Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Fli1
Z-value: 1.86
Transcription factors associated with Fli1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Fli1
|
ENSMUSG00000016087.14 | Fli1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Fli1 | mm39_v1_chr9_-_32452885_32452898 | -0.20 | 8.6e-02 | Click! |
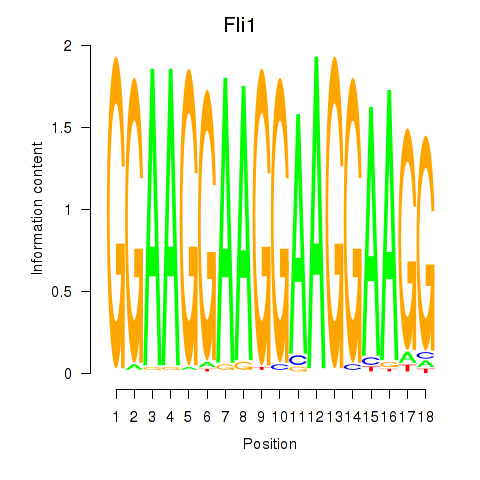
Activity profile of Fli1 motif
Sorted Z-values of Fli1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Fli1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_40175709 | 81.19 |
ENSMUST00000051846.13
|
Cyp2c70
|
cytochrome P450, family 2, subfamily c, polypeptide 70 |
| chr5_-_89583469 | 52.96 |
ENSMUST00000200534.2
|
Gc
|
vitamin D binding protein |
| chr9_-_57590926 | 38.84 |
ENSMUST00000034860.5
|
Cyp1a2
|
cytochrome P450, family 1, subfamily a, polypeptide 2 |
| chr7_-_26638802 | 36.16 |
ENSMUST00000170227.3
|
Cyp2a22
|
cytochrome P450, family 2, subfamily a, polypeptide 22 |
| chr15_-_96918203 | 32.59 |
ENSMUST00000166223.2
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr15_-_96917804 | 30.86 |
ENSMUST00000231039.2
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr8_-_25066313 | 24.39 |
ENSMUST00000121992.2
|
Ido2
|
indoleamine 2,3-dioxygenase 2 |
| chr17_-_32643131 | 23.97 |
ENSMUST00000236386.2
|
Pglyrp2
|
peptidoglycan recognition protein 2 |
| chr17_-_32643067 | 22.40 |
ENSMUST00000237130.2
|
Pglyrp2
|
peptidoglycan recognition protein 2 |
| chr14_-_55204023 | 15.59 |
ENSMUST00000124930.8
|
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha |
| chr7_-_19410749 | 14.23 |
ENSMUST00000003074.16
|
Apoc2
|
apolipoprotein C-II |
| chr7_-_13571334 | 14.03 |
ENSMUST00000108522.5
|
Sult2a1
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 1 |
| chr13_-_4573312 | 13.19 |
ENSMUST00000221564.2
ENSMUST00000078239.5 ENSMUST00000080361.13 |
Akr1c20
|
aldo-keto reductase family 1, member C20 |
| chr16_+_37400500 | 11.69 |
ENSMUST00000160847.2
|
Hgd
|
homogentisate 1, 2-dioxygenase |
| chr16_+_37400590 | 11.30 |
ENSMUST00000159787.8
|
Hgd
|
homogentisate 1, 2-dioxygenase |
| chr11_-_5865124 | 10.90 |
ENSMUST00000109823.9
ENSMUST00000109822.8 |
Gck
|
glucokinase |
| chr10_-_93375832 | 10.80 |
ENSMUST00000016034.3
|
Amdhd1
|
amidohydrolase domain containing 1 |
| chr7_-_48493388 | 10.32 |
ENSMUST00000167786.4
|
Csrp3
|
cysteine and glycine-rich protein 3 |
| chr13_+_4484305 | 9.40 |
ENSMUST00000021630.15
|
Akr1c6
|
aldo-keto reductase family 1, member C6 |
| chr14_-_55204054 | 9.31 |
ENSMUST00000226297.2
|
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha |
| chr2_+_58644922 | 8.86 |
ENSMUST00000059102.13
|
Upp2
|
uridine phosphorylase 2 |
| chr8_-_3770642 | 8.73 |
ENSMUST00000062037.7
|
Clec4g
|
C-type lectin domain family 4, member g |
| chr6_+_124470053 | 8.24 |
ENSMUST00000049124.10
|
C1rl
|
complement component 1, r subcomponent-like |
| chr19_+_46120327 | 6.45 |
ENSMUST00000043739.6
ENSMUST00000237098.2 |
Elovl3
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 3 |
| chr6_+_138117295 | 6.40 |
ENSMUST00000008684.11
|
Mgst1
|
microsomal glutathione S-transferase 1 |
| chr14_-_55204092 | 6.37 |
ENSMUST00000081857.14
|
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha |
| chr11_+_16702203 | 6.35 |
ENSMUST00000102884.10
ENSMUST00000020329.13 |
Egfr
|
epidermal growth factor receptor |
| chr7_+_13467422 | 6.20 |
ENSMUST00000086148.8
|
Sult2a2
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 2 |
| chr6_+_138117519 | 6.10 |
ENSMUST00000120939.8
ENSMUST00000204628.3 ENSMUST00000140932.2 ENSMUST00000120302.8 |
Mgst1
|
microsomal glutathione S-transferase 1 |
| chr9_+_40098375 | 5.77 |
ENSMUST00000062229.6
|
Olfr986
|
olfactory receptor 986 |
| chr6_-_146536025 | 5.22 |
ENSMUST00000037709.16
|
Tm7sf3
|
transmembrane 7 superfamily member 3 |
| chr17_-_45906768 | 4.82 |
ENSMUST00000164618.8
ENSMUST00000097317.10 ENSMUST00000170113.8 |
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr4_+_148686985 | 4.75 |
ENSMUST00000105701.9
ENSMUST00000052060.7 |
Masp2
|
mannan-binding lectin serine peptidase 2 |
| chr6_-_119394634 | 4.55 |
ENSMUST00000032272.13
|
Adipor2
|
adiponectin receptor 2 |
| chr7_-_140001552 | 4.42 |
ENSMUST00000213801.2
|
Olfr532
|
olfactory receptor 532 |
| chr4_-_150085722 | 4.39 |
ENSMUST00000153394.2
|
H6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr17_+_79359617 | 3.92 |
ENSMUST00000233916.2
|
Qpct
|
glutaminyl-peptide cyclotransferase (glutaminyl cyclase) |
| chr6_-_119394436 | 3.91 |
ENSMUST00000189710.2
|
Adipor2
|
adiponectin receptor 2 |
| chr7_-_80055168 | 3.80 |
ENSMUST00000107362.10
ENSMUST00000135306.3 |
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr19_+_12241335 | 3.78 |
ENSMUST00000214551.3
|
Olfr235
|
olfactory receptor 235 |
| chr3_+_122039274 | 3.73 |
ENSMUST00000178826.8
|
Gclm
|
glutamate-cysteine ligase, modifier subunit |
| chr15_-_76010736 | 3.61 |
ENSMUST00000054022.12
ENSMUST00000089654.4 |
BC024139
|
cDNA sequence BC024139 |
| chr3_-_89820451 | 3.53 |
ENSMUST00000029559.7
ENSMUST00000197679.5 |
Il6ra
|
interleukin 6 receptor, alpha |
| chr7_-_15978077 | 3.50 |
ENSMUST00000238893.2
ENSMUST00000171425.4 |
C5ar2
|
complement component 5a receptor 2 |
| chr18_-_20192535 | 3.34 |
ENSMUST00000075214.9
ENSMUST00000039247.11 |
Dsc2
|
desmocollin 2 |
| chr3_+_122039206 | 3.30 |
ENSMUST00000029769.14
|
Gclm
|
glutamate-cysteine ligase, modifier subunit |
| chr18_-_75830595 | 3.14 |
ENSMUST00000165559.3
|
Ctif
|
CBP80/20-dependent translation initiation factor |
| chr9_+_118307918 | 3.10 |
ENSMUST00000150633.2
|
Eomes
|
eomesodermin |
| chr12_+_87561085 | 3.07 |
ENSMUST00000110152.3
|
Eif1ad8
|
eukaryotic translation initiation factor 1A domain containing 8 |
| chr10_+_26105605 | 3.05 |
ENSMUST00000218301.2
ENSMUST00000164660.8 ENSMUST00000060716.6 |
Samd3
|
sterile alpha motif domain containing 3 |
| chr4_+_40722911 | 2.90 |
ENSMUST00000164233.8
ENSMUST00000137246.8 ENSMUST00000125442.8 |
Dnaja1
|
DnaJ heat shock protein family (Hsp40) member A1 |
| chr4_+_33078796 | 2.85 |
ENSMUST00000131920.8
|
Gabrr2
|
gamma-aminobutyric acid (GABA) C receptor, subunit rho 2 |
| chr2_-_38177182 | 2.80 |
ENSMUST00000130472.8
|
Dennd1a
|
DENN/MADD domain containing 1A |
| chr2_-_86940289 | 2.77 |
ENSMUST00000215828.3
|
Olfr259
|
olfactory receptor 259 |
| chr9_+_118307250 | 2.73 |
ENSMUST00000111763.8
|
Eomes
|
eomesodermin |
| chr19_-_56378309 | 2.69 |
ENSMUST00000166203.2
ENSMUST00000167239.8 |
Nrap
|
nebulin-related anchoring protein |
| chr7_-_19556612 | 2.66 |
ENSMUST00000120537.8
|
Bcl3
|
B cell leukemia/lymphoma 3 |
| chr10_-_127456791 | 2.64 |
ENSMUST00000118455.2
ENSMUST00000121829.8 |
Lrp1
|
low density lipoprotein receptor-related protein 1 |
| chr7_+_11905206 | 2.62 |
ENSMUST00000226953.2
ENSMUST00000227530.2 |
Vmn1r79
|
vomeronasal 1 receptor 79 |
| chr16_+_91184661 | 2.59 |
ENSMUST00000139503.2
|
Ifnar2
|
interferon (alpha and beta) receptor 2 |
| chr10_-_127457001 | 2.59 |
ENSMUST00000049149.15
|
Lrp1
|
low density lipoprotein receptor-related protein 1 |
| chr14_+_53404083 | 2.57 |
ENSMUST00000196639.2
ENSMUST00000177578.2 |
Trav14n-1
|
T cell receptor alpha variable 14N-1 |
| chr14_+_53093071 | 2.49 |
ENSMUST00000181038.3
ENSMUST00000187138.2 |
Trav14d-1
|
T cell receptor alpha variable 14D-1 |
| chr14_-_20714634 | 2.48 |
ENSMUST00000119483.2
|
Synpo2l
|
synaptopodin 2-like |
| chr19_-_56378459 | 2.41 |
ENSMUST00000040711.15
ENSMUST00000095947.11 ENSMUST00000073536.13 |
Nrap
|
nebulin-related anchoring protein |
| chr9_+_111268131 | 2.36 |
ENSMUST00000111879.5
|
Dclk3
|
doublecortin-like kinase 3 |
| chr16_+_91169671 | 2.31 |
ENSMUST00000023693.14
ENSMUST00000134491.9 ENSMUST00000117836.8 |
Ifnar2
|
interferon (alpha and beta) receptor 2 |
| chr7_-_108471586 | 2.16 |
ENSMUST00000216500.2
|
Olfr517
|
olfactory receptor 517 |
| chr7_-_6525801 | 2.11 |
ENSMUST00000213504.2
ENSMUST00000216447.2 ENSMUST00000213656.2 ENSMUST00000207820.3 |
Olfr1349
|
olfactory receptor 1349 |
| chr19_+_37674029 | 2.11 |
ENSMUST00000073391.5
|
Cyp26c1
|
cytochrome P450, family 26, subfamily c, polypeptide 1 |
| chr10_+_84753480 | 2.08 |
ENSMUST00000038523.15
ENSMUST00000214693.2 ENSMUST00000095385.5 |
Ric8b
|
RIC8 guanine nucleotide exchange factor B |
| chr5_-_92511136 | 2.07 |
ENSMUST00000077820.6
|
Cxcl11
|
chemokine (C-X-C motif) ligand 11 |
| chr4_+_40723083 | 2.02 |
ENSMUST00000149794.2
|
Dnaja1
|
DnaJ heat shock protein family (Hsp40) member A1 |
| chr1_+_182591771 | 2.01 |
ENSMUST00000193660.6
|
Susd4
|
sushi domain containing 4 |
| chr13_-_61312010 | 1.95 |
ENSMUST00000021889.6
|
Ctsr
|
cathepsin R |
| chr2_-_38177359 | 1.95 |
ENSMUST00000102787.10
|
Dennd1a
|
DENN/MADD domain containing 1A |
| chr12_+_87665639 | 1.91 |
ENSMUST00000164838.3
|
Eif1ad6
|
eukaryotic translation initiation factor 1A domain containing 6 |
| chr2_+_119378178 | 1.90 |
ENSMUST00000014221.13
ENSMUST00000119172.2 |
Chp1
|
calcineurin-like EF hand protein 1 |
| chr2_+_155453103 | 1.80 |
ENSMUST00000092995.6
|
Myh7b
|
myosin, heavy chain 7B, cardiac muscle, beta |
| chr7_-_104426677 | 1.78 |
ENSMUST00000211384.2
ENSMUST00000053464.9 |
Usp17le
|
ubiquitin specific peptidase 17-like E |
| chr5_+_127709302 | 1.71 |
ENSMUST00000118139.3
|
Glt1d1
|
glycosyltransferase 1 domain containing 1 |
| chr12_+_87783347 | 1.67 |
ENSMUST00000110149.2
|
Eif1ad2
|
eukaryotic translation initiation factor 1A domain containing 2 |
| chr7_-_104777612 | 1.67 |
ENSMUST00000214399.4
|
Olfr682-ps1
|
olfactory receptor 682, pseudogene 1 |
| chr9_-_64928927 | 1.65 |
ENSMUST00000036615.7
|
Hacd3
|
3-hydroxyacyl-CoA dehydratase 3 |
| chr15_-_76406602 | 1.63 |
ENSMUST00000096365.5
|
Scrt1
|
scratch family zinc finger 1 |
| chr5_-_90514360 | 1.63 |
ENSMUST00000168058.7
ENSMUST00000014421.15 |
Ankrd17
|
ankyrin repeat domain 17 |
| chr19_+_4264470 | 1.59 |
ENSMUST00000237171.2
|
Gm45928
|
predicted gene, 45928 |
| chr16_+_3648742 | 1.58 |
ENSMUST00000214238.2
ENSMUST00000214590.2 |
Olfr15
|
olfactory receptor 15 |
| chr16_+_91169770 | 1.55 |
ENSMUST00000089042.7
|
Ifnar2
|
interferon (alpha and beta) receptor 2 |
| chr15_+_102884874 | 1.53 |
ENSMUST00000173306.2
|
Hoxc9
|
homeobox C9 |
| chr9_-_32452885 | 1.47 |
ENSMUST00000016231.14
|
Fli1
|
Friend leukemia integration 1 |
| chr15_+_25940781 | 1.46 |
ENSMUST00000227275.2
|
Retreg1
|
reticulophagy regulator 1 |
| chr19_+_4264437 | 1.41 |
ENSMUST00000235355.2
|
Clcf1
|
cardiotrophin-like cytokine factor 1 |
| chr12_-_88290796 | 1.37 |
ENSMUST00000218054.3
|
Eif1ad15
|
eukaryotic translation initiation factor 1A domain containing 15 |
| chr9_+_118307412 | 1.35 |
ENSMUST00000035020.15
|
Eomes
|
eomesodermin |
| chrX_+_41157242 | 1.33 |
ENSMUST00000115095.9
|
Xiap
|
X-linked inhibitor of apoptosis |
| chr9_-_109397316 | 1.32 |
ENSMUST00000198112.2
ENSMUST00000198397.5 ENSMUST00000056745.12 |
Fbxw15
|
F-box and WD-40 domain protein 15 |
| chr6_+_41512010 | 1.32 |
ENSMUST00000103288.2
|
Trbj1-5
|
T cell receptor beta joining 1-5 |
| chr1_+_182591425 | 1.31 |
ENSMUST00000155229.7
ENSMUST00000153348.8 |
Susd4
|
sushi domain containing 4 |
| chr11_-_100583265 | 1.30 |
ENSMUST00000153494.2
|
Zfp385c
|
zinc finger protein 385C |
| chr5_-_106606032 | 1.30 |
ENSMUST00000086795.8
|
Barhl2
|
BarH like homeobox 2 |
| chr10_+_19810037 | 1.29 |
ENSMUST00000095806.10
ENSMUST00000120259.8 |
Map3k5
|
mitogen-activated protein kinase kinase kinase 5 |
| chr9_-_109116150 | 1.25 |
ENSMUST00000198928.2
|
Fbxw14
|
F-box and WD-40 domain protein 14 |
| chr2_+_152360167 | 1.25 |
ENSMUST00000121912.2
|
Defb28
|
defensin beta 28 |
| chr5_+_134961246 | 1.22 |
ENSMUST00000111218.8
ENSMUST00000136246.5 ENSMUST00000201847.3 |
Mettl27
|
methyltransferase like 27 |
| chr16_-_36486429 | 1.22 |
ENSMUST00000089620.11
|
Cd86
|
CD86 antigen |
| chr4_+_114914607 | 1.19 |
ENSMUST00000136946.8
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr3_+_144276154 | 1.18 |
ENSMUST00000082437.11
|
Selenof
|
selenoprotein F |
| chr9_-_8004586 | 1.17 |
ENSMUST00000086580.12
ENSMUST00000065353.13 |
Yap1
|
yes-associated protein 1 |
| chr10_+_78853158 | 1.16 |
ENSMUST00000080730.2
|
Olfr1351
|
olfactory receptor 1351 |
| chr4_+_148025316 | 1.16 |
ENSMUST00000103232.2
|
2510039O18Rik
|
RIKEN cDNA 2510039O18 gene |
| chr16_-_10884005 | 1.14 |
ENSMUST00000162323.2
|
Litaf
|
LPS-induced TN factor |
| chr16_+_48104098 | 1.13 |
ENSMUST00000096045.9
ENSMUST00000050705.4 |
Dppa4
|
developmental pluripotency associated 4 |
| chr13_-_34027389 | 1.10 |
ENSMUST00000110275.8
ENSMUST00000145221.2 |
Serpinb6e
|
serine (or cysteine) peptidase inhibitor, clade B, member 6e |
| chrX_+_108138965 | 1.09 |
ENSMUST00000033598.9
|
Sh3bgrl
|
SH3-binding domain glutamic acid-rich protein like |
| chr4_+_35152056 | 1.08 |
ENSMUST00000058595.7
|
Ifnk
|
interferon kappa |
| chr2_-_119378108 | 1.07 |
ENSMUST00000060009.9
|
Exd1
|
exonuclease 3'-5' domain containing 1 |
| chr1_+_42992109 | 1.05 |
ENSMUST00000179766.3
|
Gpr45
|
G protein-coupled receptor 45 |
| chr4_+_3167320 | 1.05 |
ENSMUST00000226198.2
|
Vmn1r2
|
vomeronasal 1 receptor 2 |
| chr17_+_37769807 | 1.05 |
ENSMUST00000214668.2
ENSMUST00000217602.2 ENSMUST00000214938.2 |
Olfr109
|
olfactory receptor 109 |
| chr12_-_118265163 | 1.05 |
ENSMUST00000221844.2
|
Sp4
|
trans-acting transcription factor 4 |
| chr3_+_93107517 | 1.03 |
ENSMUST00000098884.4
|
Flg2
|
filaggrin family member 2 |
| chr17_-_24428351 | 1.02 |
ENSMUST00000024931.6
|
Ntn3
|
netrin 3 |
| chr6_-_57668992 | 1.01 |
ENSMUST00000053386.6
|
Pyurf
|
Pigy upstream reading frame |
| chrX_+_158771429 | 0.99 |
ENSMUST00000033665.9
|
Map3k15
|
mitogen-activated protein kinase kinase kinase 15 |
| chr1_+_170060318 | 0.99 |
ENSMUST00000162752.2
|
Sh2d1b2
|
SH2 domain containing 1B2 |
| chr11_+_87486472 | 0.98 |
ENSMUST00000134216.3
|
Mtmr4
|
myotubularin related protein 4 |
| chr17_-_24863956 | 0.98 |
ENSMUST00000019684.13
|
Slc9a3r2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 |
| chr5_-_90514061 | 0.97 |
ENSMUST00000081914.13
|
Ankrd17
|
ankyrin repeat domain 17 |
| chr17_-_36014863 | 0.95 |
ENSMUST00000148065.8
|
Ddr1
|
discoidin domain receptor family, member 1 |
| chr9_+_44394080 | 0.92 |
ENSMUST00000220303.2
|
Bcl9l
|
B cell CLL/lymphoma 9-like |
| chr19_+_8713156 | 0.92 |
ENSMUST00000210512.2
ENSMUST00000049424.11 |
Wdr74
|
WD repeat domain 74 |
| chr4_-_130031421 | 0.92 |
ENSMUST00000164887.3
|
Hcrtr1
|
hypocretin (orexin) receptor 1 |
| chr11_-_53525520 | 0.92 |
ENSMUST00000020650.2
|
Il13
|
interleukin 13 |
| chr4_-_144412938 | 0.91 |
ENSMUST00000105747.2
|
Aadacl4fm4
|
AADACL4 family member 4 |
| chr7_+_11608557 | 0.91 |
ENSMUST00000227611.2
ENSMUST00000226622.2 ENSMUST00000228646.2 ENSMUST00000226855.2 ENSMUST00000228268.2 ENSMUST00000228463.2 |
Vmn1r75
|
vomeronasal 1 receptor 75 |
| chr2_-_121786573 | 0.89 |
ENSMUST00000104936.4
|
Mageb3
|
MAGE family member B3 |
| chr14_+_53497035 | 0.88 |
ENSMUST00000197614.2
|
Trav14n-2
|
T cell receptor alpha variable 14N-2 |
| chr7_+_79675727 | 0.88 |
ENSMUST00000049680.10
|
Zfp710
|
zinc finger protein 710 |
| chr5_-_90514034 | 0.86 |
ENSMUST00000218526.2
|
Ankrd17
|
ankyrin repeat domain 17 |
| chr5_-_66330394 | 0.84 |
ENSMUST00000201544.4
|
Rbm47
|
RNA binding motif protein 47 |
| chr9_-_109063792 | 0.84 |
ENSMUST00000197329.2
ENSMUST00000079548.12 |
Fbxw20
|
F-box and WD-40 domain protein 20 |
| chr9_-_108991088 | 0.83 |
ENSMUST00000199540.2
ENSMUST00000198076.5 ENSMUST00000054925.13 |
Fbxw21
|
F-box and WD-40 domain protein 21 |
| chrX_+_30768610 | 0.79 |
ENSMUST00000179532.2
|
Btbd35f29
|
BTB domain containing 35, family member 29 |
| chr12_-_75642969 | 0.79 |
ENSMUST00000021447.9
ENSMUST00000220035.2 |
Ppp2r5e
|
protein phosphatase 2, regulatory subunit B', epsilon |
| chr9_-_109324943 | 0.78 |
ENSMUST00000076617.7
|
Fbxw19
|
F-box and WD-40 domain protein 19 |
| chr14_-_56472102 | 0.77 |
ENSMUST00000015585.4
|
Gzmc
|
granzyme C |
| chr9_-_109575157 | 0.76 |
ENSMUST00000071917.4
|
Fbxw26
|
F-box and WD-40 domain protein 26 |
| chr5_-_116162415 | 0.76 |
ENSMUST00000031486.14
ENSMUST00000111999.8 |
Prkab1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
| chr14_-_101846551 | 0.75 |
ENSMUST00000100340.4
|
Tbc1d4
|
TBC1 domain family, member 4 |
| chrX_+_41156713 | 0.74 |
ENSMUST00000115094.8
|
Xiap
|
X-linked inhibitor of apoptosis |
| chr2_+_79465696 | 0.74 |
ENSMUST00000111785.9
|
Itprid2
|
ITPR interacting domain containing 2 |
| chr14_+_53791444 | 0.73 |
ENSMUST00000198297.2
|
Trav14-1
|
T cell receptor alpha variable 14-1 |
| chr11_+_17207558 | 0.72 |
ENSMUST00000000594.9
ENSMUST00000156784.2 |
C1d
|
C1D nuclear receptor co-repressor |
| chrX_+_33094635 | 0.72 |
ENSMUST00000177912.2
|
Btbd35f13
|
BTB domain containing 35, family member 13 |
| chr17_-_24863907 | 0.71 |
ENSMUST00000234505.2
|
Slc9a3r2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 |
| chr1_-_37996838 | 0.71 |
ENSMUST00000027254.10
ENSMUST00000114894.2 |
Lyg1
|
lysozyme G-like 1 |
| chr19_+_29675224 | 0.69 |
ENSMUST00000025719.5
|
Mlana
|
melan-A |
| chrX_-_33139812 | 0.68 |
ENSMUST00000105117.3
|
Btbd35f14
|
BTB domain containing 35, family member 14 |
| chr17_-_28584150 | 0.67 |
ENSMUST00000114794.2
|
Tulp1
|
tubby like protein 1 |
| chrX_-_33014777 | 0.66 |
ENSMUST00000186329.2
|
Btbd35f15
|
BTB domain containing 35, family member 15 |
| chrX_+_33814112 | 0.66 |
ENSMUST00000238270.2
|
Btbd35f8
|
BTB domain containing 35, family member 8 |
| chrX_-_31034822 | 0.66 |
ENSMUST00000238426.2
|
Btbd35f19
|
BTB domain containing 35, family member 19 |
| chr9_+_78137927 | 0.65 |
ENSMUST00000098537.4
|
Gsta1
|
glutathione S-transferase, alpha 1 (Ya) |
| chrX_+_32750842 | 0.65 |
ENSMUST00000178827.3
|
Btbd35f12
|
BTB domain containing 35, family member 12 |
| chrX_+_4855477 | 0.64 |
ENSMUST00000178143.2
|
Btbd35f27
|
BTB domain containing 35, family member 27 |
| chrX_+_32411401 | 0.63 |
ENSMUST00000178747.3
|
Btbd35f5
|
BTB domain containing 35, family member 5 |
| chr4_-_41731142 | 0.62 |
ENSMUST00000171251.8
|
Arid3c
|
AT rich interactive domain 3C (BRIGHT-like) |
| chr15_-_76406102 | 0.62 |
ENSMUST00000164703.2
|
Scrt1
|
scratch family zinc finger 1 |
| chrX_-_33580888 | 0.61 |
ENSMUST00000238632.2
|
Btbd35f9
|
BTB domain containing 35, family member 9 |
| chr7_-_126626152 | 0.61 |
ENSMUST00000206254.2
ENSMUST00000206291.2 |
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr6_-_88423464 | 0.60 |
ENSMUST00000204459.3
ENSMUST00000203213.3 ENSMUST00000205179.3 ENSMUST00000165242.4 |
Eefsec
|
eukaryotic elongation factor, selenocysteine-tRNA-specific |
| chrX_+_3942699 | 0.59 |
ENSMUST00000140300.2
|
Btbd35f3
|
BTB domain containing 35, family member 3 |
| chrX_+_4273978 | 0.59 |
ENSMUST00000105014.2
|
Btbd35f17
|
BTB domain containing 35, family member 17 |
| chr9_-_106666329 | 0.59 |
ENSMUST00000046502.7
|
Rad54l2
|
RAD54 like 2 (S. cerevisiae) |
| chrX_+_4703838 | 0.59 |
ENSMUST00000105011.4
|
Btbd35f4
|
BTB domain containing 35, family member 4 |
| chrX_+_72527208 | 0.58 |
ENSMUST00000033741.15
ENSMUST00000169489.2 |
Bgn
|
biglycan |
| chrX_+_3605258 | 0.57 |
ENSMUST00000105019.3
|
Btbd35f18
|
BTB domain containing 35, family member 18 |
| chr7_+_79676095 | 0.57 |
ENSMUST00000206039.2
|
Zfp710
|
zinc finger protein 710 |
| chr18_-_62748811 | 0.57 |
ENSMUST00000235190.2
|
Spink13
|
serine peptidase inhibitor, Kazal type 13 |
| chrX_-_3862027 | 0.56 |
ENSMUST00000185755.2
|
Btbd35f16
|
BTB domain containing 35, family member 16 |
| chrX_-_3657910 | 0.55 |
ENSMUST00000178621.2
|
Btbd35f10
|
BTB domain containing 35, family member 10 |
| chrX_+_32070863 | 0.55 |
ENSMUST00000238237.2
|
Btbd35f1
|
BTB domain containing 35, family member 1 |
| chrX_-_3150852 | 0.55 |
ENSMUST00000178080.3
|
Btbd35f24
|
BTB domain containing 35, family member 24 |
| chr5_+_134961205 | 0.55 |
ENSMUST00000047196.14
ENSMUST00000111221.9 ENSMUST00000111219.8 ENSMUST00000068617.12 |
Mettl27
|
methyltransferase like 27 |
| chr17_-_28584182 | 0.54 |
ENSMUST00000041819.14
|
Tulp1
|
tubby like protein 1 |
| chr6_+_57183497 | 0.53 |
ENSMUST00000227298.2
|
Vmn1r13
|
vomeronasal 1 receptor 13 |
| chr9_+_19624125 | 0.52 |
ENSMUST00000077023.4
|
Olfr857
|
olfactory receptor 857 |
| chr7_+_103064279 | 0.51 |
ENSMUST00000106892.2
|
Usp17lc
|
ubiquitin specific peptidase 17-like C |
| chrX_-_3398715 | 0.51 |
ENSMUST00000105020.2
|
Btbd35f11
|
BTB domain containing 35, family member 11 |
| chrX_-_8042129 | 0.50 |
ENSMUST00000143984.2
|
Tbc1d25
|
TBC1 domain family, member 25 |
| chrX_+_31839202 | 0.50 |
ENSMUST00000179991.3
|
Btbd35f2
|
BTB domain containing 35, family member 2 |
| chrX_+_3076875 | 0.49 |
ENSMUST00000189190.2
|
Btbd35f23
|
BTB domain containing 35, family member 23 |
| chr13_-_99584091 | 0.48 |
ENSMUST00000223725.2
|
Map1b
|
microtubule-associated protein 1B |
| chr7_-_141794815 | 0.48 |
ENSMUST00000211591.2
|
Gm40460
|
predicted gene, 40460 |
| chr2_+_90912710 | 0.48 |
ENSMUST00000169852.2
|
Spi1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr2_-_28511941 | 0.48 |
ENSMUST00000028156.8
ENSMUST00000164290.8 |
Gfi1b
|
growth factor independent 1B |
| chr9_-_109493720 | 0.47 |
ENSMUST00000163839.3
|
Fbxw25
|
F-box and WD-40 domain protein 25 |
| chr7_+_11884184 | 0.47 |
ENSMUST00000228664.2
ENSMUST00000228244.2 |
Vmn1r78
|
vomeronasal 1 receptor 78 |
| chr17_-_28584117 | 0.46 |
ENSMUST00000233420.2
|
Tulp1
|
tubby like protein 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.7 | 38.8 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 7.8 | 31.3 | GO:0007522 | visceral muscle development(GO:0007522) |
| 7.7 | 46.4 | GO:0032827 | negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 3.6 | 14.2 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 3.6 | 117.3 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 3.4 | 10.3 | GO:1903920 | positive regulation of actin filament severing(GO:1903920) |
| 3.1 | 9.4 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 3.1 | 12.5 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 2.6 | 23.0 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 2.5 | 53.0 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 2.4 | 7.2 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 2.2 | 24.4 | GO:0042436 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) |
| 2.2 | 10.9 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 2.2 | 10.8 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 1.3 | 3.9 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 1.3 | 5.2 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 1.2 | 3.5 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 1.2 | 3.5 | GO:1902623 | negative regulation of neutrophil migration(GO:1902623) |
| 1.1 | 61.2 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 1.1 | 6.3 | GO:0070459 | prolactin secretion(GO:0070459) regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 1.0 | 3.8 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.9 | 2.7 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.8 | 3.3 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) |
| 0.8 | 4.8 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.8 | 8.5 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.7 | 6.2 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.7 | 4.8 | GO:0015862 | uridine transport(GO:0015862) |
| 0.6 | 14.0 | GO:0051923 | sulfation(GO:0051923) |
| 0.6 | 2.4 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.6 | 3.4 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.5 | 4.9 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.5 | 6.5 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.5 | 1.9 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.4 | 1.2 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.4 | 3.3 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.4 | 4.7 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.4 | 8.7 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.4 | 0.7 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.3 | 2.1 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.3 | 1.6 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.3 | 1.0 | GO:0051466 | positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.2 | 0.9 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 0.2 | 6.4 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.2 | 4.4 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.2 | 1.5 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.2 | 0.8 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.1 | 1.2 | GO:0060242 | contact inhibition(GO:0060242) metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.1 | 0.6 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 1.7 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 | 1.2 | GO:0002667 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.1 | 0.4 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.1 | 0.5 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.1 | 0.4 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.1 | 0.5 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.6 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.1 | 1.2 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.1 | 0.5 | GO:0051572 | negative regulation of histone H3-K4 methylation(GO:0051572) |
| 0.1 | 1.3 | GO:1902170 | cellular response to reactive nitrogen species(GO:1902170) |
| 0.1 | 0.8 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.8 | GO:0035878 | nail development(GO:0035878) |
| 0.1 | 1.1 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.1 | 0.6 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 1.7 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.1 | 13.2 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.1 | 0.6 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.6 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 1.1 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 7.4 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 1.7 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.9 | GO:0045953 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) negative regulation of natural killer cell mediated cytotoxicity(GO:0045953) |
| 0.0 | 1.5 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 1.1 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 5.7 | GO:0009166 | nucleotide catabolic process(GO:0009166) |
| 0.0 | 0.5 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 6.2 | GO:0008202 | steroid metabolic process(GO:0008202) |
| 0.0 | 0.8 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.3 | GO:0036506 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 | 2.6 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.5 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.0 | 0.7 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.0 | 0.5 | GO:0061162 | establishment of monopolar cell polarity(GO:0061162) |
| 0.0 | 2.2 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 0.2 | GO:0044334 | canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) |
| 0.0 | 0.5 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.4 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 2.5 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 2.4 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.0 | 0.2 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 1.0 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.9 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 | 24.6 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.7 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.9 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 | 0.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.7 | GO:0002717 | positive regulation of natural killer cell mediated immunity(GO:0002717) positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.0 | 1.0 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 5.7 | GO:0007606 | sensory perception of chemical stimulus(GO:0007606) |
| 0.0 | 0.2 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 1.0 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.2 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.9 | GO:0001825 | blastocyst formation(GO:0001825) |
| 0.0 | 0.4 | GO:0060713 | labyrinthine layer morphogenesis(GO:0060713) |
| 0.0 | 1.3 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 1.3 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 7.0 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 1.3 | 14.2 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 1.3 | 6.3 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 1.2 | 33.1 | GO:0032982 | myosin filament(GO:0032982) |
| 0.9 | 3.5 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.8 | 8.9 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.6 | 5.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.6 | 10.9 | GO:0045180 | basal cortex(GO:0045180) |
| 0.5 | 2.7 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.4 | 1.2 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 0.3 | 1.2 | GO:0071149 | TEAD-1-YAP complex(GO:0071148) TEAD-2-YAP complex(GO:0071149) |
| 0.3 | 1.1 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.2 | 1.3 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.2 | 1.0 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.2 | 52.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.2 | 3.2 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.2 | 1.1 | GO:1990923 | PET complex(GO:1990923) |
| 0.1 | 4.7 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.1 | 12.5 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 0.4 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.1 | 3.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 3.5 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 1.0 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 9.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 3.3 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 97.6 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 1.5 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 1.0 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.7 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.4 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) RISC-loading complex(GO:0070578) |
| 0.0 | 0.3 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.2 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 1.7 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 49.8 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
| 0.0 | 0.5 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.9 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.8 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 61.6 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 3.4 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.5 | GO:0005776 | autophagosome(GO:0005776) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.2 | 53.0 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 8.1 | 24.4 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 7.7 | 46.4 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 6.5 | 38.8 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 4.5 | 31.3 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 3.7 | 117.3 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 3.1 | 9.4 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 2.4 | 14.2 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 2.3 | 7.0 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 2.3 | 14.0 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 1.7 | 8.5 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 1.6 | 6.3 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 1.5 | 8.9 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 1.5 | 10.3 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 1.5 | 4.4 | GO:0047936 | glucose 1-dehydrogenase [NAD(P)] activity(GO:0047936) |
| 1.3 | 6.4 | GO:0004905 | interferon receptor activity(GO:0004904) type I interferon receptor activity(GO:0004905) type I interferon binding(GO:0019962) |
| 1.2 | 10.8 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 1.2 | 3.5 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 1.2 | 3.5 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 1.1 | 10.9 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) |
| 0.7 | 23.0 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.5 | 4.9 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.5 | 6.5 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.5 | 13.2 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.5 | 13.2 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.4 | 61.2 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.4 | 2.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.3 | 5.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.3 | 1.6 | GO:0102344 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.3 | 8.7 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.3 | 4.7 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.2 | 3.8 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.2 | 0.9 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.2 | 5.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.2 | 4.8 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 0.4 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.1 | 1.0 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 4.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 5.7 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.1 | 2.2 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.1 | 1.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 5.0 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 1.9 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.5 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.6 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 1.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.5 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 1.7 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 2.4 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.7 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0001132 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.0 | 1.0 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.5 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.8 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 1.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 12.9 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.3 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 6.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 2.9 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 0.7 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 14.9 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 1.9 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 1.8 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 1.1 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 1.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 1.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 12.2 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.2 | 13.2 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 0.9 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 14.0 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 5.2 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 2.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 2.7 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 4.9 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 3.8 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 1.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 8.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.1 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 1.7 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.5 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 1.2 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.5 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.9 | 38.8 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 1.7 | 24.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 1.3 | 53.0 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 1.0 | 14.0 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.9 | 61.2 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.7 | 14.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.6 | 4.7 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.5 | 31.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.4 | 8.9 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.4 | 20.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.3 | 10.9 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.3 | 3.8 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.3 | 6.4 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.2 | 6.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.2 | 4.7 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.1 | 33.8 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 4.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 1.8 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 1.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 0.8 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 2.4 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 1.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 2.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.8 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 1.0 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 1.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |