Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
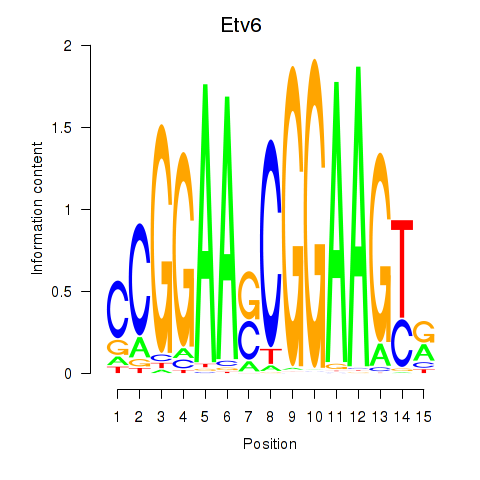
Results for Etv6
Z-value: 1.02
Transcription factors associated with Etv6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Etv6
|
ENSMUSG00000030199.17 | Etv6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Etv6 | mm39_v1_chr6_+_134012916_134012936 | 0.09 | 4.4e-01 | Click! |
Activity profile of Etv6 motif
Sorted Z-values of Etv6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Etv6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_64940004 | 3.70 |
ENSMUST00000167773.2
|
Dpp8
|
dipeptidylpeptidase 8 |
| chr9_+_64939695 | 3.29 |
ENSMUST00000034960.14
|
Dpp8
|
dipeptidylpeptidase 8 |
| chr7_-_108682491 | 2.92 |
ENSMUST00000120876.2
ENSMUST00000055993.13 |
Ric3
|
RIC3 acetylcholine receptor chaperone |
| chr8_-_22888604 | 2.84 |
ENSMUST00000033871.8
|
Slc25a15
|
solute carrier family 25 (mitochondrial carrier ornithine transporter), member 15 |
| chr11_+_58221569 | 2.71 |
ENSMUST00000073128.7
|
Sh3bp5l
|
SH3 binding domain protein 5 like |
| chr7_+_126808016 | 2.51 |
ENSMUST00000206204.2
ENSMUST00000206772.2 |
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr5_-_149559636 | 2.38 |
ENSMUST00000201452.4
|
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr12_+_109425769 | 2.38 |
ENSMUST00000173812.2
|
Dlk1
|
delta like non-canonical Notch ligand 1 |
| chr19_+_44994905 | 2.33 |
ENSMUST00000026227.3
|
Twnk
|
twinkle mtDNA helicase |
| chr11_-_35725317 | 2.21 |
ENSMUST00000018992.4
|
Rars
|
arginyl-tRNA synthetase |
| chr5_-_149559737 | 2.21 |
ENSMUST00000200805.4
|
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr3_-_88332401 | 2.15 |
ENSMUST00000168755.7
ENSMUST00000193433.6 ENSMUST00000195657.6 ENSMUST00000057935.9 |
Slc25a44
|
solute carrier family 25, member 44 |
| chrX_-_93256244 | 2.15 |
ENSMUST00000113922.2
|
Eif2s3x
|
eukaryotic translation initiation factor 2, subunit 3, structural gene X-linked |
| chr14_+_52122439 | 2.14 |
ENSMUST00000167984.2
|
Mettl17
|
methyltransferase like 17 |
| chr2_-_10084866 | 2.12 |
ENSMUST00000130067.2
ENSMUST00000139810.8 |
Atp5c1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| chr11_-_96807192 | 2.09 |
ENSMUST00000144731.8
ENSMUST00000127048.8 |
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr11_+_58221538 | 2.08 |
ENSMUST00000116376.9
|
Sh3bp5l
|
SH3 binding domain protein 5 like |
| chr5_-_149559792 | 2.05 |
ENSMUST00000202361.4
ENSMUST00000202089.4 ENSMUST00000200825.2 ENSMUST00000201559.4 |
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr2_-_10085133 | 2.03 |
ENSMUST00000145530.8
ENSMUST00000026887.14 ENSMUST00000114896.8 |
Atp5c1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| chr6_-_120799641 | 2.00 |
ENSMUST00000205049.3
|
Atp6v1e1
|
ATPase, H+ transporting, lysosomal V1 subunit E1 |
| chr19_-_6134703 | 1.95 |
ENSMUST00000161548.8
|
Zfpl1
|
zinc finger like protein 1 |
| chr1_-_179373826 | 1.95 |
ENSMUST00000027769.6
|
Tfb2m
|
transcription factor B2, mitochondrial |
| chr17_-_26063391 | 1.93 |
ENSMUST00000176591.8
|
Rhot2
|
ras homolog family member T2 |
| chr2_-_90735171 | 1.87 |
ENSMUST00000005647.4
|
Ndufs3
|
NADH:ubiquinone oxidoreductase core subunit S3 |
| chr15_+_58805605 | 1.83 |
ENSMUST00000022980.5
|
Ndufb9
|
NADH:ubiquinone oxidoreductase subunit B9 |
| chr6_-_120799500 | 1.83 |
ENSMUST00000204699.2
|
Atp6v1e1
|
ATPase, H+ transporting, lysosomal V1 subunit E1 |
| chrX_-_168103266 | 1.82 |
ENSMUST00000033717.9
ENSMUST00000112115.2 |
Hccs
|
holocytochrome c synthetase |
| chr15_-_58805537 | 1.75 |
ENSMUST00000226931.2
ENSMUST00000228538.3 ENSMUST00000110155.3 |
Tatdn1
|
TatD DNase domain containing 1 |
| chr9_+_46184362 | 1.73 |
ENSMUST00000156440.8
ENSMUST00000114552.4 |
Zpr1
|
ZPR1 zinc finger |
| chr19_+_46587523 | 1.71 |
ENSMUST00000138302.9
ENSMUST00000099376.11 |
Wbp1l
|
WW domain binding protein 1 like |
| chr19_+_10872587 | 1.67 |
ENSMUST00000025642.14
|
Prpf19
|
pre-mRNA processing factor 19 |
| chr11_+_118319319 | 1.63 |
ENSMUST00000017590.9
|
C1qtnf1
|
C1q and tumor necrosis factor related protein 1 |
| chr3_-_65299967 | 1.63 |
ENSMUST00000119896.2
|
Ssr3
|
signal sequence receptor, gamma |
| chr2_-_10085299 | 1.62 |
ENSMUST00000114897.9
|
Atp5c1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| chr7_+_44499374 | 1.61 |
ENSMUST00000141311.8
ENSMUST00000107880.9 ENSMUST00000208384.2 |
Akt1s1
|
AKT1 substrate 1 (proline-rich) |
| chr9_+_107440445 | 1.61 |
ENSMUST00000010198.5
|
Tusc2
|
tumor suppressor 2, mitochondrial calcium regulator |
| chr13_-_74356778 | 1.61 |
ENSMUST00000035934.7
|
Exoc3
|
exocyst complex component 3 |
| chr14_+_52122299 | 1.61 |
ENSMUST00000047899.13
ENSMUST00000164902.8 |
Mettl17
|
methyltransferase like 17 |
| chr1_+_4878460 | 1.58 |
ENSMUST00000131119.2
|
Lypla1
|
lysophospholipase 1 |
| chr16_+_3690232 | 1.57 |
ENSMUST00000151988.8
|
Naa60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit |
| chr4_-_118347249 | 1.56 |
ENSMUST00000047421.6
|
Tie1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
| chrX_+_7750558 | 1.55 |
ENSMUST00000208640.2
ENSMUST00000207114.2 ENSMUST00000208633.2 ENSMUST00000208397.2 ENSMUST00000153620.3 ENSMUST00000123277.8 |
Slc35a2
|
solute carrier family 35 (UDP-galactose transporter), member A2 |
| chr1_-_58625350 | 1.54 |
ENSMUST00000038372.14
ENSMUST00000097724.10 |
Fam126b
|
family with sequence similarity 126, member B |
| chr6_-_120799672 | 1.54 |
ENSMUST00000019354.11
|
Atp6v1e1
|
ATPase, H+ transporting, lysosomal V1 subunit E1 |
| chr11_-_96807233 | 1.53 |
ENSMUST00000130774.2
|
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr14_-_101878106 | 1.52 |
ENSMUST00000100339.9
|
Commd6
|
COMM domain containing 6 |
| chr11_-_58221145 | 1.52 |
ENSMUST00000186859.2
ENSMUST00000065533.3 |
Zfp672
|
zinc finger protein 672 |
| chr5_-_149559667 | 1.52 |
ENSMUST00000074846.14
|
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chrX_+_20570145 | 1.51 |
ENSMUST00000033383.3
|
Usp11
|
ubiquitin specific peptidase 11 |
| chr7_+_44499818 | 1.49 |
ENSMUST00000136232.2
ENSMUST00000207223.2 |
Akt1s1
|
AKT1 substrate 1 (proline-rich) |
| chr11_-_120348762 | 1.49 |
ENSMUST00000137632.2
ENSMUST00000044007.3 |
Oxld1
|
oxidoreductase like domain containing 1 |
| chr11_+_88095222 | 1.48 |
ENSMUST00000118784.8
ENSMUST00000139170.8 ENSMUST00000107915.10 ENSMUST00000144070.3 |
Mrps23
|
mitochondrial ribosomal protein S23 |
| chr11_-_50101592 | 1.46 |
ENSMUST00000143379.2
ENSMUST00000015981.12 ENSMUST00000102774.11 |
Sqstm1
|
sequestosome 1 |
| chr17_+_47022384 | 1.45 |
ENSMUST00000002840.9
|
Pex6
|
peroxisomal biogenesis factor 6 |
| chr2_+_155360015 | 1.44 |
ENSMUST00000103142.12
|
Acss2
|
acyl-CoA synthetase short-chain family member 2 |
| chr8_-_78337297 | 1.44 |
ENSMUST00000141202.2
ENSMUST00000152168.8 |
Tmem184c
|
transmembrane protein 184C |
| chr11_+_94544593 | 1.41 |
ENSMUST00000025278.8
|
Mrpl27
|
mitochondrial ribosomal protein L27 |
| chr8_+_87350672 | 1.38 |
ENSMUST00000034141.18
ENSMUST00000122188.10 |
Lonp2
|
lon peptidase 2, peroxisomal |
| chr2_-_168072295 | 1.38 |
ENSMUST00000154111.8
|
Dpm1
|
dolichol-phosphate (beta-D) mannosyltransferase 1 |
| chr2_+_37333275 | 1.31 |
ENSMUST00000133434.8
ENSMUST00000061179.12 |
Rabgap1
|
RAB GTPase activating protein 1 |
| chr11_+_120348919 | 1.30 |
ENSMUST00000058370.14
ENSMUST00000175970.8 ENSMUST00000176120.2 |
Ccdc137
|
coiled-coil domain containing 137 |
| chr7_-_46569617 | 1.27 |
ENSMUST00000210664.2
ENSMUST00000156335.9 |
Tsg101
|
tumor susceptibility gene 101 |
| chr8_+_125739772 | 1.27 |
ENSMUST00000212603.2
|
Tsnax
|
translin-associated factor X |
| chr12_-_44256843 | 1.27 |
ENSMUST00000220421.2
|
Dnajb9
|
DnaJ heat shock protein family (Hsp40) member B9 |
| chr2_-_168072493 | 1.26 |
ENSMUST00000109193.8
|
Dpm1
|
dolichol-phosphate (beta-D) mannosyltransferase 1 |
| chr8_-_107783282 | 1.25 |
ENSMUST00000034391.4
ENSMUST00000095517.12 |
Cog8
|
component of oligomeric golgi complex 8 |
| chr15_+_87509413 | 1.25 |
ENSMUST00000068088.8
|
Tafa5
|
TAFA chemokine like family member 5 |
| chr11_-_62349334 | 1.24 |
ENSMUST00000141447.2
|
Ncor1
|
nuclear receptor co-repressor 1 |
| chr14_-_70757601 | 1.24 |
ENSMUST00000022693.9
|
Bmp1
|
bone morphogenetic protein 1 |
| chr11_-_70537878 | 1.23 |
ENSMUST00000014750.15
|
Slc25a11
|
solute carrier family 25 (mitochondrial carrier oxoglutarate carrier), member 11 |
| chr3_-_94794256 | 1.22 |
ENSMUST00000005923.7
|
Psmb4
|
proteasome (prosome, macropain) subunit, beta type 4 |
| chr7_+_112026712 | 1.21 |
ENSMUST00000106643.8
ENSMUST00000033030.14 |
Parva
|
parvin, alpha |
| chr8_+_110944575 | 1.21 |
ENSMUST00000056972.6
|
Cmtr2
|
cap methyltransferase 2 |
| chr11_-_79853200 | 1.21 |
ENSMUST00000108241.8
ENSMUST00000043152.6 |
Utp6
|
UTP6 small subunit processome component |
| chr17_+_21165573 | 1.21 |
ENSMUST00000007708.14
|
Ppp2r1a
|
protein phosphatase 2, regulatory subunit A, alpha |
| chr12_+_55201889 | 1.21 |
ENSMUST00000110708.4
|
Srp54b
|
signal recognition particle 54B |
| chr3_-_65300000 | 1.20 |
ENSMUST00000029414.12
|
Ssr3
|
signal sequence receptor, gamma |
| chr6_-_124942170 | 1.19 |
ENSMUST00000148485.2
ENSMUST00000129976.8 |
Cops7a
|
COP9 signalosome subunit 7A |
| chr19_+_3332901 | 1.19 |
ENSMUST00000025745.10
ENSMUST00000025743.7 |
Mrpl21
|
mitochondrial ribosomal protein L21 |
| chr19_+_10872778 | 1.19 |
ENSMUST00000179297.3
|
Prpf19
|
pre-mRNA processing factor 19 |
| chr5_+_114238386 | 1.19 |
ENSMUST00000200119.3
|
Usp30
|
ubiquitin specific peptidase 30 |
| chr9_+_108385247 | 1.18 |
ENSMUST00000207810.2
ENSMUST00000207862.2 ENSMUST00000208581.2 ENSMUST00000134939.9 ENSMUST00000207713.2 |
Qars
|
glutaminyl-tRNA synthetase |
| chr11_-_106146868 | 1.18 |
ENSMUST00000021048.7
|
Ftsj3
|
FtsJ RNA methyltransferase homolog 3 (E. coli) |
| chr7_-_144024451 | 1.16 |
ENSMUST00000033407.13
|
Cttn
|
cortactin |
| chr11_+_88095206 | 1.16 |
ENSMUST00000024486.14
|
Mrps23
|
mitochondrial ribosomal protein S23 |
| chr4_-_108637979 | 1.15 |
ENSMUST00000106657.8
|
Zfyve9
|
zinc finger, FYVE domain containing 9 |
| chrX_-_93256291 | 1.15 |
ENSMUST00000050328.15
|
Eif2s3x
|
eukaryotic translation initiation factor 2, subunit 3, structural gene X-linked |
| chr15_+_102898966 | 1.15 |
ENSMUST00000001703.8
|
Hoxc8
|
homeobox C8 |
| chr7_+_23811739 | 1.14 |
ENSMUST00000120006.8
ENSMUST00000005413.4 |
Zfp112
|
zinc finger protein 112 |
| chr14_+_55829165 | 1.14 |
ENSMUST00000019443.15
|
Rnf31
|
ring finger protein 31 |
| chr11_+_106146966 | 1.14 |
ENSMUST00000021049.9
|
Psmc5
|
protease (prosome, macropain) 26S subunit, ATPase 5 |
| chr8_-_73324877 | 1.14 |
ENSMUST00000058733.9
ENSMUST00000167290.8 |
Smim7
|
small integral membrane protein 7 |
| chr12_-_44257109 | 1.13 |
ENSMUST00000015049.5
|
Dnajb9
|
DnaJ heat shock protein family (Hsp40) member B9 |
| chr1_+_167135933 | 1.13 |
ENSMUST00000195015.6
|
Tmco1
|
transmembrane and coiled-coil domains 1 |
| chr2_+_90735077 | 1.13 |
ENSMUST00000111464.8
ENSMUST00000090682.4 |
Kbtbd4
|
kelch repeat and BTB (POZ) domain containing 4 |
| chr14_-_101877870 | 1.12 |
ENSMUST00000168587.3
|
Commd6
|
COMM domain containing 6 |
| chr14_-_55909314 | 1.12 |
ENSMUST00000163750.8
|
Nedd8
|
neural precursor cell expressed, developmentally down-regulated gene 8 |
| chr19_-_6134903 | 1.12 |
ENSMUST00000160977.8
ENSMUST00000159859.2 ENSMUST00000025707.9 ENSMUST00000160712.8 ENSMUST00000237738.2 |
Zfpl1
|
zinc finger like protein 1 |
| chr9_+_107440484 | 1.12 |
ENSMUST00000193418.2
|
Tusc2
|
tumor suppressor 2, mitochondrial calcium regulator |
| chr4_+_118266582 | 1.12 |
ENSMUST00000144577.2
|
Med8
|
mediator complex subunit 8 |
| chr17_-_26063488 | 1.11 |
ENSMUST00000176709.2
|
Rhot2
|
ras homolog family member T2 |
| chr4_-_118346952 | 1.11 |
ENSMUST00000184261.8
|
Tie1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
| chr17_+_15616464 | 1.10 |
ENSMUST00000055352.8
|
Fam120b
|
family with sequence similarity 120, member B |
| chr6_-_124942366 | 1.10 |
ENSMUST00000129446.8
ENSMUST00000032220.15 |
Cops7a
|
COP9 signalosome subunit 7A |
| chr2_-_132657891 | 1.10 |
ENSMUST00000039554.7
|
Trmt6
|
tRNA methyltransferase 6 |
| chr1_-_58625431 | 1.09 |
ENSMUST00000161000.2
ENSMUST00000161600.8 |
Fam126b
|
family with sequence similarity 126, member B |
| chr11_+_96932395 | 1.08 |
ENSMUST00000054252.5
|
Mrpl10
|
mitochondrial ribosomal protein L10 |
| chr14_+_47536075 | 1.08 |
ENSMUST00000227554.2
|
Mapk1ip1l
|
mitogen-activated protein kinase 1 interacting protein 1-like |
| chr5_-_110417682 | 1.05 |
ENSMUST00000196381.2
ENSMUST00000059229.16 ENSMUST00000112505.9 |
Pgam5
|
phosphoglycerate mutase family member 5 |
| chr8_+_114362181 | 1.05 |
ENSMUST00000179926.9
|
Mon1b
|
MON1 homolog B, secretory traffciking associated |
| chr4_+_44012638 | 1.05 |
ENSMUST00000107847.10
ENSMUST00000170241.8 ENSMUST00000107849.10 ENSMUST00000107851.10 ENSMUST00000107845.4 |
Clta
|
clathrin, light polypeptide (Lca) |
| chr8_-_78337226 | 1.05 |
ENSMUST00000034030.15
|
Tmem184c
|
transmembrane protein 184C |
| chr4_-_108637700 | 1.04 |
ENSMUST00000106658.8
|
Zfyve9
|
zinc finger, FYVE domain containing 9 |
| chr18_+_34994253 | 1.04 |
ENSMUST00000165033.2
|
Egr1
|
early growth response 1 |
| chr16_-_31767250 | 1.03 |
ENSMUST00000202722.2
|
0610012G03Rik
|
RIKEN cDNA 0610012G03 gene |
| chr5_+_147797425 | 1.02 |
ENSMUST00000201376.4
ENSMUST00000201120.2 |
Pomp
|
proteasome maturation protein |
| chr12_+_55126999 | 1.02 |
ENSMUST00000220578.2
ENSMUST00000221655.2 |
Srp54a
|
signal recognition particle 54A |
| chr19_+_8848924 | 1.02 |
ENSMUST00000238036.2
|
Ubxn1
|
UBX domain protein 1 |
| chr17_+_46421908 | 1.01 |
ENSMUST00000024763.10
ENSMUST00000123646.2 |
Mrps18a
|
mitochondrial ribosomal protein S18A |
| chr12_-_102724024 | 1.01 |
ENSMUST00000179306.2
ENSMUST00000173969.2 ENSMUST00000179263.2 |
Gm28051
Gon7
|
predicted gene, 28051 GON7 subunit of KEOPS complex |
| chr2_-_94236991 | 1.01 |
ENSMUST00000111237.9
ENSMUST00000094801.5 ENSMUST00000111238.8 |
Ttc17
|
tetratricopeptide repeat domain 17 |
| chr15_+_9140614 | 1.01 |
ENSMUST00000227556.3
|
Lmbrd2
|
LMBR1 domain containing 2 |
| chr15_+_9140685 | 1.00 |
ENSMUST00000090380.6
|
Lmbrd2
|
LMBR1 domain containing 2 |
| chr19_+_8848876 | 0.99 |
ENSMUST00000166407.9
|
Ubxn1
|
UBX domain protein 1 |
| chr11_+_96932379 | 0.99 |
ENSMUST00000001485.10
|
Mrpl10
|
mitochondrial ribosomal protein L10 |
| chr9_-_108338111 | 0.99 |
ENSMUST00000193895.6
|
Klhdc8b
|
kelch domain containing 8B |
| chr19_+_8849000 | 0.98 |
ENSMUST00000096255.7
|
Ubxn1
|
UBX domain protein 1 |
| chr6_-_124942457 | 0.97 |
ENSMUST00000112439.9
|
Cops7a
|
COP9 signalosome subunit 7A |
| chr13_-_4329421 | 0.96 |
ENSMUST00000021632.5
|
Akr1c12
|
aldo-keto reductase family 1, member C12 |
| chr1_+_58625539 | 0.96 |
ENSMUST00000027193.9
|
Ndufb3
|
NADH:ubiquinone oxidoreductase subunit B3 |
| chr5_+_124690908 | 0.95 |
ENSMUST00000071057.14
ENSMUST00000111438.2 |
Ddx55
|
DEAD box helicase 55 |
| chr4_-_98271469 | 0.95 |
ENSMUST00000143116.2
ENSMUST00000030292.12 ENSMUST00000102793.11 |
Tm2d1
|
TM2 domain containing 1 |
| chr10_-_31485180 | 0.95 |
ENSMUST00000081989.8
|
Rnf217
|
ring finger protein 217 |
| chrX_-_106859842 | 0.94 |
ENSMUST00000120722.2
|
2610002M06Rik
|
RIKEN cDNA 2610002M06 gene |
| chr4_-_136613498 | 0.93 |
ENSMUST00000046384.9
|
C1qb
|
complement component 1, q subcomponent, beta polypeptide |
| chr16_+_13721016 | 0.93 |
ENSMUST00000128757.8
|
Mpv17l
|
Mpv17 transgene, kidney disease mutant-like |
| chr9_-_21061196 | 0.93 |
ENSMUST00000215296.2
ENSMUST00000019615.11 |
Cdc37
|
cell division cycle 37 |
| chr2_+_155359868 | 0.92 |
ENSMUST00000029135.15
ENSMUST00000065973.9 |
Acss2
|
acyl-CoA synthetase short-chain family member 2 |
| chr4_+_31964082 | 0.92 |
ENSMUST00000037607.11
ENSMUST00000080933.13 ENSMUST00000108183.8 ENSMUST00000108184.3 |
Map3k7
|
mitogen-activated protein kinase kinase kinase 7 |
| chr1_+_167136217 | 0.91 |
ENSMUST00000193446.6
|
Tmco1
|
transmembrane and coiled-coil domains 1 |
| chr5_-_110928436 | 0.91 |
ENSMUST00000149208.2
ENSMUST00000031483.15 ENSMUST00000086643.12 ENSMUST00000170468.8 ENSMUST00000031481.13 |
Pus1
|
pseudouridine synthase 1 |
| chr12_+_55276953 | 0.91 |
ENSMUST00000218879.2
|
Srp54c
|
signal recognition particle 54C |
| chr4_-_155170738 | 0.90 |
ENSMUST00000030914.4
|
Rer1
|
retention in endoplasmic reticulum sorting receptor 1 |
| chr11_-_120358239 | 0.89 |
ENSMUST00000076921.7
|
Arl16
|
ADP-ribosylation factor-like 16 |
| chr3_-_95778679 | 0.88 |
ENSMUST00000142437.2
ENSMUST00000067298.5 |
Mrps21
|
mitochondrial ribosomal protein S21 |
| chr1_-_36283326 | 0.88 |
ENSMUST00000046875.14
|
Uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chrX_+_41157242 | 0.86 |
ENSMUST00000115095.9
|
Xiap
|
X-linked inhibitor of apoptosis |
| chr12_-_81579614 | 0.86 |
ENSMUST00000169158.2
ENSMUST00000164431.2 ENSMUST00000163402.8 ENSMUST00000166664.2 ENSMUST00000164386.8 |
Synj2bp
Gm20498
|
synaptojanin 2 binding protein predicted gene 20498 |
| chr7_+_5083212 | 0.85 |
ENSMUST00000098845.10
ENSMUST00000146317.8 ENSMUST00000153169.2 ENSMUST00000045277.7 |
Epn1
|
epsin 1 |
| chr15_+_5215000 | 0.85 |
ENSMUST00000118193.8
ENSMUST00000022751.15 |
Ttc33
|
tetratricopeptide repeat domain 33 |
| chr2_+_160730076 | 0.84 |
ENSMUST00000109457.3
|
Lpin3
|
lipin 3 |
| chr9_-_44632680 | 0.84 |
ENSMUST00000148929.2
ENSMUST00000123406.8 |
Phldb1
|
pleckstrin homology like domain, family B, member 1 |
| chr11_-_76969230 | 0.84 |
ENSMUST00000102494.8
|
Nsrp1
|
nuclear speckle regulatory protein 1 |
| chr4_+_124696336 | 0.84 |
ENSMUST00000138807.8
ENSMUST00000030723.3 |
Mtf1
|
metal response element binding transcription factor 1 |
| chr15_-_57755753 | 0.83 |
ENSMUST00000022993.7
|
Derl1
|
Der1-like domain family, member 1 |
| chr10_+_127126643 | 0.83 |
ENSMUST00000026475.15
ENSMUST00000139091.2 ENSMUST00000230446.2 |
Ddit3
Ddit3
|
DNA-damage inducible transcript 3 DNA-damage inducible transcript 3 |
| chr9_+_99457829 | 0.83 |
ENSMUST00000066650.12
ENSMUST00000148987.8 |
Dbr1
|
debranching RNA lariats 1 |
| chr8_+_105066980 | 0.82 |
ENSMUST00000211885.2
|
Cmtm3
|
CKLF-like MARVEL transmembrane domain containing 3 |
| chrX_-_99669507 | 0.81 |
ENSMUST00000059099.7
|
Pdzd11
|
PDZ domain containing 11 |
| chr1_+_134422366 | 0.81 |
ENSMUST00000047978.9
|
Rabif
|
RAB interacting factor |
| chr13_+_46822992 | 0.81 |
ENSMUST00000099547.4
|
Fam8a1
|
family with sequence similarity 8, member A1 |
| chr5_+_114238368 | 0.81 |
ENSMUST00000031588.12
|
Usp30
|
ubiquitin specific peptidase 30 |
| chr8_-_125589697 | 0.81 |
ENSMUST00000034465.9
|
2810004N23Rik
|
RIKEN cDNA 2810004N23 gene |
| chr19_+_8897732 | 0.81 |
ENSMUST00000096243.7
|
B3gat3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chr9_-_79667129 | 0.80 |
ENSMUST00000034881.8
|
Cox7a2
|
cytochrome c oxidase subunit 7A2 |
| chr11_+_50101717 | 0.80 |
ENSMUST00000147468.8
|
Mgat4b
|
mannoside acetylglucosaminyltransferase 4, isoenzyme B |
| chr9_+_20492593 | 0.80 |
ENSMUST00000115557.10
|
Zfp846
|
zinc finger protein 846 |
| chr19_-_27407206 | 0.80 |
ENSMUST00000076219.6
|
Pum3
|
pumilio RNA-binding family member 3 |
| chr10_+_128641663 | 0.78 |
ENSMUST00000218511.2
|
Dnajc14
|
DnaJ heat shock protein family (Hsp40) member C14 |
| chr6_-_35516700 | 0.77 |
ENSMUST00000201026.2
ENSMUST00000031866.9 |
Mtpn
|
myotrophin |
| chr11_+_105017251 | 0.76 |
ENSMUST00000021030.8
|
Mettl2
|
methyltransferase like 2 |
| chr11_-_96807273 | 0.75 |
ENSMUST00000103152.11
|
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr7_+_79910948 | 0.75 |
ENSMUST00000117989.2
|
Ngrn
|
neugrin, neurite outgrowth associated |
| chr18_+_53995260 | 0.73 |
ENSMUST00000237880.2
|
Csnk1g3
|
casein kinase 1, gamma 3 |
| chr14_+_55909816 | 0.72 |
ENSMUST00000227178.2
ENSMUST00000227914.2 |
Gmpr2
|
guanosine monophosphate reductase 2 |
| chr5_+_124767114 | 0.72 |
ENSMUST00000037865.13
|
Atp6v0a2
|
ATPase, H+ transporting, lysosomal V0 subunit A2 |
| chr1_+_171246593 | 0.72 |
ENSMUST00000171362.2
|
Tstd1
|
thiosulfate sulfurtransferase (rhodanese)-like domain containing 1 |
| chr2_+_145776720 | 0.71 |
ENSMUST00000152515.8
ENSMUST00000138774.8 ENSMUST00000130168.8 ENSMUST00000133433.8 ENSMUST00000118002.2 |
Cfap61
|
cilia and flagella associated protein 61 |
| chr18_+_62681982 | 0.70 |
ENSMUST00000055725.12
ENSMUST00000162365.8 |
Spink10
|
serine peptidase inhibitor, Kazal type 10 |
| chr4_-_133005039 | 0.70 |
ENSMUST00000105907.9
|
Tmem222
|
transmembrane protein 222 |
| chr15_-_81931783 | 0.69 |
ENSMUST00000080622.9
|
Snu13
|
SNU13 homolog, small nuclear ribonucleoprotein (U4/U6.U5) |
| chr7_-_46569662 | 0.69 |
ENSMUST00000143413.3
ENSMUST00000014546.15 |
Tsg101
|
tumor susceptibility gene 101 |
| chr2_+_160730019 | 0.69 |
ENSMUST00000109455.9
ENSMUST00000040872.13 |
Lpin3
|
lipin 3 |
| chr15_+_76215711 | 0.68 |
ENSMUST00000169378.2
|
Gpaa1
|
GPI anchor attachment protein 1 |
| chr16_-_87229485 | 0.68 |
ENSMUST00000039449.9
|
Ltn1
|
listerin E3 ubiquitin protein ligase 1 |
| chr9_+_117869642 | 0.68 |
ENSMUST00000134433.8
|
Azi2
|
5-azacytidine induced gene 2 |
| chr15_+_12824925 | 0.68 |
ENSMUST00000090292.13
|
Drosha
|
drosha, ribonuclease type III |
| chr7_+_81412695 | 0.67 |
ENSMUST00000133034.2
|
Ramac
|
RNA guanine-7 methyltransferase activating subunit |
| chr11_+_118319029 | 0.66 |
ENSMUST00000124861.2
|
C1qtnf1
|
C1q and tumor necrosis factor related protein 1 |
| chr13_+_4241149 | 0.66 |
ENSMUST00000021634.4
|
Akr1c13
|
aldo-keto reductase family 1, member C13 |
| chr8_+_105066924 | 0.66 |
ENSMUST00000212081.2
|
Cmtm3
|
CKLF-like MARVEL transmembrane domain containing 3 |
| chr12_+_65012564 | 0.66 |
ENSMUST00000066296.9
ENSMUST00000223166.2 |
Togaram1
|
TOG array regulator of axonemal microtubules 1 |
| chr7_-_80597479 | 0.66 |
ENSMUST00000026818.12
ENSMUST00000117383.8 ENSMUST00000119980.2 |
Sec11a
|
SEC11 homolog A, signal peptidase complex subunit |
| chr14_-_55909527 | 0.65 |
ENSMUST00000010520.10
|
Nedd8
|
neural precursor cell expressed, developmentally down-regulated gene 8 |
| chrX_-_133442596 | 0.65 |
ENSMUST00000054213.5
|
Timm8a1
|
translocase of inner mitochondrial membrane 8A1 |
| chr9_+_44966464 | 0.65 |
ENSMUST00000114664.8
|
Mpzl3
|
myelin protein zero-like 3 |
| chr12_-_55126882 | 0.65 |
ENSMUST00000021406.6
|
2700097O09Rik
|
RIKEN cDNA 2700097O09 gene |
| chr3_-_5641171 | 0.64 |
ENSMUST00000071280.8
ENSMUST00000195855.6 ENSMUST00000165309.8 ENSMUST00000164828.8 |
Pex2
|
peroxisomal biogenesis factor 2 |
| chr14_-_30723292 | 0.63 |
ENSMUST00000228736.2
ENSMUST00000226374.2 |
Spcs1
|
signal peptidase complex subunit 1 homolog (S. cerevisiae) |
| chr8_+_111573646 | 0.62 |
ENSMUST00000172668.8
ENSMUST00000034203.17 ENSMUST00000174398.8 |
Cog4
|
component of oligomeric golgi complex 4 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0000349 | generation of catalytic spliceosome for first transesterification step(GO:0000349) |
| 0.9 | 2.8 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.9 | 2.7 | GO:0032618 | interleukin-15 production(GO:0032618) response to immune response of other organism involved in symbiotic interaction(GO:0052564) response to host immune response(GO:0052572) |
| 0.8 | 8.2 | GO:1903753 | negative regulation of p38MAPK cascade(GO:1903753) |
| 0.8 | 2.4 | GO:0019542 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate biosynthetic process(GO:0019542) |
| 0.5 | 1.6 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.5 | 4.4 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.5 | 1.5 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.5 | 2.0 | GO:2000395 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.5 | 1.5 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.4 | 2.2 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.4 | 3.0 | GO:2000157 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.4 | 1.2 | GO:0046038 | GMP catabolic process(GO:0046038) |
| 0.4 | 1.2 | GO:1903537 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.4 | 2.0 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.4 | 1.5 | GO:1902530 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.4 | 2.6 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.3 | 1.0 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.3 | 1.7 | GO:0071931 | Cajal body organization(GO:0030576) positive regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071931) |
| 0.3 | 1.9 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.3 | 1.2 | GO:0072362 | regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.3 | 1.1 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.3 | 1.3 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.3 | 2.3 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.3 | 0.8 | GO:0002946 | tRNA C5-cytosine methylation(GO:0002946) |
| 0.2 | 2.3 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.2 | 0.8 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.2 | 1.2 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.2 | 3.2 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.2 | 6.8 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.2 | 1.6 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.2 | 3.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.2 | 0.8 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.2 | 1.8 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.2 | 2.0 | GO:0006983 | ER overload response(GO:0006983) |
| 0.2 | 4.5 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.2 | 0.9 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.2 | 1.2 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.2 | 0.5 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) |
| 0.2 | 0.9 | GO:1901026 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.2 | 0.5 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.2 | 0.9 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 0.6 | GO:0046881 | positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.1 | 0.8 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 0.4 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.1 | 3.1 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 0.8 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.1 | 0.5 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.4 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.1 | 3.0 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.1 | 2.3 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 2.2 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.1 | 1.5 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 0.6 | GO:0032055 | negative regulation of translation in response to stress(GO:0032055) positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.1 | 1.8 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.6 | GO:0035553 | oxidative single-stranded DNA demethylation(GO:0035552) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 | 2.6 | GO:0072663 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.1 | 1.6 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 1.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 1.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 4.8 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 | 0.3 | GO:1904430 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) negative regulation of t-circle formation(GO:1904430) |
| 0.1 | 1.2 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.1 | 1.2 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.1 | 1.1 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.1 | 2.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 0.9 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.1 | 0.9 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.1 | 0.9 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.1 | 1.2 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.1 | 4.0 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.1 | 1.8 | GO:0017004 | cytochrome complex assembly(GO:0017004) |
| 0.1 | 2.7 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.1 | 0.8 | GO:1904261 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.1 | 1.1 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 0.7 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.1 | 0.4 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.1 | 0.2 | GO:0007225 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.1 | 2.5 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.1 | 1.8 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 2.3 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 1.1 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.0 | 0.8 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.4 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.6 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 1.9 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 1.1 | GO:0035357 | peroxisome proliferator activated receptor signaling pathway(GO:0035357) |
| 0.0 | 0.7 | GO:1900004 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.5 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.4 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.4 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 1.0 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 1.1 | GO:0070266 | necroptotic process(GO:0070266) |
| 0.0 | 2.6 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.9 | GO:0051084 | 'de novo' posttranslational protein folding(GO:0051084) |
| 0.0 | 0.3 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.0 | 1.2 | GO:0061036 | positive regulation of cartilage development(GO:0061036) |
| 0.0 | 0.5 | GO:0046473 | phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 1.5 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.8 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.6 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.4 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.0 | 0.1 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 1.6 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.6 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) |
| 0.0 | 0.3 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.0 | 0.6 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.3 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.6 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 1.3 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 1.2 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.5 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 1.9 | GO:0045333 | cellular respiration(GO:0045333) |
| 0.0 | 0.1 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.6 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.6 | GO:0015992 | proton transport(GO:0015992) |
| 0.0 | 0.3 | GO:1903427 | negative regulation of reactive oxygen species biosynthetic process(GO:1903427) |
| 0.0 | 0.8 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.3 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 0.5 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 1.6 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.2 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 2.4 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 0.5 | GO:0071174 | mitotic spindle checkpoint(GO:0071174) |
| 0.0 | 0.3 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.6 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.6 | 5.8 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.5 | 3.9 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.5 | 1.5 | GO:0044753 | amphisome(GO:0044753) |
| 0.4 | 1.2 | GO:1903095 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.4 | 1.2 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.3 | 1.0 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.3 | 5.4 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.3 | 3.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.3 | 0.8 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) CHOP-ATF3 complex(GO:1990622) |
| 0.3 | 3.1 | GO:0048500 | signal recognition particle(GO:0048500) |
| 0.2 | 2.3 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.2 | 1.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.2 | 1.7 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.2 | 3.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.2 | 0.6 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.2 | 1.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.2 | 3.0 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.2 | 1.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.2 | 0.9 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.2 | 2.9 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.2 | 5.7 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 2.0 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 4.5 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 1.9 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 1.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 1.1 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 0.5 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 1.0 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 0.7 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 4.7 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 0.4 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.1 | 1.7 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 0.6 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.7 | GO:0001651 | dense fibrillar component(GO:0001651) box C/D snoRNP complex(GO:0031428) |
| 0.1 | 5.0 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 0.6 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 1.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 1.8 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 3.2 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.2 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 1.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.8 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.8 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.3 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 1.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.7 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.4 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 1.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.2 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.6 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 0.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 1.1 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.0 | 3.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.4 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.4 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 2.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.5 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 1.1 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 1.5 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 10.5 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 6.5 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 1.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.9 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.9 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.0 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.0 | GO:0071796 | K6-linked polyubiquitin binding(GO:0071796) |
| 0.7 | 7.8 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.7 | 2.6 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.6 | 1.9 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.6 | 2.4 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.5 | 2.8 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.5 | 2.4 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.4 | 2.2 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.4 | 1.2 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.4 | 1.2 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.4 | 1.2 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.4 | 1.1 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.4 | 2.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.3 | 1.4 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.3 | 1.3 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 0.3 | 5.8 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.3 | 2.1 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.3 | 5.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 1.2 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.2 | 0.7 | GO:0030622 | U4atac snRNA binding(GO:0030622) |
| 0.2 | 0.6 | GO:0051747 | cytosine C-5 DNA demethylase activity(GO:0051747) |
| 0.2 | 1.2 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.2 | 1.9 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.2 | 1.2 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.2 | 0.5 | GO:1990955 | G-rich single-stranded DNA binding(GO:1990955) |
| 0.2 | 1.8 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.2 | 4.4 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.2 | 2.0 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 0.6 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.1 | 2.9 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.6 | GO:0000010 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.1 | 1.3 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.1 | 0.8 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 0.5 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 0.5 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 1.8 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 3.8 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 0.9 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 7.0 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.1 | 0.5 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.1 | 0.8 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.1 | 1.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.8 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.6 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 0.9 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 0.3 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 1.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.8 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 1.6 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 0.9 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 1.0 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 1.9 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 1.6 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.1 | 0.5 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.1 | 2.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 1.5 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.3 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.1 | 1.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.8 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 1.7 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.8 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.8 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.0 | 0.9 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 1.1 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 2.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 2.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.9 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.4 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 3.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 4.8 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 1.2 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.4 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 1.5 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.2 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 2.7 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 2.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.3 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 2.3 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.5 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 0.8 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.4 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.4 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 2.4 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.3 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 2.0 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 1.0 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 1.1 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.2 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.4 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.3 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.6 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 1.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 1.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 3.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.5 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.6 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.0 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 2.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 2.4 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 4.1 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.5 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.6 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 1.2 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 2.4 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.2 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 3.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.8 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.2 | 2.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 1.5 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.2 | 6.7 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 2.6 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 0.9 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 3.1 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 2.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 2.9 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 0.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 1.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 2.2 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 1.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 5.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 3.1 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 0.9 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 2.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 0.4 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.1 | 1.0 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 0.8 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 2.9 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 1.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 2.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.2 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 2.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.1 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 1.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.9 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.7 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 1.2 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 1.0 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 1.5 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.8 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.8 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.5 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.5 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.9 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.0 | 0.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.3 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 1.2 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.0 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 2.8 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |