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.2 | 2.4 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.2 | 1.8 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.2 | 1.0 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.2 | 0.8 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.2 | 0.8 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.2 | 0.9 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.2 | 3.3 | GO:0070513 | death domain binding(GO:0070513) |
| 0.2 | 0.6 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.1 | 0.9 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 3.0 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 0.4 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.1 | 1.0 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 1.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 1.4 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 3.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 5.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.5 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 1.7 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 2.7 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 2.4 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 1.4 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 0.3 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.1 | 0.7 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.1 | 3.4 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 2.7 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 0.7 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.1 | 0.6 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.1 | 0.4 | GO:0001002 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.1 | 1.7 | GO:0015923 | alpha-mannosidase activity(GO:0004559) mannosidase activity(GO:0015923) |
| 0.1 | 1.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 1.2 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 14.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.9 | GO:0008061 | chitin binding(GO:0008061) |
| 0.1 | 3.7 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 0.4 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 0.3 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.1 | 1.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 2.6 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 0.1 | 1.8 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 5.8 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 0.2 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 2.7 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.7 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 0.2 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.1 | 10.8 | GO:0020037 | heme binding(GO:0020037) |
| 0.1 | 3.0 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 5.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 21.1 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 1.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.3 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 2.5 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.4 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 1.9 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.4 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 1.0 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.1 | 3.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 3.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 2.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 2.2 | GO:0016645 | oxidoreductase activity, acting on the CH-NH group of donors(GO:0016645) |
| 0.1 | 0.5 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 0.4 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.4 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.4 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.0 | 1.5 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 1.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.7 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.3 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 1.9 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.2 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 2.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.2 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.9 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.0 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.4 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.5 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 1.1 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 1.7 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.2 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.6 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 1.4 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.7 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 1.4 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.7 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 2.1 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 1.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.5 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.7 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 1.1 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.2 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 2.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.8 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.4 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 1.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.8 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 12.8 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 1.3 | GO:0004536 | deoxyribonuclease activity(GO:0004536) |
| 0.0 | 3.8 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.5 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.0 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.9 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 1.0 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 1.3 | GO:0101005 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.5 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.4 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 6.8 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 7.8 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 2.0 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.6 | GO:0005518 | collagen binding(GO:0005518) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.2 | 3.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 10.2 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 0.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 8.0 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.1 | 3.6 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 2.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 4.2 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 2.2 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 3.6 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 7.3 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 5.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 3.1 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 2.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 1.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 0.5 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 2.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.7 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.9 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 1.7 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 2.1 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.4 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.7 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 2.0 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.9 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.3 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 5.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.8 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 1.2 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 2.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.0 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.3 | 4.0 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.3 | 4.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.2 | 3.1 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.2 | 2.0 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.2 | 0.9 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.2 | 5.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 2.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 3.9 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.2 | 5.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 4.6 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 2.5 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 2.9 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 1.4 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 2.2 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 2.8 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 2.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 1.5 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 2.8 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 4.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 8.1 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 2.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 1.7 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 3.2 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 1.8 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 1.0 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 2.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 3.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 4.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 3.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 4.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.7 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 1.9 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 1.8 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 2.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.0 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.7 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 5.6 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 4.4 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 3.3 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.5 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.0 | 1.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.5 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.7 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.1 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.4 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.3 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.3 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.4 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |