Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Evx2
Z-value: 0.79
Transcription factors associated with Evx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Evx2
|
ENSMUSG00000001815.16 | Evx2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Evx2 | mm39_v1_chr2_-_74489763_74489901 | 0.10 | 3.8e-01 | Click! |
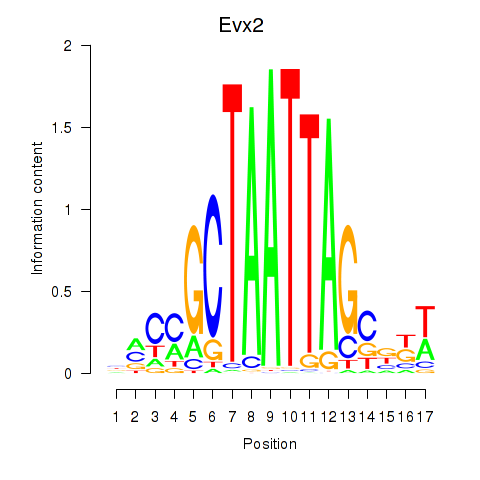
Activity profile of Evx2 motif
Sorted Z-values of Evx2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Evx2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_69415741 | 7.65 |
ENSMUST00000103354.3
|
Igkv4-59
|
immunoglobulin kappa variable 4-59 |
| chr6_-_69394425 | 5.68 |
ENSMUST00000199160.2
|
Igkv4-61
|
immunoglobulin kappa chain variable 4-61 |
| chr6_-_68887922 | 5.12 |
ENSMUST00000103337.3
|
Igkv4-86
|
immunoglobulin kappa variable 4-86 |
| chr6_-_69282389 | 5.01 |
ENSMUST00000103350.3
|
Igkv4-68
|
immunoglobulin kappa variable 4-68 |
| chr6_-_69553484 | 4.92 |
ENSMUST00000103357.4
|
Igkv4-57
|
immunoglobulin kappa variable 4-57 |
| chr6_-_69245427 | 4.84 |
ENSMUST00000103348.3
|
Igkv4-70
|
immunoglobulin kappa chain variable 4-70 |
| chr13_+_51799268 | 4.82 |
ENSMUST00000075853.6
|
Cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr5_-_65090893 | 4.67 |
ENSMUST00000197315.5
|
Tlr1
|
toll-like receptor 1 |
| chr6_-_68994064 | 4.67 |
ENSMUST00000103341.4
|
Igkv4-80
|
immunoglobulin kappa variable 4-80 |
| chr6_-_69204417 | 4.40 |
ENSMUST00000103346.3
|
Igkv4-72
|
immunoglobulin kappa chain variable 4-72 |
| chr6_-_69162381 | 4.32 |
ENSMUST00000103344.3
|
Igkv4-74
|
immunoglobulin kappa variable 4-74 |
| chr6_-_68887957 | 4.31 |
ENSMUST00000200454.2
|
Igkv4-86
|
immunoglobulin kappa variable 4-86 |
| chr2_-_168608949 | 3.98 |
ENSMUST00000075044.10
|
Sall4
|
spalt like transcription factor 4 |
| chr4_-_131802561 | 3.97 |
ENSMUST00000105970.8
ENSMUST00000105975.8 |
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr10_-_128361731 | 3.73 |
ENSMUST00000026427.8
|
Esyt1
|
extended synaptotagmin-like protein 1 |
| chr6_-_68968278 | 3.69 |
ENSMUST00000197966.2
|
Igkv4-81
|
immunoglobulin kappa variable 4-81 |
| chr6_-_69355456 | 3.68 |
ENSMUST00000196595.2
|
Igkv4-63
|
immunoglobulin kappa variable 4-63 |
| chr4_-_131802606 | 3.65 |
ENSMUST00000146021.8
|
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr6_+_41928559 | 3.15 |
ENSMUST00000031898.5
|
Sval1
|
seminal vesicle antigen-like 1 |
| chr1_+_36800874 | 2.94 |
ENSMUST00000027291.7
|
Zap70
|
zeta-chain (TCR) associated protein kinase |
| chr7_-_48493388 | 2.88 |
ENSMUST00000167786.4
|
Csrp3
|
cysteine and glycine-rich protein 3 |
| chr2_-_168609110 | 2.88 |
ENSMUST00000029061.12
ENSMUST00000103074.2 |
Sall4
|
spalt like transcription factor 4 |
| chr17_+_88748139 | 2.87 |
ENSMUST00000112238.9
ENSMUST00000155640.2 |
Foxn2
|
forkhead box N2 |
| chr3_-_15491482 | 2.79 |
ENSMUST00000099201.9
ENSMUST00000194144.3 ENSMUST00000192700.3 |
Sirpb1a
|
signal-regulatory protein beta 1A |
| chr6_-_69678271 | 2.75 |
ENSMUST00000103363.2
|
Igkv4-50
|
immunoglobulin kappa variable 4-50 |
| chr14_-_59602882 | 2.70 |
ENSMUST00000160425.8
ENSMUST00000095157.11 |
Phf11d
|
PHD finger protein 11D |
| chr6_-_68784692 | 2.56 |
ENSMUST00000103334.4
|
Igkv4-90
|
immunoglobulin kappa chain variable 4-90 |
| chr11_+_116734104 | 2.41 |
ENSMUST00000106370.10
|
Mettl23
|
methyltransferase like 23 |
| chr2_-_154916367 | 2.39 |
ENSMUST00000137242.2
ENSMUST00000054607.16 |
Ahcy
|
S-adenosylhomocysteine hydrolase |
| chr15_-_9529898 | 2.38 |
ENSMUST00000228782.2
ENSMUST00000003981.6 |
Il7r
|
interleukin 7 receptor |
| chr2_-_27365633 | 2.36 |
ENSMUST00000138693.8
ENSMUST00000113941.9 ENSMUST00000077737.13 |
Brd3
|
bromodomain containing 3 |
| chr6_-_69020489 | 2.31 |
ENSMUST00000103342.4
|
Igkv4-79
|
immunoglobulin kappa variable 4-79 |
| chr16_+_91184661 | 2.31 |
ENSMUST00000139503.2
|
Ifnar2
|
interferon (alpha and beta) receptor 2 |
| chr14_+_62529924 | 2.30 |
ENSMUST00000166879.8
|
Rnaseh2b
|
ribonuclease H2, subunit B |
| chr10_-_80537805 | 2.28 |
ENSMUST00000218090.2
|
Mob3a
|
MOB kinase activator 3A |
| chr11_-_65160810 | 2.27 |
ENSMUST00000108695.9
|
Myocd
|
myocardin |
| chr14_+_19801333 | 2.27 |
ENSMUST00000022340.5
|
Nid2
|
nidogen 2 |
| chr11_-_65160767 | 2.23 |
ENSMUST00000102635.10
|
Myocd
|
myocardin |
| chr14_-_59602859 | 2.13 |
ENSMUST00000161031.2
|
Phf11d
|
PHD finger protein 11D |
| chr6_-_69261303 | 2.12 |
ENSMUST00000103349.2
|
Igkv4-69
|
immunoglobulin kappa variable 4-69 |
| chr10_+_26648473 | 2.04 |
ENSMUST00000039557.9
|
Arhgap18
|
Rho GTPase activating protein 18 |
| chrX_-_56384089 | 2.02 |
ENSMUST00000033468.11
|
Arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr3_-_15640045 | 1.98 |
ENSMUST00000192382.6
ENSMUST00000195778.3 ENSMUST00000091319.7 |
Sirpb1b
|
signal-regulatory protein beta 1B |
| chr5_-_90788323 | 1.97 |
ENSMUST00000202784.4
ENSMUST00000031317.10 ENSMUST00000201370.2 |
Rassf6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr2_+_118877610 | 1.90 |
ENSMUST00000153300.8
ENSMUST00000028799.12 |
Knl1
|
kinetochore scaffold 1 |
| chr1_-_171854818 | 1.82 |
ENSMUST00000138714.2
ENSMUST00000027837.13 ENSMUST00000111264.8 |
Vangl2
|
VANGL planar cell polarity 2 |
| chr11_+_60428788 | 1.81 |
ENSMUST00000044250.4
|
Alkbh5
|
alkB homolog 5, RNA demethylase |
| chr18_-_64688271 | 1.80 |
ENSMUST00000235459.2
|
Atp8b1
|
ATPase, class I, type 8B, member 1 |
| chr9_-_85631361 | 1.78 |
ENSMUST00000039213.15
|
Ibtk
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr6_-_69377328 | 1.74 |
ENSMUST00000198345.2
|
Igkv4-62
|
immunoglobulin kappa variable 4-62 |
| chr10_-_80537652 | 1.70 |
ENSMUST00000003438.11
|
Mob3a
|
MOB kinase activator 3A |
| chr9_-_107556823 | 1.65 |
ENSMUST00000010205.9
|
Gnat1
|
guanine nucleotide binding protein, alpha transducing 1 |
| chr4_-_132073048 | 1.58 |
ENSMUST00000084250.11
|
Rcc1
|
regulator of chromosome condensation 1 |
| chr12_-_55061117 | 1.50 |
ENSMUST00000172875.8
|
Baz1a
|
bromodomain adjacent to zinc finger domain 1A |
| chr19_+_5138562 | 1.33 |
ENSMUST00000238093.2
ENSMUST00000025811.6 ENSMUST00000237025.2 |
Yif1a
|
Yip1 interacting factor homolog A (S. cerevisiae) |
| chr10_+_39488930 | 1.32 |
ENSMUST00000019987.7
|
Traf3ip2
|
TRAF3 interacting protein 2 |
| chr8_-_35962641 | 1.28 |
ENSMUST00000033927.8
|
Eri1
|
exoribonuclease 1 |
| chr2_-_150097511 | 1.28 |
ENSMUST00000063463.6
|
Gm21994
|
predicted gene 21994 |
| chr5_-_116162415 | 1.27 |
ENSMUST00000031486.14
ENSMUST00000111999.8 |
Prkab1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
| chr19_+_47056161 | 1.24 |
ENSMUST00000026027.7
|
Taf5
|
TATA-box binding protein associated factor 5 |
| chr5_-_90788460 | 1.19 |
ENSMUST00000202704.4
|
Rassf6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr6_+_116627635 | 0.94 |
ENSMUST00000204555.2
|
Depp1
|
DEPP1 autophagy regulator |
| chr6_-_51443602 | 0.92 |
ENSMUST00000203253.2
|
Hnrnpa2b1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr2_-_111820618 | 0.82 |
ENSMUST00000216948.2
ENSMUST00000214935.2 ENSMUST00000217452.2 ENSMUST00000215045.2 |
Olfr1309
|
olfactory receptor 1309 |
| chr4_-_52856572 | 0.80 |
ENSMUST00000051520.3
|
Olfr273
|
olfactory receptor 273 |
| chr3_+_94745009 | 0.78 |
ENSMUST00000107266.8
ENSMUST00000042402.12 ENSMUST00000107269.2 |
Pogz
|
pogo transposable element with ZNF domain |
| chr4_-_132072988 | 0.77 |
ENSMUST00000030726.13
|
Rcc1
|
regulator of chromosome condensation 1 |
| chr14_+_53521353 | 0.76 |
ENSMUST00000103625.3
|
Trav3n-3
|
T cell receptor alpha variable 3N-3 |
| chr17_-_37430949 | 0.72 |
ENSMUST00000214994.2
ENSMUST00000216341.2 |
Olfr92
|
olfactory receptor 92 |
| chr2_+_167345009 | 0.65 |
ENSMUST00000078050.7
|
Rnf114
|
ring finger protein 114 |
| chr10_-_128425519 | 0.65 |
ENSMUST00000082059.7
|
Erbb3
|
erb-b2 receptor tyrosine kinase 3 |
| chr10_+_70010839 | 0.55 |
ENSMUST00000156001.8
ENSMUST00000135607.2 |
Ccdc6
|
coiled-coil domain containing 6 |
| chr3_+_94744844 | 0.54 |
ENSMUST00000107270.9
|
Pogz
|
pogo transposable element with ZNF domain |
| chr8_-_31658775 | 0.52 |
ENSMUST00000033983.6
|
Mak16
|
MAK16 homolog |
| chr19_-_13828056 | 0.49 |
ENSMUST00000208493.3
|
Olfr1501
|
olfactory receptor 1501 |
| chr13_+_38388904 | 0.46 |
ENSMUST00000091641.13
ENSMUST00000178564.2 |
Snrnp48
|
small nuclear ribonucleoprotein 48 (U11/U12) |
| chr6_+_122603369 | 0.45 |
ENSMUST00000049644.9
|
Dppa3
|
developmental pluripotency-associated 3 |
| chr8_+_93581946 | 0.42 |
ENSMUST00000046290.3
ENSMUST00000210099.2 |
Lpcat2
|
lysophosphatidylcholine acyltransferase 2 |
| chr11_+_58468556 | 0.40 |
ENSMUST00000203418.4
|
Olfr325
|
olfactory receptor 325 |
| chr10_-_128885867 | 0.39 |
ENSMUST00000216460.2
|
Olfr765
|
olfactory receptor 765 |
| chr5_+_33493529 | 0.38 |
ENSMUST00000202113.2
|
Maea
|
macrophage erythroblast attacher |
| chr19_-_41921676 | 0.37 |
ENSMUST00000075280.12
ENSMUST00000112123.4 |
Exosc1
|
exosome component 1 |
| chr14_+_73411249 | 0.36 |
ENSMUST00000166875.2
|
Rcbtb2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr2_-_84545504 | 0.36 |
ENSMUST00000035840.6
|
Zdhhc5
|
zinc finger, DHHC domain containing 5 |
| chr4_+_115747862 | 0.36 |
ENSMUST00000132221.3
ENSMUST00000165938.2 |
6430628N08Rik
|
RIKEN cDNA 6430628N08 gene |
| chr1_-_92423800 | 0.35 |
ENSMUST00000204766.2
ENSMUST00000204009.3 |
Olfr1415
|
olfactory receptor 1415 |
| chr18_-_88945571 | 0.34 |
ENSMUST00000147313.2
|
Socs6
|
suppressor of cytokine signaling 6 |
| chr19_-_13827773 | 0.34 |
ENSMUST00000215350.2
|
Olfr1501
|
olfactory receptor 1501 |
| chrX_-_74207359 | 0.33 |
ENSMUST00000101433.9
|
Smim9
|
small integral membrane protein 9 |
| chr6_-_69377081 | 0.32 |
ENSMUST00000177795.2
|
Igkv4-62
|
immunoglobulin kappa variable 4-62 |
| chr3_-_130503041 | 0.30 |
ENSMUST00000043937.9
|
Ostc
|
oligosaccharyltransferase complex subunit (non-catalytic) |
| chr8_+_14938022 | 0.29 |
ENSMUST00000123990.2
ENSMUST00000027554.8 |
Cln8
|
CLN8 transmembrane ER and ERGIC protein |
| chr14_+_65612788 | 0.27 |
ENSMUST00000224687.2
|
Zfp395
|
zinc finger protein 395 |
| chr7_+_12700030 | 0.27 |
ENSMUST00000210619.2
|
Zfp324
|
zinc finger protein 324 |
| chr3_+_121085471 | 0.18 |
ENSMUST00000199554.2
|
Alg14
|
asparagine-linked glycosylation 14 |
| chr7_-_86016045 | 0.16 |
ENSMUST00000213255.2
ENSMUST00000216700.2 ENSMUST00000213869.2 |
Olfr305
|
olfactory receptor 305 |
| chr16_+_3648742 | 0.15 |
ENSMUST00000214238.2
ENSMUST00000214590.2 |
Olfr15
|
olfactory receptor 15 |
| chr2_-_86257093 | 0.13 |
ENSMUST00000217481.2
|
Olfr1062
|
olfactory receptor 1062 |
| chr11_-_61157986 | 0.12 |
ENSMUST00000066277.10
ENSMUST00000074127.14 ENSMUST00000108715.3 |
Aldh3a2
|
aldehyde dehydrogenase family 3, subfamily A2 |
| chr1_-_144427302 | 0.11 |
ENSMUST00000184189.3
|
Rgs21
|
regulator of G-protein signalling 21 |
| chr4_-_42661893 | 0.11 |
ENSMUST00000108006.4
|
Il11ra2
|
interleukin 11 receptor, alpha chain 2 |
| chrX_+_57075981 | 0.11 |
ENSMUST00000088627.11
|
Zic3
|
zinc finger protein of the cerebellum 3 |
| chr3_+_121085500 | 0.08 |
ENSMUST00000198341.2
|
Alg14
|
asparagine-linked glycosylation 14 |
| chr17_+_38143840 | 0.04 |
ENSMUST00000213857.2
|
Olfr125
|
olfactory receptor 125 |
| chr11_-_6217718 | 0.04 |
ENSMUST00000004507.11
ENSMUST00000151446.2 |
Ddx56
|
DEAD box helicase 56 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.7 | GO:0042495 | detection of triacyl bacterial lipopeptide(GO:0042495) |
| 1.5 | 4.5 | GO:1900239 | regulation of phenotypic switching(GO:1900239) negative regulation of vascular associated smooth muscle cell migration(GO:1904753) regulation of cardiac vascular smooth muscle cell differentiation(GO:2000722) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 1.0 | 2.9 | GO:1903920 | positive regulation of actin filament severing(GO:1903920) |
| 0.6 | 1.8 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.6 | 2.4 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.5 | 2.9 | GO:0043366 | beta selection(GO:0043366) |
| 0.4 | 7.6 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.4 | 2.4 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.3 | 0.9 | GO:1990428 | miRNA transport(GO:1990428) |
| 0.3 | 1.8 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.3 | 1.6 | GO:0051342 | sensory perception of umami taste(GO:0050917) regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) |
| 0.3 | 71.4 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.3 | 6.9 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.2 | 1.8 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.1 | 1.3 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.1 | 0.4 | GO:0048822 | enucleate erythrocyte development(GO:0048822) |
| 0.1 | 0.5 | GO:1901536 | negative regulation of DNA demethylation(GO:1901536) |
| 0.1 | 1.9 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.1 | 2.3 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 1.3 | GO:0035878 | nail development(GO:0035878) |
| 0.1 | 0.4 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.1 | 2.3 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.1 | 1.3 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 4.8 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.1 | 0.3 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.1 | 2.3 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.1 | GO:0006714 | sesquiterpenoid metabolic process(GO:0006714) phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 0.0 | 2.8 | GO:0050766 | positive regulation of phagocytosis(GO:0050766) |
| 0.0 | 2.9 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.5 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 2.0 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.1 | GO:0071910 | determination of pancreatic left/right asymmetry(GO:0035469) determination of liver left/right asymmetry(GO:0071910) |
| 0.0 | 0.6 | GO:0014037 | Schwann cell differentiation(GO:0014037) |
| 0.0 | 1.2 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 1.3 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.7 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.6 | 1.8 | GO:0060187 | cell pole(GO:0060187) |
| 0.4 | 2.3 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.4 | 1.5 | GO:0008623 | CHRAC(GO:0008623) |
| 0.2 | 3.7 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.2 | 1.3 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.1 | 7.6 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.1 | 2.9 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 0.4 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 0.9 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 4.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 1.2 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.1 | 1.9 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 6.4 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 0.5 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 1.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.4 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 1.6 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 1.3 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 65.6 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.8 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 4.5 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.5 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 2.3 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.7 | GO:0042497 | triacyl lipopeptide binding(GO:0042497) |
| 0.8 | 4.8 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.5 | 2.3 | GO:0004905 | interferon receptor activity(GO:0004904) type I interferon receptor activity(GO:0004905) type I interferon binding(GO:0019962) |
| 0.4 | 2.9 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.3 | 2.4 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.3 | 2.3 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.3 | 2.3 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.3 | 1.3 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.3 | 1.8 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.2 | 7.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.2 | 1.8 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.2 | 1.8 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 0.4 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.1 | 4.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.9 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 2.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.6 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.1 | 1.3 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 1.6 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 2.9 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 4.6 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 2.4 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 1.2 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 2.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 2.4 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.6 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 4.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.6 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 6.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 2.3 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 2.9 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 1.8 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 2.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 4.5 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 1.4 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.6 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 1.3 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 2.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.4 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.7 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.3 | 2.9 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 2.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 2.3 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 1.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 2.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 2.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.6 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 1.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 3.6 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 1.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 0.3 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |