Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Fos
Z-value: 1.77
Transcription factors associated with Fos
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Fos
|
ENSMUSG00000021250.14 | Fos |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Fos | mm39_v1_chr12_+_85520652_85520672 | 0.37 | 1.5e-03 | Click! |
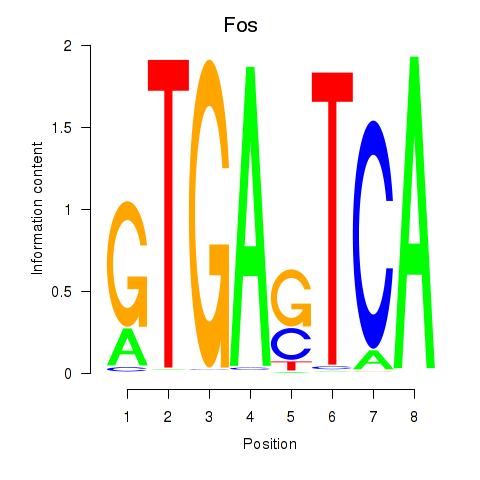
Activity profile of Fos motif
Sorted Z-values of Fos motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Fos
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.0 | 28.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 4.1 | 12.4 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 3.6 | 17.8 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 3.4 | 16.9 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 3.0 | 21.2 | GO:0044359 | modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 2.5 | 76.5 | GO:0032060 | bleb assembly(GO:0032060) |
| 2.3 | 13.7 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 2.2 | 13.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 2.1 | 8.5 | GO:1901003 | negative regulation of fermentation(GO:1901003) |
| 2.0 | 6.1 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 1.7 | 10.4 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 1.7 | 10.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 1.6 | 6.3 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 1.5 | 7.4 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 1.4 | 11.2 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 1.4 | 12.6 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 1.4 | 6.8 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 1.4 | 8.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 1.3 | 5.4 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 1.2 | 17.1 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 1.2 | 5.9 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 1.2 | 1.2 | GO:0002649 | regulation of tolerance induction to self antigen(GO:0002649) |
| 1.1 | 44.3 | GO:0031424 | keratinization(GO:0031424) |
| 1.1 | 6.6 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 1.1 | 5.4 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 1.0 | 4.1 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 1.0 | 15.4 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 1.0 | 2.9 | GO:0046108 | uridine catabolic process(GO:0006218) uridine metabolic process(GO:0046108) |
| 0.9 | 2.8 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.9 | 6.6 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.9 | 10.3 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.9 | 7.3 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.9 | 5.5 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.9 | 7.0 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.9 | 9.4 | GO:0015705 | iodide transport(GO:0015705) |
| 0.7 | 4.4 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.7 | 6.6 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.7 | 2.2 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.7 | 6.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.7 | 10.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.7 | 2.7 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.7 | 2.0 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.7 | 2.0 | GO:0071349 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.7 | 4.0 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.6 | 13.3 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.6 | 1.3 | GO:0002461 | tolerance induction dependent upon immune response(GO:0002461) |
| 0.6 | 4.4 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.6 | 6.7 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.6 | 6.3 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.6 | 2.3 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.6 | 1.7 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.5 | 4.3 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.5 | 3.7 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.5 | 8.9 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.5 | 7.8 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.5 | 8.1 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.5 | 2.5 | GO:0046292 | spermatid nucleus elongation(GO:0007290) formaldehyde metabolic process(GO:0046292) |
| 0.5 | 4.9 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.5 | 1.4 | GO:0002343 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 0.4 | 1.7 | GO:2000852 | regulation of corticosterone secretion(GO:2000852) |
| 0.4 | 5.8 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.4 | 1.8 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.4 | 3.2 | GO:0009137 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.3 | 1.0 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.3 | 1.4 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.3 | 6.5 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.3 | 5.9 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.3 | 1.0 | GO:0060875 | subpallium cell proliferation in forebrain(GO:0022012) lateral ganglionic eminence cell proliferation(GO:0022018) lambdoid suture morphogenesis(GO:0060366) sagittal suture morphogenesis(GO:0060367) anterior semicircular canal development(GO:0060873) lateral semicircular canal development(GO:0060875) |
| 0.3 | 0.9 | GO:0061723 | glycophagy(GO:0061723) |
| 0.3 | 16.1 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.3 | 1.4 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.3 | 17.6 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.3 | 6.9 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.3 | 3.8 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.3 | 3.5 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.3 | 1.1 | GO:0071336 | regulation of hair follicle cell proliferation(GO:0071336) positive regulation of progesterone secretion(GO:2000872) |
| 0.3 | 5.3 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.3 | 21.9 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.3 | 0.8 | GO:0015881 | creatine transport(GO:0015881) |
| 0.3 | 17.5 | GO:0032720 | negative regulation of tumor necrosis factor production(GO:0032720) |
| 0.3 | 7.4 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.3 | 0.5 | GO:1903233 | regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) |
| 0.2 | 2.5 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.2 | 2.5 | GO:0032264 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.2 | 28.2 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.2 | 2.5 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.2 | 4.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.2 | 7.5 | GO:0043616 | keratinocyte proliferation(GO:0043616) |
| 0.2 | 1.0 | GO:0002268 | follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
| 0.2 | 1.4 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.2 | 7.9 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.2 | 7.3 | GO:0032967 | positive regulation of collagen metabolic process(GO:0010714) positive regulation of collagen biosynthetic process(GO:0032967) positive regulation of multicellular organismal metabolic process(GO:0044253) |
| 0.2 | 3.2 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.2 | 1.3 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.2 | 0.6 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.2 | 2.8 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.2 | 0.6 | GO:0044650 | adhesion of symbiont to host cell(GO:0044650) |
| 0.2 | 2.4 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.2 | 3.3 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.2 | 7.4 | GO:0033198 | response to ATP(GO:0033198) |
| 0.2 | 1.1 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.2 | 1.3 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.2 | 2.7 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.2 | 0.7 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.2 | 3.7 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.2 | 2.9 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.2 | 2.9 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.2 | 4.5 | GO:0043486 | histone exchange(GO:0043486) |
| 0.2 | 0.9 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.2 | 1.5 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.2 | 0.6 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.1 | 2.1 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 1.2 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.1 | 5.0 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) |
| 0.1 | 6.6 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 0.6 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 1.6 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.1 | 0.4 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.1 | 1.7 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 5.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 3.7 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.1 | 2.3 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 0.4 | GO:2000705 | regulation of dense core granule biogenesis(GO:2000705) |
| 0.1 | 6.0 | GO:0034113 | heterotypic cell-cell adhesion(GO:0034113) |
| 0.1 | 1.4 | GO:0035815 | positive regulation of renal sodium excretion(GO:0035815) |
| 0.1 | 2.8 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.1 | 0.6 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 0.5 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.1 | 12.6 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 0.9 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.1 | 1.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.1 | 2.4 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 2.4 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.1 | 2.9 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.1 | 3.8 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.1 | 0.4 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.1 | 3.3 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 4.7 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.1 | 0.6 | GO:1990839 | response to endothelin(GO:1990839) |
| 0.1 | 13.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 1.1 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.1 | 2.7 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.1 | 0.3 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.1 | 0.1 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.1 | 5.4 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 1.0 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 | 0.8 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 0.9 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.1 | 1.2 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.1 | 2.3 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.1 | 2.4 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 3.5 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.1 | 1.3 | GO:1903427 | negative regulation of reactive oxygen species biosynthetic process(GO:1903427) |
| 0.1 | 17.0 | GO:0042060 | wound healing(GO:0042060) |
| 0.1 | 1.0 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.1 | 0.5 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.1 | 0.4 | GO:0002887 | negative regulation of myeloid leukocyte mediated immunity(GO:0002887) |
| 0.1 | 5.6 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.1 | 10.6 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.1 | 4.3 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.3 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 1.5 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 1.5 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.3 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.0 | 2.0 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 1.0 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 1.2 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.2 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.6 | GO:0051770 | positive regulation of nitric-oxide synthase biosynthetic process(GO:0051770) |
| 0.0 | 2.5 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.7 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.9 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 0.1 | GO:0043622 | cortical microtubule organization(GO:0043622) |
| 0.0 | 0.9 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.2 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.0 | 1.6 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 1.2 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 1.0 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 | 1.8 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 3.2 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 1.0 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 1.0 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 3.5 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.0 | 0.1 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.0 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.4 | GO:0030488 | tRNA methylation(GO:0030488) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.5 | 19.6 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 4.5 | 13.6 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 2.4 | 7.3 | GO:0038045 | large latent transforming growth factor-beta complex(GO:0038045) |
| 2.3 | 7.0 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 2.2 | 6.6 | GO:0031904 | endosome lumen(GO:0031904) |
| 2.0 | 10.2 | GO:0005914 | spot adherens junction(GO:0005914) |
| 1.8 | 5.5 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 1.5 | 10.2 | GO:0005638 | lamin filament(GO:0005638) |
| 1.3 | 3.8 | GO:0034677 | integrin alpha7-beta1 complex(GO:0034677) |
| 1.1 | 6.6 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 1.0 | 11.2 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.9 | 3.8 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.9 | 17.6 | GO:0042599 | lamellar body(GO:0042599) |
| 0.8 | 42.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.7 | 21.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.6 | 17.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.6 | 10.3 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.5 | 15.1 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.4 | 7.4 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.3 | 2.4 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.3 | 2.4 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.3 | 3.7 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.3 | 7.0 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.3 | 1.8 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.3 | 7.6 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.3 | 29.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.3 | 13.1 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 1.2 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.2 | 7.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.2 | 7.7 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.2 | 13.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.2 | 1.4 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.2 | 5.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 0.6 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.2 | 1.0 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.2 | 5.0 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 2.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 14.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.2 | 3.2 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 2.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 0.6 | GO:1990590 | Lewy body core(GO:1990037) ATF4-CREB1 transcription factor complex(GO:1990589) ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.1 | 0.9 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 1.0 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 13.3 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 0.6 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 21.7 | GO:0031674 | I band(GO:0031674) |
| 0.1 | 2.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 18.0 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 5.5 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 4.2 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 5.5 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 0.6 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 5.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.3 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.1 | 0.9 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 5.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 0.9 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 1.8 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 0.6 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 4.2 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.5 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 2.2 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.6 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 1.9 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 2.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 3.3 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 1.5 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 6.8 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 2.8 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 1.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 3.7 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 4.9 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 3.0 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 8.9 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 32.2 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 3.5 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 9.0 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 5.1 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 3.1 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.3 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.0 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 1.9 | GO:0043296 | apical junction complex(GO:0043296) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 23.0 | GO:1990254 | keratin filament binding(GO:1990254) |
| 2.7 | 13.7 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 2.7 | 13.5 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 2.6 | 7.9 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 1.8 | 7.4 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 1.8 | 7.3 | GO:0050436 | microfibril binding(GO:0050436) |
| 1.8 | 10.8 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 1.7 | 17.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 1.7 | 11.8 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 1.6 | 8.0 | GO:0051373 | FATZ binding(GO:0051373) |
| 1.6 | 9.4 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 1.5 | 15.4 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 1.5 | 13.6 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 1.3 | 5.3 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 1.2 | 7.4 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 1.1 | 5.4 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.9 | 18.8 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.9 | 16.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.9 | 3.5 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.9 | 6.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.8 | 13.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.8 | 7.4 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.8 | 23.6 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.7 | 2.9 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.7 | 6.3 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.7 | 4.0 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.7 | 5.9 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.6 | 6.5 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.6 | 12.6 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.6 | 25.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.5 | 5.5 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.5 | 8.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.5 | 1.5 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.5 | 2.9 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.5 | 3.3 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.5 | 15.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.4 | 9.0 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.4 | 2.0 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.4 | 3.7 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.4 | 2.0 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.4 | 1.2 | GO:0005135 | erythropoietin receptor binding(GO:0005128) interleukin-3 receptor binding(GO:0005135) |
| 0.4 | 2.5 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.4 | 6.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.4 | 3.2 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.4 | 5.0 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.4 | 7.0 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.3 | 1.4 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.3 | 5.4 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.3 | 9.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.3 | 2.8 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.3 | 2.4 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.3 | 8.9 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.3 | 8.5 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.3 | 19.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.3 | 0.8 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.3 | 7.0 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.2 | 5.0 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.2 | 2.5 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.2 | 3.5 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.2 | 6.2 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.2 | 4.1 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.2 | 1.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.2 | 2.8 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 16.5 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.2 | 8.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 4.8 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.2 | 1.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.2 | 5.3 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 3.6 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 1.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 1.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 3.9 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 1.3 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.1 | 1.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 1.9 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 2.1 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 2.9 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 1.5 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 2.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 1.0 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 6.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 6.5 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 2.7 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.7 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 5.5 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 5.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.6 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 1.0 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.3 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 1.2 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 2.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.6 | GO:0002135 | CTP binding(GO:0002135) |
| 0.1 | 5.4 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 2.7 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 12.1 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 1.8 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 2.7 | GO:0005539 | glycosaminoglycan binding(GO:0005539) |
| 0.1 | 5.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 1.4 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 8.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 3.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 40.1 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 0.6 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 7.2 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.1 | 1.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 6.5 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 1.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 1.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.5 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 8.1 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 1.5 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 1.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.6 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 1.2 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 2.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 3.5 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.5 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.8 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.4 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 1.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 3.2 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.0 | 1.5 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 1.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.5 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 6.9 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 1.0 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.2 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 2.3 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 1.0 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.4 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.3 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 1.9 | GO:0005244 | voltage-gated ion channel activity(GO:0005244) voltage-gated channel activity(GO:0022832) |
| 0.0 | 0.4 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 29.8 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.5 | 37.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.4 | 18.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.3 | 2.4 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.2 | 4.9 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.2 | 8.0 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.2 | 12.3 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.2 | 10.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.2 | 21.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.2 | 10.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.2 | 14.3 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.2 | 9.0 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.2 | 5.0 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.2 | 7.8 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.2 | 3.7 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.2 | 6.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.2 | 4.4 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.2 | 2.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 14.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 3.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 4.2 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 4.8 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 4.5 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 9.9 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 18.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 2.8 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 4.1 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 1.3 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 8.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 18.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 1.0 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 10.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 2.7 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.6 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 1.2 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.5 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.7 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 1.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.1 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.7 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 1.0 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 2.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.6 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 5.3 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 0.4 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.6 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.6 | PID CDC42 PATHWAY | CDC42 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 7.0 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.4 | 10.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.4 | 7.9 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.4 | 4.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.3 | 37.9 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.3 | 11.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.3 | 6.7 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.3 | 5.0 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.3 | 6.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.3 | 14.1 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.3 | 5.8 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.2 | 0.7 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.2 | 10.2 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.2 | 3.8 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.2 | 2.0 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.2 | 13.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.2 | 10.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.2 | 12.3 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 6.6 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 2.9 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 6.4 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 1.6 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 7.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 2.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 1.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 2.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 2.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 1.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 4.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 3.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 1.5 | REACTOME GABA B RECEPTOR ACTIVATION | Genes involved in GABA B receptor activation |
| 0.1 | 6.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 0.9 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.1 | 3.5 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 2.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 2.2 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 1.7 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 3.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 3.7 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.6 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 1.3 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.4 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 1.9 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 1.1 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 2.9 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 2.3 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 3.8 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 2.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |