Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Figla
Z-value: 1.01
Transcription factors associated with Figla
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Figla
|
ENSMUSG00000030001.5 | Figla |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Figla | mm39_v1_chr6_+_85994173_85994214 | 0.35 | 3.0e-03 | Click! |
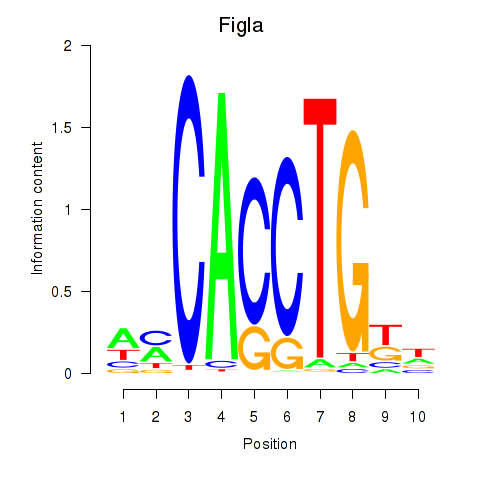
Activity profile of Figla motif
Sorted Z-values of Figla motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Figla
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_152626273 | 6.06 |
ENSMUST00000068875.5
|
Apobec4
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 4 (putative) |
| chr4_-_128154709 | 5.92 |
ENSMUST00000053830.5
|
Hmgb4
|
high-mobility group box 4 |
| chr11_-_99501015 | 5.87 |
ENSMUST00000076478.2
|
Gm11937
|
predicted gene 11937 |
| chr6_-_124519240 | 5.80 |
ENSMUST00000159463.8
ENSMUST00000162844.2 ENSMUST00000160505.8 ENSMUST00000162443.8 |
C1s1
|
complement component 1, s subcomponent 1 |
| chr11_-_99519057 | 5.78 |
ENSMUST00000081007.7
|
Krtap4-1
|
keratin associated protein 4-1 |
| chr14_-_20231871 | 5.66 |
ENSMUST00000024011.10
|
Kcnk5
|
potassium channel, subfamily K, member 5 |
| chrX_+_100427331 | 5.52 |
ENSMUST00000119190.2
|
Gjb1
|
gap junction protein, beta 1 |
| chr11_+_99755302 | 5.39 |
ENSMUST00000092694.4
|
Gm11559
|
predicted gene 11559 |
| chr8_+_106032205 | 5.20 |
ENSMUST00000109375.4
ENSMUST00000212033.2 |
Elmo3
|
engulfment and cell motility 3 |
| chr2_+_122479770 | 5.13 |
ENSMUST00000047498.15
ENSMUST00000110512.4 |
AA467197
|
expressed sequence AA467197 |
| chr6_-_3494587 | 5.03 |
ENSMUST00000049985.15
|
Hepacam2
|
HEPACAM family member 2 |
| chr10_+_127702326 | 5.03 |
ENSMUST00000092058.4
|
Rdh16f2
|
RDH16 family member 2 |
| chr11_-_120441952 | 4.95 |
ENSMUST00000026121.3
|
Ppp1r27
|
protein phosphatase 1, regulatory subunit 27 |
| chr11_+_115714853 | 4.86 |
ENSMUST00000103032.11
ENSMUST00000133250.8 ENSMUST00000177736.8 |
Llgl2
|
LLGL2 scribble cell polarity complex component |
| chr11_+_96820220 | 4.80 |
ENSMUST00000062172.6
|
Prr15l
|
proline rich 15-like |
| chr7_+_30673212 | 4.70 |
ENSMUST00000129773.2
|
Fam187b
|
family with sequence similarity 187, member B |
| chr2_-_91025492 | 4.61 |
ENSMUST00000111354.2
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr11_-_5900019 | 4.54 |
ENSMUST00000102920.4
|
Gck
|
glucokinase |
| chr15_+_88703786 | 4.45 |
ENSMUST00000024042.5
|
Creld2
|
cysteine-rich with EGF-like domains 2 |
| chr16_-_10614679 | 4.36 |
ENSMUST00000023144.6
|
Prm1
|
protamine 1 |
| chr9_-_108443916 | 4.33 |
ENSMUST00000194381.2
|
Ndufaf3
|
NADH:ubiquinone oxidoreductase complex assembly factor 3 |
| chr7_+_27770655 | 4.30 |
ENSMUST00000138392.8
ENSMUST00000076648.8 |
Fcgbp
|
Fc fragment of IgG binding protein |
| chr6_-_135224881 | 4.25 |
ENSMUST00000087729.12
|
Gsg1
|
germ cell associated 1 |
| chr2_-_91025208 | 4.25 |
ENSMUST00000111355.8
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr4_-_49681954 | 4.24 |
ENSMUST00000029991.3
|
Ppp3r2
|
protein phosphatase 3, regulatory subunit B, alpha isoform (calcineurin B, type II) |
| chr7_-_30672824 | 4.23 |
ENSMUST00000147431.2
ENSMUST00000098553.11 ENSMUST00000108116.10 |
Lsr
|
lipolysis stimulated lipoprotein receptor |
| chr11_+_96820091 | 4.22 |
ENSMUST00000054311.6
ENSMUST00000107636.4 |
Prr15l
|
proline rich 15-like |
| chr16_+_4825216 | 4.18 |
ENSMUST00000185147.8
|
Smim22
|
small integral membrane protein 22 |
| chr8_+_55053809 | 4.16 |
ENSMUST00000033917.7
|
Spata4
|
spermatogenesis associated 4 |
| chr8_-_71834543 | 4.14 |
ENSMUST00000002466.9
|
Nr2f6
|
nuclear receptor subfamily 2, group F, member 6 |
| chr7_-_30672747 | 4.13 |
ENSMUST00000205961.2
|
Lsr
|
lipolysis stimulated lipoprotein receptor |
| chr11_-_67812960 | 4.09 |
ENSMUST00000021288.10
ENSMUST00000108677.2 |
Usp43
|
ubiquitin specific peptidase 43 |
| chr5_-_66238313 | 4.06 |
ENSMUST00000202700.4
ENSMUST00000094757.9 ENSMUST00000113724.6 |
Rbm47
|
RNA binding motif protein 47 |
| chr16_+_14523696 | 4.06 |
ENSMUST00000023356.8
|
Snai2
|
snail family zinc finger 2 |
| chr17_+_47747657 | 4.00 |
ENSMUST00000150819.3
|
AI661453
|
expressed sequence AI661453 |
| chr9_-_50657800 | 4.00 |
ENSMUST00000239417.2
ENSMUST00000034564.4 |
2310030G06Rik
|
RIKEN cDNA 2310030G06 gene |
| chr7_-_30672889 | 3.99 |
ENSMUST00000001279.15
|
Lsr
|
lipolysis stimulated lipoprotein receptor |
| chr2_+_164611812 | 3.94 |
ENSMUST00000088248.13
ENSMUST00000001439.7 |
Ube2c
|
ubiquitin-conjugating enzyme E2C |
| chr1_+_171265103 | 3.90 |
ENSMUST00000043839.5
|
F11r
|
F11 receptor |
| chr12_-_40088024 | 3.83 |
ENSMUST00000101472.4
|
Arl4a
|
ADP-ribosylation factor-like 4A |
| chr11_-_99482165 | 3.82 |
ENSMUST00000104930.2
|
Krtap1-3
|
keratin associated protein 1-3 |
| chr2_-_91025380 | 3.81 |
ENSMUST00000111356.8
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr7_+_34818709 | 3.81 |
ENSMUST00000205391.2
ENSMUST00000042985.11 |
Cebpa
|
CCAAT/enhancer binding protein (C/EBP), alpha |
| chr2_-_52225763 | 3.80 |
ENSMUST00000238288.2
ENSMUST00000238749.2 |
Neb
|
nebulin |
| chr6_+_29694181 | 3.78 |
ENSMUST00000046750.14
ENSMUST00000115250.4 |
Tspan33
|
tetraspanin 33 |
| chr11_-_99494134 | 3.73 |
ENSMUST00000072306.4
|
Gm11938
|
predicted gene 11938 |
| chr11_+_99805555 | 3.69 |
ENSMUST00000105053.2
|
Gm11565
|
predicted gene 11565 |
| chr10_+_75729237 | 3.64 |
ENSMUST00000009236.6
ENSMUST00000217811.2 |
Derl3
|
Der1-like domain family, member 3 |
| chr17_-_32639936 | 3.63 |
ENSMUST00000170392.9
ENSMUST00000237165.2 ENSMUST00000235892.2 ENSMUST00000114455.3 |
Pglyrp2
|
peptidoglycan recognition protein 2 |
| chr11_+_45871135 | 3.60 |
ENSMUST00000049038.4
|
Sox30
|
SRY (sex determining region Y)-box 30 |
| chr2_+_153334710 | 3.52 |
ENSMUST00000109783.2
|
4930404H24Rik
|
RIKEN cDNA 4930404H24 gene |
| chr7_-_24145107 | 3.42 |
ENSMUST00000205776.2
|
Irgc1
|
immunity-related GTPase family, cinema 1 |
| chr7_+_126811831 | 3.27 |
ENSMUST00000127710.3
|
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr11_+_117700479 | 3.25 |
ENSMUST00000026649.14
ENSMUST00000177131.8 ENSMUST00000120928.2 ENSMUST00000175737.2 ENSMUST00000132298.2 |
Syngr2
Gm20708
|
synaptogyrin 2 predicted gene 20708 |
| chr19_-_38113056 | 3.22 |
ENSMUST00000236283.2
|
Rbp4
|
retinol binding protein 4, plasma |
| chr16_+_4825170 | 3.14 |
ENSMUST00000178155.9
|
Smim22
|
small integral membrane protein 22 |
| chr17_-_43813664 | 3.10 |
ENSMUST00000024707.9
ENSMUST00000117137.8 |
Mep1a
|
meprin 1 alpha |
| chr10_+_97528915 | 3.07 |
ENSMUST00000060703.6
|
Ccer1
|
coiled-coil glutamate-rich protein 1 |
| chr11_+_101875095 | 3.04 |
ENSMUST00000176722.8
ENSMUST00000175972.2 |
Cfap97d1
|
CFAP97 domain containing 1 |
| chr6_-_120271520 | 3.03 |
ENSMUST00000057283.8
ENSMUST00000212457.2 |
B4galnt3
|
beta-1,4-N-acetyl-galactosaminyl transferase 3 |
| chr17_+_57369231 | 3.01 |
ENSMUST00000097299.10
ENSMUST00000169543.8 ENSMUST00000163763.2 |
Crb3
|
crumbs family member 3 |
| chr9_-_63509747 | 3.01 |
ENSMUST00000080527.12
ENSMUST00000042322.11 |
Iqch
|
IQ motif containing H |
| chr12_-_40087393 | 3.00 |
ENSMUST00000146905.2
|
Arl4a
|
ADP-ribosylation factor-like 4A |
| chr6_-_52194440 | 2.96 |
ENSMUST00000048715.9
|
Hoxa7
|
homeobox A7 |
| chr10_+_108168520 | 2.96 |
ENSMUST00000218332.2
|
Pawr
|
PRKC, apoptosis, WT1, regulator |
| chr11_-_82761954 | 2.93 |
ENSMUST00000108173.10
ENSMUST00000071152.14 |
Rffl
|
ring finger and FYVE like domain containing protein |
| chr3_+_106393348 | 2.92 |
ENSMUST00000183271.2
|
Dennd2d
|
DENN/MADD domain containing 2D |
| chr2_-_92201342 | 2.92 |
ENSMUST00000176810.8
ENSMUST00000090582.11 ENSMUST00000068586.13 |
Large2
|
LARGE xylosyl- and glucuronyltransferase 2 |
| chr9_-_63509699 | 2.91 |
ENSMUST00000171243.2
ENSMUST00000163982.8 ENSMUST00000163624.8 |
Iqch
|
IQ motif containing H |
| chr17_+_47747540 | 2.91 |
ENSMUST00000037701.13
|
AI661453
|
expressed sequence AI661453 |
| chr11_-_68817875 | 2.89 |
ENSMUST00000038932.14
ENSMUST00000125134.2 |
Odf4
|
outer dense fiber of sperm tails 4 |
| chr15_+_99615396 | 2.88 |
ENSMUST00000023760.13
ENSMUST00000162194.2 |
Gpd1
|
glycerol-3-phosphate dehydrogenase 1 (soluble) |
| chr15_-_89263790 | 2.88 |
ENSMUST00000238996.2
|
Odf3b
|
outer dense fiber of sperm tails 3B |
| chr16_+_4825146 | 2.83 |
ENSMUST00000184439.8
|
Smim22
|
small integral membrane protein 22 |
| chr10_+_127595639 | 2.75 |
ENSMUST00000128247.2
|
Rdh16f1
|
RDH16 family member 1 |
| chr2_+_167922924 | 2.74 |
ENSMUST00000052125.7
|
Pard6b
|
par-6 family cell polarity regulator beta |
| chr5_+_20112771 | 2.69 |
ENSMUST00000200443.2
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr9_-_108444561 | 2.64 |
ENSMUST00000074208.6
|
Ndufaf3
|
NADH:ubiquinone oxidoreductase complex assembly factor 3 |
| chr9_-_114673158 | 2.63 |
ENSMUST00000047013.4
|
Cmtm8
|
CKLF-like MARVEL transmembrane domain containing 8 |
| chr3_+_51324022 | 2.61 |
ENSMUST00000192419.6
|
Naa15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| chr11_-_69576363 | 2.59 |
ENSMUST00000018896.14
|
Tnfsf13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr11_-_100036792 | 2.58 |
ENSMUST00000007317.8
|
Krt19
|
keratin 19 |
| chr1_+_59555423 | 2.58 |
ENSMUST00000114243.8
|
Gm973
|
predicted gene 973 |
| chr11_+_53661251 | 2.57 |
ENSMUST00000138913.8
ENSMUST00000123376.8 ENSMUST00000019043.13 ENSMUST00000133291.3 |
Irf1
|
interferon regulatory factor 1 |
| chr2_-_168607166 | 2.56 |
ENSMUST00000137536.2
|
Sall4
|
spalt like transcription factor 4 |
| chr10_-_21036792 | 2.53 |
ENSMUST00000188495.8
|
Myb
|
myeloblastosis oncogene |
| chr15_-_89263128 | 2.52 |
ENSMUST00000227834.2
|
Odf3b
|
outer dense fiber of sperm tails 3B |
| chr9_+_107424488 | 2.51 |
ENSMUST00000010188.9
|
Zmynd10
|
zinc finger, MYND domain containing 10 |
| chr16_+_4653271 | 2.44 |
ENSMUST00000100211.11
|
4930562C15Rik
|
RIKEN cDNA 4930562C15 gene |
| chr11_-_99511257 | 2.44 |
ENSMUST00000073853.3
|
Gm11562
|
predicted gene 11562 |
| chr12_+_78795647 | 2.42 |
ENSMUST00000219667.2
|
Mpp5
|
membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5) |
| chr6_+_54303837 | 2.37 |
ENSMUST00000059138.6
|
Prr15
|
proline rich 15 |
| chr8_+_123148759 | 2.32 |
ENSMUST00000050963.4
|
Il17c
|
interleukin 17C |
| chr13_-_100922910 | 2.28 |
ENSMUST00000174038.2
ENSMUST00000091295.14 ENSMUST00000072119.15 |
Ccnb1
|
cyclin B1 |
| chr17_-_56607286 | 2.26 |
ENSMUST00000097303.3
|
Arrdc5
|
arrestin domain containing 5 |
| chr15_-_89263448 | 2.22 |
ENSMUST00000049968.9
|
Odf3b
|
outer dense fiber of sperm tails 3B |
| chrX_+_21581135 | 2.20 |
ENSMUST00000033414.8
|
Slc6a14
|
solute carrier family 6 (neurotransmitter transporter), member 14 |
| chr15_+_25622611 | 2.19 |
ENSMUST00000110457.8
ENSMUST00000137601.8 |
Myo10
|
myosin X |
| chr3_-_20329823 | 2.19 |
ENSMUST00000011607.6
|
Cpb1
|
carboxypeptidase B1 (tissue) |
| chr7_+_18828519 | 2.18 |
ENSMUST00000049454.6
|
Six5
|
sine oculis-related homeobox 5 |
| chr5_-_28415020 | 2.18 |
ENSMUST00000118882.2
|
Cnpy1
|
canopy FGF signaling regulator 1 |
| chr1_-_156168522 | 2.17 |
ENSMUST00000212747.2
|
Axdnd1
|
axonemal dynein light chain domain containing 1 |
| chr11_+_69983531 | 2.16 |
ENSMUST00000124721.2
|
Asgr2
|
asialoglycoprotein receptor 2 |
| chr14_-_21798694 | 2.14 |
ENSMUST00000183943.2
|
Dusp13
|
dual specificity phosphatase 13 |
| chr6_-_52194414 | 2.14 |
ENSMUST00000140316.2
|
Hoxa7
|
homeobox A7 |
| chrX_-_52203405 | 2.10 |
ENSMUST00000114843.9
|
Plac1
|
placental specific protein 1 |
| chr4_+_106418224 | 2.07 |
ENSMUST00000047973.4
|
Dhcr24
|
24-dehydrocholesterol reductase |
| chr6_+_39358036 | 2.06 |
ENSMUST00000031986.5
|
Rab19
|
RAB19, member RAS oncogene family |
| chr11_+_69983459 | 2.05 |
ENSMUST00000102572.8
|
Asgr2
|
asialoglycoprotein receptor 2 |
| chr9_-_50679408 | 2.03 |
ENSMUST00000177546.8
ENSMUST00000176238.2 |
1110032A03Rik
|
RIKEN cDNA 1110032A03 gene |
| chr14_-_21798678 | 2.02 |
ENSMUST00000075040.9
|
Dusp13
|
dual specificity phosphatase 13 |
| chr10_-_21036824 | 2.01 |
ENSMUST00000020158.9
|
Myb
|
myeloblastosis oncogene |
| chr19_+_36325683 | 2.01 |
ENSMUST00000225920.2
|
Pcgf5
|
polycomb group ring finger 5 |
| chr9_+_107445101 | 1.99 |
ENSMUST00000192887.6
ENSMUST00000195752.6 |
Hyal2
|
hyaluronoglucosaminidase 2 |
| chr10_+_50770836 | 1.99 |
ENSMUST00000219436.2
|
Sim1
|
single-minded family bHLH transcription factor 1 |
| chr14_-_20319242 | 1.95 |
ENSMUST00000024155.9
|
Kcnk16
|
potassium channel, subfamily K, member 16 |
| chr2_+_152753231 | 1.95 |
ENSMUST00000028970.8
|
Mylk2
|
myosin, light polypeptide kinase 2, skeletal muscle |
| chr12_+_31315227 | 1.94 |
ENSMUST00000169088.8
|
Lamb1
|
laminin B1 |
| chr5_-_30401416 | 1.91 |
ENSMUST00000125367.4
|
Adgrf3
|
adhesion G protein-coupled receptor F3 |
| chr3_+_40663285 | 1.90 |
ENSMUST00000091184.9
|
Slc25a31
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 31 |
| chr1_-_192880260 | 1.89 |
ENSMUST00000161367.2
|
Traf3ip3
|
TRAF3 interacting protein 3 |
| chr11_-_48836975 | 1.89 |
ENSMUST00000104958.2
|
Psme2b
|
protease (prosome, macropain) activator subunit 2B |
| chr9_-_97252011 | 1.87 |
ENSMUST00000035026.5
|
Trim42
|
tripartite motif-containing 42 |
| chr10_-_63257568 | 1.86 |
ENSMUST00000054760.6
|
Gm7075
|
predicted gene 7075 |
| chr11_-_99742434 | 1.85 |
ENSMUST00000107437.2
|
Krtap4-16
|
keratin associated protein 4-16 |
| chr3_+_65435825 | 1.85 |
ENSMUST00000047906.10
|
Tiparp
|
TCDD-inducible poly(ADP-ribose) polymerase |
| chr17_-_56607250 | 1.84 |
ENSMUST00000233911.2
|
Arrdc5
|
arrestin domain containing 5 |
| chr9_+_51027300 | 1.84 |
ENSMUST00000114431.4
ENSMUST00000213117.2 |
Btg4
|
BTG anti-proliferation factor 4 |
| chr11_+_69983479 | 1.83 |
ENSMUST00000143772.8
|
Asgr2
|
asialoglycoprotein receptor 2 |
| chr8_+_120121612 | 1.81 |
ENSMUST00000098367.5
|
Mlycd
|
malonyl-CoA decarboxylase |
| chr14_-_47426863 | 1.81 |
ENSMUST00000089959.7
|
Gch1
|
GTP cyclohydrolase 1 |
| chr5_-_28415166 | 1.81 |
ENSMUST00000117098.2
|
Cnpy1
|
canopy FGF signaling regulator 1 |
| chr9_-_44253630 | 1.80 |
ENSMUST00000097558.5
|
Hmbs
|
hydroxymethylbilane synthase |
| chr8_+_93084253 | 1.79 |
ENSMUST00000210246.2
ENSMUST00000034184.12 |
Irx5
|
Iroquois homeobox 5 |
| chr7_+_79675727 | 1.75 |
ENSMUST00000049680.10
|
Zfp710
|
zinc finger protein 710 |
| chr11_+_72326391 | 1.74 |
ENSMUST00000100903.3
|
Ggt6
|
gamma-glutamyltransferase 6 |
| chr5_+_123887759 | 1.74 |
ENSMUST00000031366.12
|
Kntc1
|
kinetochore associated 1 |
| chr3_+_102981326 | 1.73 |
ENSMUST00000090715.13
ENSMUST00000155034.6 |
Ampd1
|
adenosine monophosphate deaminase 1 |
| chr7_+_48993197 | 1.73 |
ENSMUST00000207743.2
|
Nav2
|
neuron navigator 2 |
| chr17_+_57369490 | 1.72 |
ENSMUST00000163628.2
|
Crb3
|
crumbs family member 3 |
| chr13_+_113346193 | 1.71 |
ENSMUST00000038144.9
|
Esm1
|
endothelial cell-specific molecule 1 |
| chr2_-_52225146 | 1.70 |
ENSMUST00000075301.10
|
Neb
|
nebulin |
| chr17_+_35413415 | 1.70 |
ENSMUST00000025262.6
ENSMUST00000173600.2 |
Ltb
|
lymphotoxin B |
| chr4_+_122889407 | 1.69 |
ENSMUST00000144998.2
|
Mycl
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived |
| chr5_-_138170077 | 1.68 |
ENSMUST00000155902.8
ENSMUST00000148879.8 |
Mcm7
|
minichromosome maintenance complex component 7 |
| chr12_+_31315270 | 1.65 |
ENSMUST00000002979.16
ENSMUST00000239496.2 ENSMUST00000170495.3 |
Lamb1
|
laminin B1 |
| chr7_+_27879650 | 1.64 |
ENSMUST00000172467.8
|
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chrX_-_73325318 | 1.64 |
ENSMUST00000239458.2
ENSMUST00000019232.10 |
Dnase1l1
|
deoxyribonuclease 1-like 1 |
| chr8_+_46338557 | 1.63 |
ENSMUST00000210422.2
|
Pdlim3
|
PDZ and LIM domain 3 |
| chr1_+_165288606 | 1.63 |
ENSMUST00000027853.6
|
Mpc2
|
mitochondrial pyruvate carrier 2 |
| chr9_-_44253588 | 1.63 |
ENSMUST00000215091.2
|
Hmbs
|
hydroxymethylbilane synthase |
| chr4_-_137157824 | 1.63 |
ENSMUST00000102522.5
|
Cela3b
|
chymotrypsin-like elastase family, member 3B |
| chr11_-_99505668 | 1.62 |
ENSMUST00000074926.6
|
Krtap2-4
|
keratin associated protein 2-4 |
| chr11_-_99920694 | 1.58 |
ENSMUST00000073890.4
|
Krt33b
|
keratin 33B |
| chr15_-_65784103 | 1.56 |
ENSMUST00000079776.14
|
Oc90
|
otoconin 90 |
| chr9_+_43978369 | 1.56 |
ENSMUST00000177054.8
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr3_+_102981352 | 1.55 |
ENSMUST00000176440.2
|
Ampd1
|
adenosine monophosphate deaminase 1 |
| chr13_+_21364330 | 1.55 |
ENSMUST00000223065.2
|
Trim27
|
tripartite motif-containing 27 |
| chr4_+_107825529 | 1.53 |
ENSMUST00000106713.5
ENSMUST00000238795.2 |
Slc1a7
|
solute carrier family 1 (glutamate transporter), member 7 |
| chr1_-_155617773 | 1.53 |
ENSMUST00000027740.14
|
Lhx4
|
LIM homeobox protein 4 |
| chr5_+_146168020 | 1.52 |
ENSMUST00000161181.8
ENSMUST00000161652.8 ENSMUST00000031640.15 ENSMUST00000159467.2 |
Cdk8
|
cyclin-dependent kinase 8 |
| chr10_-_75768302 | 1.51 |
ENSMUST00000120281.8
ENSMUST00000000924.13 |
Mmp11
|
matrix metallopeptidase 11 |
| chr11_+_53324126 | 1.51 |
ENSMUST00000018382.7
|
Gdf9
|
growth differentiation factor 9 |
| chr15_+_59520199 | 1.50 |
ENSMUST00000067543.8
|
Trib1
|
tribbles pseudokinase 1 |
| chr9_+_57468217 | 1.49 |
ENSMUST00000045791.11
ENSMUST00000216986.2 |
Scamp2
|
secretory carrier membrane protein 2 |
| chr6_-_93889483 | 1.48 |
ENSMUST00000205116.3
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr8_-_123349473 | 1.47 |
ENSMUST00000212966.2
ENSMUST00000093059.3 |
Pabpn1l
|
poly(A)binding protein nuclear 1-like |
| chr5_+_75236250 | 1.47 |
ENSMUST00000040477.4
ENSMUST00000160104.3 |
Gsx2
|
GS homeobox 2 |
| chr4_+_116414855 | 1.46 |
ENSMUST00000030460.15
|
Gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr8_+_70953766 | 1.45 |
ENSMUST00000127983.2
|
Crlf1
|
cytokine receptor-like factor 1 |
| chr14_-_104705420 | 1.43 |
ENSMUST00000053016.10
|
Pou4f1
|
POU domain, class 4, transcription factor 1 |
| chr4_+_150233362 | 1.42 |
ENSMUST00000059893.8
|
Slc2a7
|
solute carrier family 2 (facilitated glucose transporter), member 7 |
| chr6_-_125357756 | 1.41 |
ENSMUST00000042647.7
|
Plekhg6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6 |
| chr7_+_78564062 | 1.41 |
ENSMUST00000205981.2
|
Isg20
|
interferon-stimulated protein |
| chr14_-_31552335 | 1.41 |
ENSMUST00000228037.2
|
Ankrd28
|
ankyrin repeat domain 28 |
| chr7_+_78563964 | 1.41 |
ENSMUST00000120331.4
|
Isg20
|
interferon-stimulated protein |
| chr3_-_32546380 | 1.40 |
ENSMUST00000164954.3
|
Kcnmb3
|
potassium large conductance calcium-activated channel, subfamily M, beta member 3 |
| chr14_+_54406112 | 1.39 |
ENSMUST00000103694.3
ENSMUST00000199452.2 |
Traj49
|
T cell receptor alpha joining 49 |
| chr19_-_11243530 | 1.38 |
ENSMUST00000169159.3
|
Ms4a1
|
membrane-spanning 4-domains, subfamily A, member 1 |
| chr10_-_128579879 | 1.38 |
ENSMUST00000026414.9
|
Dgka
|
diacylglycerol kinase, alpha |
| chr13_-_95615229 | 1.37 |
ENSMUST00000022186.5
|
S100z
|
S100 calcium binding protein, zeta |
| chr15_+_59520493 | 1.36 |
ENSMUST00000118228.2
|
Trib1
|
tribbles pseudokinase 1 |
| chr4_-_137137088 | 1.33 |
ENSMUST00000024200.7
|
Cela3a
|
chymotrypsin-like elastase family, member 3A |
| chr15_-_89263466 | 1.32 |
ENSMUST00000228111.2
|
Odf3b
|
outer dense fiber of sperm tails 3B |
| chr4_-_126362372 | 1.30 |
ENSMUST00000097888.10
ENSMUST00000239399.2 |
Ago1
|
argonaute RISC catalytic subunit 1 |
| chr15_-_65784246 | 1.30 |
ENSMUST00000060522.11
|
Oc90
|
otoconin 90 |
| chr7_-_142233270 | 1.29 |
ENSMUST00000162317.2
ENSMUST00000125933.2 ENSMUST00000105931.8 ENSMUST00000105930.8 ENSMUST00000105933.8 ENSMUST00000105932.2 ENSMUST00000000220.3 |
Ins2
|
insulin II |
| chr7_-_80051455 | 1.29 |
ENSMUST00000120753.3
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr2_-_110144869 | 1.28 |
ENSMUST00000133608.2
|
Bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase 1 (gamma-butyrobetaine hydroxylase) |
| chr2_-_122441719 | 1.27 |
ENSMUST00000028624.9
|
Gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr13_+_47347301 | 1.26 |
ENSMUST00000110111.4
|
Rnf144b
|
ring finger protein 144B |
| chr8_-_48443525 | 1.26 |
ENSMUST00000057561.9
|
Wwc2
|
WW, C2 and coiled-coil domain containing 2 |
| chr9_+_107431776 | 1.24 |
ENSMUST00000010211.7
|
Rassf1
|
Ras association (RalGDS/AF-6) domain family member 1 |
| chr7_+_44866635 | 1.22 |
ENSMUST00000097216.5
ENSMUST00000209343.2 ENSMUST00000209678.2 |
Tead2
|
TEA domain family member 2 |
| chr11_+_43419586 | 1.21 |
ENSMUST00000050574.7
|
Ccnjl
|
cyclin J-like |
| chr11_-_99932373 | 1.20 |
ENSMUST00000056362.3
|
Krt34
|
keratin 34 |
| chr2_-_28453374 | 1.19 |
ENSMUST00000028161.6
|
Cel
|
carboxyl ester lipase |
| chr15_-_78290038 | 1.18 |
ENSMUST00000058659.9
|
Tst
|
thiosulfate sulfurtransferase, mitochondrial |
| chr2_+_132688558 | 1.17 |
ENSMUST00000028835.13
ENSMUST00000110122.10 |
Crls1
|
cardiolipin synthase 1 |
| chr8_-_44080313 | 1.16 |
ENSMUST00000056023.5
|
Adam34l
|
a disintegrin and metallopeptidase domain 34 like |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 12.3 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 3.2 | 12.7 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 1.4 | 4.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 1.2 | 4.9 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 1.1 | 3.4 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 1.0 | 3.0 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) positive regulation of hindgut contraction(GO:0060450) regulation of relaxation of smooth muscle(GO:1901080) positive regulation of relaxation of smooth muscle(GO:1901082) |
| 1.0 | 2.9 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 1.0 | 2.9 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.9 | 4.5 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.9 | 3.6 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.9 | 2.6 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.7 | 2.9 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.6 | 3.2 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.6 | 2.6 | GO:0035740 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.6 | 3.6 | GO:0032824 | negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.6 | 1.8 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.6 | 2.3 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 0.5 | 1.6 | GO:1902361 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.5 | 2.0 | GO:0019064 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.5 | 3.9 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.5 | 1.9 | GO:0031448 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.5 | 2.8 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.5 | 5.5 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.5 | 1.8 | GO:0014916 | regulation of lung blood pressure(GO:0014916) |
| 0.4 | 5.8 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.4 | 2.2 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.4 | 1.3 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.4 | 4.7 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.4 | 1.7 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.4 | 5.1 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.4 | 1.3 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.4 | 1.2 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.4 | 1.5 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.4 | 1.5 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.4 | 7.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.4 | 1.4 | GO:0048880 | proprioception(GO:0019230) sensory system development(GO:0048880) |
| 0.3 | 2.1 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.3 | 4.3 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.3 | 3.6 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.3 | 3.3 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.3 | 4.5 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.3 | 1.3 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.3 | 1.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.3 | 4.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.3 | 2.6 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.3 | 4.2 | GO:0007320 | insemination(GO:0007320) |
| 0.3 | 2.6 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.3 | 1.0 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.3 | 1.5 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.2 | 1.7 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.2 | 2.2 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.2 | 1.7 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.2 | 2.9 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.2 | 0.7 | GO:0035638 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.2 | 0.9 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.2 | 0.7 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.2 | 2.6 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.2 | 3.9 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.2 | 4.5 | GO:0050961 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.2 | 0.6 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.2 | 1.8 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.2 | 2.5 | GO:2000980 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.2 | 2.5 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.2 | 0.8 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.2 | 0.8 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.2 | 1.1 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.2 | 0.7 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.2 | 2.1 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.2 | 8.1 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.2 | 2.3 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.2 | 0.8 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.2 | 0.7 | GO:1901250 | regulation of lung goblet cell differentiation(GO:1901249) negative regulation of lung goblet cell differentiation(GO:1901250) |
| 0.2 | 1.3 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.2 | 1.4 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 1.5 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 0.7 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 2.2 | GO:0014842 | regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.1 | 3.9 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.1 | 0.9 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.1 | 1.3 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 0.7 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 1.7 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.1 | 2.4 | GO:0008105 | asymmetric protein localization(GO:0008105) |
| 0.1 | 1.2 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.1 | 0.8 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.1 | 0.8 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 1.2 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.1 | 2.7 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.1 | 0.9 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.1 | 1.0 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 2.6 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 4.4 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.1 | 1.4 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.1 | 8.0 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.1 | 1.2 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 0.6 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.1 | 1.5 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.1 | 0.3 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.1 | 1.5 | GO:0090209 | negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.1 | 0.4 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 2.6 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.1 | 1.1 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.1 | 2.1 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.1 | 2.9 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 0.8 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.1 | 4.9 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.1 | 1.5 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 0.4 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 | 0.4 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 1.2 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.0 | 0.6 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 1.2 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.4 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.0 | 0.6 | GO:1900040 | regulation of interleukin-2 secretion(GO:1900040) |
| 0.0 | 0.5 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 1.1 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 5.6 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 0.4 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.7 | GO:0046643 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.0 | 0.4 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.6 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 1.4 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 2.0 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.5 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.2 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.5 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 1.6 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 1.0 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.4 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 2.9 | GO:0007043 | cell-cell junction assembly(GO:0007043) |
| 0.0 | 3.0 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 1.1 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.7 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 1.8 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 2.8 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.8 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.4 | GO:0018195 | peptidyl-arginine modification(GO:0018195) |
| 0.0 | 0.3 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 2.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 1.0 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.3 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.5 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.1 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.0 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 1.3 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 12.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 1.3 | 3.8 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 1.2 | 3.6 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.6 | 1.7 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.6 | 2.9 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.6 | 2.3 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.5 | 3.6 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.3 | 3.8 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.3 | 6.0 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.3 | 1.2 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.3 | 1.5 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.3 | 2.6 | GO:1990357 | terminal web(GO:1990357) |
| 0.3 | 0.8 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.3 | 5.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.3 | 2.4 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.2 | 4.5 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 2.6 | GO:0031415 | NatA complex(GO:0031415) |
| 0.2 | 5.7 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 2.9 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.2 | 3.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.2 | 2.9 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 4.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 0.4 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.1 | 4.4 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 0.5 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.1 | 1.3 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) RISC-loading complex(GO:0070578) |
| 0.1 | 0.8 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.5 | GO:0035363 | histone locus body(GO:0035363) |
| 0.1 | 6.1 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.1 | 0.5 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 0.4 | GO:0031251 | PAN complex(GO:0031251) |
| 0.1 | 0.7 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 2.0 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 2.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 1.3 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 1.8 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.4 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 11.9 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 1.3 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.1 | 1.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 1.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 2.8 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.9 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 1.1 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 1.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.6 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 1.5 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.7 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 1.1 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 1.2 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 1.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.6 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.8 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 1.3 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 1.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 1.2 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 5.1 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 4.8 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.6 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 7.6 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 1.1 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 5.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 2.6 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 1.5 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 7.5 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 1.0 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 1.8 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 2.9 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 2.4 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 1.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 3.4 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 3.6 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.4 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.4 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 12.7 | GO:0032810 | sterol response element binding(GO:0032810) |
| 1.1 | 3.4 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 1.1 | 3.2 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 1.0 | 2.9 | GO:0016901 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.9 | 4.5 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.6 | 3.6 | GO:0016019 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.6 | 1.7 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.6 | 2.8 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.5 | 4.8 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.5 | 2.0 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.5 | 12.3 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.5 | 4.2 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.5 | 4.5 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) |
| 0.4 | 2.2 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.4 | 2.3 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.3 | 7.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.3 | 0.3 | GO:0031762 | alpha-1A adrenergic receptor binding(GO:0031691) alpha-1B adrenergic receptor binding(GO:0031692) follicle-stimulating hormone receptor binding(GO:0031762) |
| 0.3 | 3.3 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.3 | 2.9 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.3 | 5.7 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.3 | 2.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.3 | 2.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 1.6 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.2 | 1.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.2 | 3.8 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) |
| 0.2 | 1.8 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.2 | 1.0 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.2 | 1.2 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.2 | 3.0 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.2 | 5.1 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 2.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 6.1 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.1 | 10.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 1.0 | GO:0070404 | NADH binding(GO:0070404) |
| 0.1 | 1.7 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 0.4 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 1.5 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 2.6 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 0.8 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 4.0 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 2.9 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 4.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 0.9 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.1 | 1.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 1.0 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.1 | 3.0 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.1 | 1.3 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 1.5 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 2.9 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 0.4 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 2.9 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 4.7 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.1 | 2.2 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.7 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.1 | 4.2 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 1.2 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.1 | 2.1 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 1.2 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.1 | 1.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 1.1 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 6.4 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 1.5 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 1.4 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 2.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.4 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 11.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.8 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.7 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 0.3 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 4.1 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 1.1 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.4 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.6 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 1.2 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 2.1 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 2.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.7 | GO:0042165 | neurotransmitter binding(GO:0042165) |
| 0.0 | 1.1 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 3.3 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 8.1 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 1.8 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.2 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 1.3 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.0 | 1.3 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 4.8 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 2.5 | GO:0004536 | deoxyribonuclease activity(GO:0004536) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.6 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 2.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 8.7 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 1.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.3 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.5 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 2.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 12.2 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.4 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 2.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 3.2 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.8 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.5 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.0 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 1.7 | GO:0008201 | heparin binding(GO:0008201) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.6 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 11.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 3.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 8.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.6 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 5.2 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 2.2 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.1 | 2.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 2.3 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 2.6 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 3.0 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 3.2 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.8 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 2.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.1 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 2.2 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 1.0 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.6 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 1.3 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.7 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.7 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 2.2 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 1.4 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.2 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.2 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 3.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 13.8 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.3 | 6.2 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.2 | 5.7 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 2.9 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.2 | 1.2 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.2 | 2.0 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 16.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 4.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 3.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.0 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 3.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 2.0 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 1.5 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 1.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 1.8 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 1.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 1.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 1.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 2.6 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 2.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 2.7 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 1.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 3.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 2.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 0.9 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 1.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 2.2 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 2.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.9 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 4.1 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.5 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 1.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.7 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 2.2 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 2.6 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.6 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 1.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.4 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.8 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.5 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.7 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.5 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |