Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Fosl2_Bach2
Z-value: 1.27
Transcription factors associated with Fosl2_Bach2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
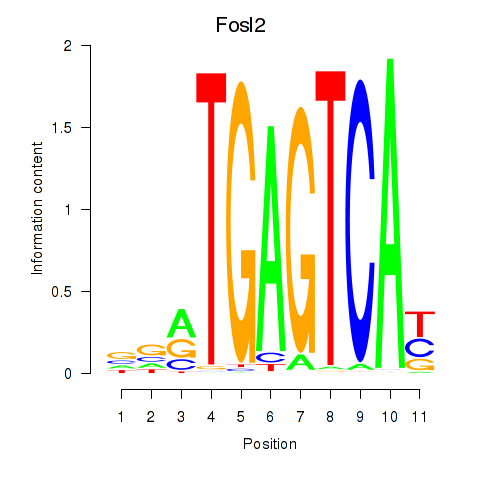
Fosl2
|
ENSMUSG00000029135.11 | Fosl2 |
|
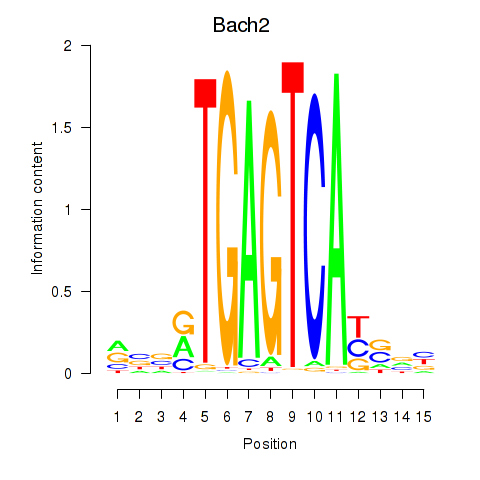
Bach2
|
ENSMUSG00000040270.17 | Bach2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Fosl2 | mm39_v1_chr5_+_32293145_32293201 | 0.71 | 2.6e-12 | Click! |
| Bach2 | mm39_v1_chr4_+_32238950_32238964 | -0.36 | 2.0e-03 | Click! |
Activity profile of Fosl2_Bach2 motif
Sorted Z-values of Fosl2_Bach2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Fosl2_Bach2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 15.6 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 3.2 | 9.6 | GO:0006218 | uridine catabolic process(GO:0006218) uridine metabolic process(GO:0046108) |
| 2.5 | 10.0 | GO:2000852 | regulation of corticosterone secretion(GO:2000852) |
| 2.3 | 6.9 | GO:0015881 | creatine transport(GO:0015881) |
| 2.1 | 8.4 | GO:1901003 | negative regulation of fermentation(GO:1901003) |
| 2.1 | 6.2 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 1.9 | 5.8 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 1.9 | 7.7 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 1.9 | 9.3 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 1.6 | 4.7 | GO:0097185 | amiloride transport(GO:0015898) cellular response to copper ion starvation(GO:0035874) response to azide(GO:0097184) cellular response to azide(GO:0097185) |
| 1.5 | 25.3 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 1.5 | 4.4 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 1.4 | 6.9 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 1.3 | 7.6 | GO:0002159 | desmosome assembly(GO:0002159) |
| 1.3 | 7.6 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 1.3 | 12.5 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 1.1 | 6.8 | GO:0014826 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) |
| 1.1 | 5.6 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 1.1 | 7.6 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 1.0 | 9.3 | GO:0002934 | desmosome organization(GO:0002934) |
| 1.0 | 3.0 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 1.0 | 2.9 | GO:0002777 | antimicrobial peptide biosynthetic process(GO:0002777) antibacterial peptide biosynthetic process(GO:0002780) |
| 0.9 | 5.5 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.9 | 2.7 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.9 | 4.4 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.9 | 5.3 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.9 | 4.3 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.8 | 4.9 | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) |
| 0.8 | 11.4 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.8 | 8.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.8 | 4.8 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.8 | 52.6 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.8 | 2.3 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.8 | 3.0 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.7 | 11.7 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.7 | 4.4 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.7 | 2.2 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.7 | 7.9 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.7 | 10.0 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.7 | 3.5 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.7 | 10.0 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.6 | 3.8 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.6 | 5.6 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.6 | 3.1 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.6 | 2.4 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.6 | 2.3 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.6 | 1.7 | GO:0002661 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.6 | 1.7 | GO:0036363 | transforming growth factor beta activation(GO:0036363) |
| 0.6 | 3.4 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.6 | 2.2 | GO:0009838 | abscission(GO:0009838) |
| 0.5 | 7.2 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.5 | 4.4 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.5 | 1.4 | GO:1904793 | regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.5 | 0.9 | GO:1904306 | positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 0.4 | 3.6 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.4 | 3.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.4 | 5.7 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.4 | 1.7 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.4 | 4.7 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.4 | 6.1 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.4 | 2.4 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.4 | 2.3 | GO:0051410 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.4 | 4.8 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.4 | 6.6 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.4 | 1.1 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.3 | 2.8 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.3 | 1.0 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.3 | 2.7 | GO:1901748 | peptide modification(GO:0031179) leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.3 | 1.0 | GO:0060540 | lung lobe formation(GO:0060464) diaphragm morphogenesis(GO:0060540) |
| 0.3 | 1.0 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.3 | 10.3 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.3 | 1.6 | GO:1905169 | protein localization to phagocytic vesicle(GO:1905161) regulation of protein localization to phagocytic vesicle(GO:1905169) positive regulation of protein localization to phagocytic vesicle(GO:1905171) |
| 0.3 | 16.4 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.3 | 1.9 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.3 | 1.8 | GO:0031394 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.3 | 2.0 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) tolerance induction dependent upon immune response(GO:0002461) |
| 0.3 | 12.5 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.3 | 0.8 | GO:0042275 | error-free postreplication DNA repair(GO:0042275) |
| 0.3 | 2.5 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.3 | 1.4 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.3 | 2.4 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.3 | 1.3 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.3 | 0.8 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.2 | 2.0 | GO:0009099 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.2 | 1.2 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.2 | 5.0 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.2 | 3.9 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.2 | 1.6 | GO:1901909 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.2 | 2.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.2 | 4.6 | GO:0031424 | keratinization(GO:0031424) |
| 0.2 | 1.1 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.2 | 3.2 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.2 | 4.4 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.2 | 4.3 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.2 | 1.4 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.2 | 5.9 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.2 | 4.9 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.2 | 3.5 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.2 | 1.1 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.2 | 0.9 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.2 | 28.4 | GO:0007586 | digestion(GO:0007586) |
| 0.2 | 0.7 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.2 | 1.8 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.2 | 2.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.2 | 3.8 | GO:0007614 | short-term memory(GO:0007614) locomotion involved in locomotory behavior(GO:0031987) |
| 0.1 | 8.5 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 5.8 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 6.1 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 1.1 | GO:2000286 | regulation of endosome size(GO:0051036) receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.1 | 1.0 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.1 | 1.1 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.1 | 1.6 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 1.3 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 10.9 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.1 | 0.4 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.1 | 3.0 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.1 | 1.2 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.1 | 0.5 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.1 | 3.6 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.1 | 2.8 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.1 | 0.3 | GO:0051461 | positive regulation of corticotropin secretion(GO:0051461) |
| 0.1 | 3.4 | GO:0033198 | response to ATP(GO:0033198) |
| 0.1 | 4.8 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 0.8 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 0.3 | GO:0071336 | regulation of hair follicle cell proliferation(GO:0071336) |
| 0.1 | 0.6 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.1 | 0.3 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.1 | 3.9 | GO:0043616 | keratinocyte proliferation(GO:0043616) |
| 0.1 | 4.9 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 1.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 5.6 | GO:0045806 | negative regulation of endocytosis(GO:0045806) |
| 0.1 | 3.7 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.1 | 3.6 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 2.2 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 7.6 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.1 | 1.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 1.5 | GO:1901028 | regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901028) |
| 0.1 | 5.0 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.1 | 0.5 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.1 | 0.5 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.1 | 2.4 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 1.3 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 0.7 | GO:2000580 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.7 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 | 2.4 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 0.5 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.1 | 0.9 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.2 | GO:1903027 | regulation of opsonization(GO:1903027) positive regulation of opsonization(GO:1903028) |
| 0.1 | 6.0 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.1 | 0.5 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.1 | 0.3 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 | 0.4 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.1 | 4.1 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.1 | 0.5 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.1 | 1.3 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 2.0 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.2 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.9 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.0 | 4.2 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 1.7 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.6 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.4 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.6 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.3 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.6 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.1 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.0 | 2.3 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 5.5 | GO:0006665 | sphingolipid metabolic process(GO:0006665) |
| 0.0 | 1.1 | GO:0040020 | regulation of meiotic nuclear division(GO:0040020) |
| 0.0 | 6.1 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 1.5 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.4 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 1.3 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.1 | GO:0030397 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 | 0.5 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.0 | 2.4 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 1.0 | GO:0043588 | skin development(GO:0043588) |
| 0.0 | 2.5 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 3.4 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 | 0.1 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.0 | 0.9 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 | 0.6 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 1.7 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.0 | 1.0 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 1.8 | GO:0050830 | defense response to Gram-positive bacterium(GO:0050830) |
| 0.0 | 0.5 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.1 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.0 | 0.4 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 4.2 | GO:0050808 | synapse organization(GO:0050808) |
| 0.0 | 0.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.5 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.0 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 3.6 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 0.2 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 | 0.8 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) |
| 0.0 | 0.3 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.0 | 0.5 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 4.1 | GO:0008544 | epidermis development(GO:0008544) |
| 0.0 | 0.9 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 1.1 | GO:0055072 | iron ion homeostasis(GO:0055072) |
| 0.0 | 0.3 | GO:0043276 | anoikis(GO:0043276) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) telomerase RNA stabilization(GO:0090669) |
| 0.0 | 0.4 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.2 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 1.9 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.0 | 1.1 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.3 | GO:0030224 | monocyte differentiation(GO:0030224) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 12.0 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 1.8 | 5.4 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 1.5 | 7.6 | GO:0005914 | spot adherens junction(GO:0005914) |
| 1.3 | 9.4 | GO:0097513 | myosin II filament(GO:0097513) |
| 1.3 | 5.2 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 1.2 | 6.2 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 1.2 | 11.0 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 1.2 | 3.5 | GO:0034677 | integrin alpha7-beta1 complex(GO:0034677) |
| 1.0 | 7.1 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.9 | 3.8 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.9 | 51.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.8 | 23.8 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.8 | 2.3 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.8 | 5.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.5 | 10.6 | GO:0042599 | lamellar body(GO:0042599) |
| 0.4 | 6.0 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.4 | 7.9 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.4 | 2.0 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.4 | 4.4 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.4 | 1.1 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.3 | 1.7 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.3 | 3.0 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.3 | 6.9 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.2 | 2.4 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.2 | 5.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.2 | 1.4 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.2 | 3.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 5.0 | GO:0043218 | compact myelin(GO:0043218) |
| 0.2 | 3.4 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.2 | 2.4 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.2 | 3.0 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 3.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 1.8 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 2.2 | GO:0090543 | Flemming body(GO:0090543) |
| 0.1 | 1.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 6.7 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 2.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 1.1 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 3.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 10.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 1.0 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 3.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 4.4 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 6.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 0.7 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.1 | 1.7 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 1.9 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 4.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 5.9 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 0.5 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.9 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 5.4 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 2.3 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 0.4 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 1.0 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 12.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 6.8 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 0.8 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.1 | 12.1 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 0.7 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 0.8 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 1.8 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 4.3 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 0.6 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 8.5 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 8.2 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 6.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 0.9 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 2.8 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 8.5 | GO:0031253 | cell projection membrane(GO:0031253) |
| 0.0 | 5.3 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 17.3 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.0 | 2.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 6.1 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.6 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 3.0 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 1.0 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.5 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 7.3 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.3 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 1.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 2.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.6 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 2.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 5.0 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.9 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 1.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 3.5 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 4.4 | GO:0034702 | ion channel complex(GO:0034702) |
| 0.0 | 0.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 4.8 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 4.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.7 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.4 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 1.1 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 1.7 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 4.3 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.6 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.9 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 2.2 | 21.6 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 1.7 | 6.8 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 1.6 | 9.6 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 1.6 | 4.7 | GO:0052598 | diamine oxidase activity(GO:0052597) histamine oxidase activity(GO:0052598) methylputrescine oxidase activity(GO:0052599) propane-1,3-diamine oxidase activity(GO:0052600) |
| 1.4 | 8.6 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 1.3 | 8.0 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 1.1 | 6.5 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.9 | 2.7 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.9 | 7.0 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.9 | 7.7 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.8 | 12.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.8 | 5.8 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.8 | 10.8 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.8 | 8.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.8 | 4.8 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.7 | 18.0 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.6 | 10.7 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.6 | 3.0 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.6 | 2.4 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.6 | 1.7 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.6 | 2.3 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.5 | 4.8 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.4 | 4.9 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.4 | 3.1 | GO:0050294 | alcohol sulfotransferase activity(GO:0004027) steroid sulfotransferase activity(GO:0050294) |
| 0.4 | 14.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.4 | 8.7 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.4 | 6.0 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.4 | 11.9 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.4 | 3.8 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.4 | 12.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.4 | 13.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.4 | 6.1 | GO:0019841 | retinol binding(GO:0019841) |
| 0.4 | 3.4 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.4 | 6.6 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.3 | 1.4 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.3 | 12.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.3 | 6.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.3 | 1.2 | GO:0051381 | histamine binding(GO:0051381) |
| 0.3 | 4.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.3 | 6.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.3 | 4.9 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.3 | 11.5 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.3 | 8.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.3 | 7.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.3 | 2.2 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.3 | 4.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.3 | 5.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.2 | 3.0 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.2 | 2.0 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.2 | 1.7 | GO:0002135 | CTP binding(GO:0002135) |
| 0.2 | 33.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 3.7 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.2 | 9.6 | GO:0043236 | laminin binding(GO:0043236) |
| 0.2 | 1.6 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.2 | 2.7 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.2 | 4.4 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.2 | 2.0 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.2 | 1.5 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.2 | 1.1 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
| 0.2 | 1.1 | GO:0043546 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) molybdopterin cofactor binding(GO:0043546) |
| 0.2 | 5.7 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.2 | 2.4 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 3.0 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.2 | 0.8 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.2 | 2.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.2 | 6.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.2 | 2.4 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.2 | 1.1 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.2 | 6.2 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.2 | 2.4 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.2 | 1.5 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.2 | 0.7 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.2 | 4.8 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.2 | 2.8 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.2 | 1.4 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 3.7 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 1.9 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.1 | 0.4 | GO:0005128 | erythropoietin receptor binding(GO:0005128) interleukin-3 receptor binding(GO:0005135) |
| 0.1 | 10.1 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 4.7 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 3.5 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.1 | 4.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 7.6 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 5.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 2.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 1.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 5.1 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 0.8 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 0.5 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.7 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 1.3 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.1 | 5.4 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 0.4 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.6 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 6.4 | GO:0016667 | oxidoreductase activity, acting on a sulfur group of donors(GO:0016667) |
| 0.1 | 0.3 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 2.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 2.3 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.1 | 0.3 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.1 | 0.5 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 13.0 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.1 | 1.9 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 1.6 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 0.5 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.1 | 1.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 2.4 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.1 | 3.6 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 3.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 3.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.7 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 0.3 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.1 | 1.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 1.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 1.0 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 5.1 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 0.6 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.1 | 1.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 22.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 2.0 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 2.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 1.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 3.5 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 2.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.4 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 2.9 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 1.3 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 2.0 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.8 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 1.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.0 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.0 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 4.9 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.3 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 3.6 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 3.2 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.1 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.1 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.0 | 0.6 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 0.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.0 | 12.5 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 1.0 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 2.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.4 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.6 | GO:0072509 | divalent inorganic cation transmembrane transporter activity(GO:0072509) |
| 0.0 | 8.9 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 3.7 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.9 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 5.9 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 1.2 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.9 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 2.0 | GO:0005244 | voltage-gated ion channel activity(GO:0005244) voltage-gated channel activity(GO:0022832) |
| 0.0 | 5.3 | GO:0005198 | structural molecule activity(GO:0005198) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 22.0 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 10.2 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.2 | 13.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 21.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 12.8 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.2 | 9.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 8.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.2 | 3.3 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 15.6 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 5.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 8.8 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 1.4 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.1 | 2.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 20.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 2.3 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 3.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 0.8 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 1.0 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 1.8 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 1.7 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 1.6 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 10.6 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 2.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 1.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 4.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 2.8 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 1.7 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 2.2 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 2.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.5 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 1.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.7 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.4 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 6.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.9 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 31.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.5 | 10.9 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.5 | 11.0 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.3 | 9.0 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.3 | 8.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.3 | 11.1 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.2 | 3.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 2.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 4.9 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.2 | 13.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 6.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 1.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 23.1 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.2 | 7.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.2 | 3.0 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 6.1 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.1 | 3.7 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 1.7 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 1.6 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 4.2 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 5.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 3.5 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 2.1 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.1 | 1.8 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 5.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 4.0 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 2.0 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 0.7 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 0.8 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.1 | 7.4 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.1 | 1.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.8 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.1 | 1.9 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 2.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 12.4 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 1.0 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 2.6 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 0.8 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 3.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.8 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 1.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.1 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.4 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 1.1 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.4 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.2 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 1.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.7 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.3 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.8 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |