Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
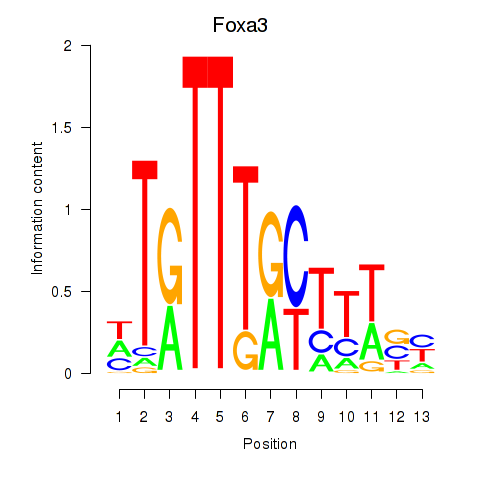
Results for Foxa3
Z-value: 1.51
Transcription factors associated with Foxa3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxa3
|
ENSMUSG00000040891.7 | Foxa3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxa3 | mm39_v1_chr7_-_18757461_18757477 | 0.79 | 3.0e-16 | Click! |
Activity profile of Foxa3 motif
Sorted Z-values of Foxa3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxa3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_46139878 | 47.44 |
ENSMUST00000034588.9
ENSMUST00000132155.2 |
Apoa1
|
apolipoprotein A-I |
| chr10_+_127734384 | 25.43 |
ENSMUST00000047134.8
|
Sdr9c7
|
4short chain dehydrogenase/reductase family 9C, member 7 |
| chr4_+_104623505 | 24.99 |
ENSMUST00000031663.10
ENSMUST00000065072.7 |
C8b
|
complement component 8, beta polypeptide |
| chr15_+_54274151 | 20.90 |
ENSMUST00000036737.4
|
Colec10
|
collectin sub-family member 10 |
| chr12_+_8062331 | 18.30 |
ENSMUST00000171239.2
|
Apob
|
apolipoprotein B |
| chr12_+_8027640 | 16.95 |
ENSMUST00000171271.8
ENSMUST00000037811.13 |
Apob
|
apolipoprotein B |
| chr12_+_8027767 | 16.81 |
ENSMUST00000037520.14
|
Apob
|
apolipoprotein B |
| chr15_-_96929086 | 16.48 |
ENSMUST00000230086.2
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr17_-_31363245 | 14.91 |
ENSMUST00000024826.8
|
Tff2
|
trefoil factor 2 (spasmolytic protein 1) |
| chr16_+_17149235 | 14.76 |
ENSMUST00000023450.15
ENSMUST00000231884.2 |
Serpind1
|
serine (or cysteine) peptidase inhibitor, clade D, member 1 |
| chr17_-_12894716 | 14.15 |
ENSMUST00000024596.10
|
Slc22a1
|
solute carrier family 22 (organic cation transporter), member 1 |
| chr3_+_106020545 | 13.97 |
ENSMUST00000079132.12
ENSMUST00000139086.2 |
Chia1
|
chitinase, acidic 1 |
| chr5_-_87054796 | 13.52 |
ENSMUST00000031181.16
ENSMUST00000113333.2 |
Ugt2b34
|
UDP glucuronosyltransferase 2 family, polypeptide B34 |
| chr18_-_32271224 | 13.32 |
ENSMUST00000234657.2
ENSMUST00000234386.2 ENSMUST00000234651.2 |
Proc
|
protein C |
| chr19_+_30210320 | 12.60 |
ENSMUST00000025797.7
|
Mbl2
|
mannose-binding lectin (protein C) 2 |
| chr4_-_63072367 | 12.15 |
ENSMUST00000030041.5
|
Ambp
|
alpha 1 microglobulin/bikunin precursor |
| chr12_-_81014849 | 12.02 |
ENSMUST00000095572.5
|
Slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 1 |
| chr3_+_138121245 | 11.94 |
ENSMUST00000161312.8
ENSMUST00000013458.9 |
Adh4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr19_-_20704896 | 11.73 |
ENSMUST00000025656.4
|
Aldh1a7
|
aldehyde dehydrogenase family 1, subfamily A7 |
| chr17_+_12597490 | 10.64 |
ENSMUST00000014578.7
|
Plg
|
plasminogen |
| chr14_-_30665232 | 10.45 |
ENSMUST00000006704.17
ENSMUST00000163118.2 |
Itih1
|
inter-alpha trypsin inhibitor, heavy chain 1 |
| chr3_+_137983250 | 10.37 |
ENSMUST00000004232.10
|
Adh1
|
alcohol dehydrogenase 1 (class I) |
| chr12_-_103739847 | 10.21 |
ENSMUST00000078869.6
|
Serpina1d
|
serine (or cysteine) peptidase inhibitor, clade A, member 1D |
| chr12_-_81014755 | 10.19 |
ENSMUST00000218342.2
|
Slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 1 |
| chr7_-_105249308 | 10.05 |
ENSMUST00000210531.2
ENSMUST00000033185.10 |
Hpx
|
hemopexin |
| chr4_-_6275629 | 9.96 |
ENSMUST00000029905.2
|
Cyp7a1
|
cytochrome P450, family 7, subfamily a, polypeptide 1 |
| chr6_-_23132977 | 9.77 |
ENSMUST00000031707.14
|
Aass
|
aminoadipate-semialdehyde synthase |
| chr17_+_25097199 | 9.55 |
ENSMUST00000050714.8
|
Igfals
|
insulin-like growth factor binding protein, acid labile subunit |
| chr19_+_20579322 | 9.44 |
ENSMUST00000087638.4
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1 |
| chr8_-_62576140 | 9.38 |
ENSMUST00000034052.14
ENSMUST00000034054.9 |
Anxa10
|
annexin A10 |
| chr12_-_84497718 | 8.97 |
ENSMUST00000085192.7
ENSMUST00000220491.2 |
Aldh6a1
|
aldehyde dehydrogenase family 6, subfamily A1 |
| chr11_+_78389913 | 8.91 |
ENSMUST00000017488.5
|
Vtn
|
vitronectin |
| chrX_+_138464065 | 8.88 |
ENSMUST00000113027.8
|
Rnf128
|
ring finger protein 128 |
| chr9_-_99599312 | 8.52 |
ENSMUST00000112882.9
ENSMUST00000131922.2 |
Cldn18
|
claudin 18 |
| chr17_-_31383976 | 8.39 |
ENSMUST00000235870.2
|
Tff1
|
trefoil factor 1 |
| chr9_-_44714263 | 8.18 |
ENSMUST00000044694.8
|
Ttc36
|
tetratricopeptide repeat domain 36 |
| chr7_-_144761806 | 7.83 |
ENSMUST00000208788.2
|
Smim38
|
small integral membrane protein 38 |
| chr2_+_58645189 | 7.43 |
ENSMUST00000102755.4
ENSMUST00000230627.2 ENSMUST00000229923.2 |
Upp2
|
uridine phosphorylase 2 |
| chr2_-_134396268 | 7.40 |
ENSMUST00000028704.3
|
Hao1
|
hydroxyacid oxidase 1, liver |
| chr9_-_103099262 | 7.37 |
ENSMUST00000170904.2
|
Trf
|
transferrin |
| chr15_+_4756684 | 6.67 |
ENSMUST00000161997.8
ENSMUST00000022788.15 |
C6
|
complement component 6 |
| chr2_+_58644922 | 6.57 |
ENSMUST00000059102.13
|
Upp2
|
uridine phosphorylase 2 |
| chr1_+_88093726 | 6.51 |
ENSMUST00000097659.5
|
Ugt1a5
|
UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr3_-_146302343 | 6.35 |
ENSMUST00000029836.9
|
Dnase2b
|
deoxyribonuclease II beta |
| chr15_+_4756657 | 6.21 |
ENSMUST00000162585.8
|
C6
|
complement component 6 |
| chr2_-_104573179 | 6.14 |
ENSMUST00000028595.8
|
Depdc7
|
DEP domain containing 7 |
| chrX_+_149377416 | 6.07 |
ENSMUST00000112713.3
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr6_+_146934082 | 5.72 |
ENSMUST00000036194.6
|
Rep15
|
RAB15 effector protein |
| chr1_+_172525613 | 5.62 |
ENSMUST00000038495.5
|
Crp
|
C-reactive protein, pentraxin-related |
| chr6_+_34575435 | 5.59 |
ENSMUST00000079391.10
ENSMUST00000142512.8 ENSMUST00000115027.8 ENSMUST00000115026.8 |
Cald1
|
caldesmon 1 |
| chr11_-_100036792 | 5.29 |
ENSMUST00000007317.8
|
Krt19
|
keratin 19 |
| chr11_-_69696428 | 5.28 |
ENSMUST00000051025.5
|
Tmem102
|
transmembrane protein 102 |
| chr9_-_71070506 | 5.21 |
ENSMUST00000074465.9
|
Aqp9
|
aquaporin 9 |
| chr9_-_99592116 | 5.18 |
ENSMUST00000035048.12
|
Cldn18
|
claudin 18 |
| chr8_+_46944000 | 5.16 |
ENSMUST00000110372.9
ENSMUST00000130563.2 |
Acsl1
|
acyl-CoA synthetase long-chain family member 1 |
| chr9_-_99592058 | 5.15 |
ENSMUST00000136429.8
|
Cldn18
|
claudin 18 |
| chr1_+_165591315 | 5.14 |
ENSMUST00000111432.10
|
Creg1
|
cellular repressor of E1A-stimulated genes 1 |
| chr2_+_67948057 | 4.96 |
ENSMUST00000112346.3
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr15_+_3300249 | 4.83 |
ENSMUST00000082424.12
ENSMUST00000159158.9 ENSMUST00000159216.10 ENSMUST00000160311.3 |
Selenop
|
selenoprotein P |
| chr9_+_98372575 | 4.71 |
ENSMUST00000035029.3
|
Rbp2
|
retinol binding protein 2, cellular |
| chr5_+_90708962 | 4.65 |
ENSMUST00000094615.8
ENSMUST00000200765.2 |
Albfm1
|
albumin superfamily member 1 |
| chr4_-_57916283 | 4.63 |
ENSMUST00000063816.6
|
D630039A03Rik
|
RIKEN cDNA D630039A03 gene |
| chr8_+_46984016 | 4.60 |
ENSMUST00000152423.2
|
Acsl1
|
acyl-CoA synthetase long-chain family member 1 |
| chr19_+_44980565 | 4.51 |
ENSMUST00000179305.2
|
Sema4g
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G |
| chr19_-_58442866 | 4.43 |
ENSMUST00000169850.8
|
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chrX_+_139808351 | 4.39 |
ENSMUST00000033806.5
|
Vsig1
|
V-set and immunoglobulin domain containing 1 |
| chr18_+_36797113 | 4.21 |
ENSMUST00000036765.8
|
Eif4ebp3
|
eukaryotic translation initiation factor 4E binding protein 3 |
| chr10_-_53952686 | 4.21 |
ENSMUST00000220088.2
|
Man1a
|
mannosidase 1, alpha |
| chr4_+_148686985 | 3.99 |
ENSMUST00000105701.9
ENSMUST00000052060.7 |
Masp2
|
mannan-binding lectin serine peptidase 2 |
| chr9_+_53212871 | 3.94 |
ENSMUST00000051014.2
|
Exph5
|
exophilin 5 |
| chr1_+_157353696 | 3.93 |
ENSMUST00000111700.8
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr19_+_29929208 | 3.66 |
ENSMUST00000136850.2
|
Il33
|
interleukin 33 |
| chr5_-_28672091 | 3.61 |
ENSMUST00000002708.5
|
Shh
|
sonic hedgehog |
| chr11_+_77656414 | 3.55 |
ENSMUST00000164315.2
|
Myo18a
|
myosin XVIIIA |
| chr12_+_119407145 | 3.49 |
ENSMUST00000048880.7
|
Macc1
|
metastasis associated in colon cancer 1 |
| chr1_+_93301596 | 3.45 |
ENSMUST00000058682.11
ENSMUST00000186641.7 |
Ano7
|
anoctamin 7 |
| chr17_-_79292856 | 3.32 |
ENSMUST00000118991.2
|
Prkd3
|
protein kinase D3 |
| chr12_+_119291343 | 3.31 |
ENSMUST00000221917.2
|
Macc1
|
metastasis associated in colon cancer 1 |
| chr3_+_85878376 | 3.31 |
ENSMUST00000238443.2
|
Sh3d19
|
SH3 domain protein D19 |
| chr2_-_91025492 | 3.28 |
ENSMUST00000111354.2
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr8_-_65582206 | 3.06 |
ENSMUST00000098713.5
|
Smim31
|
small integral membrane protein 31 |
| chr3_-_92922976 | 3.04 |
ENSMUST00000107301.2
ENSMUST00000029521.5 |
Crct1
|
cysteine-rich C-terminal 1 |
| chr19_-_58444336 | 3.01 |
ENSMUST00000131877.2
|
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr3_+_20011405 | 2.96 |
ENSMUST00000108325.9
|
Cp
|
ceruloplasmin |
| chr11_-_69553390 | 2.92 |
ENSMUST00000129224.8
ENSMUST00000155200.8 |
Mpdu1
|
mannose-P-dolichol utilization defect 1 |
| chr3_+_20011251 | 2.86 |
ENSMUST00000108328.8
|
Cp
|
ceruloplasmin |
| chr4_+_150938376 | 2.85 |
ENSMUST00000073600.9
|
Errfi1
|
ERBB receptor feedback inhibitor 1 |
| chr2_-_77349909 | 2.81 |
ENSMUST00000111830.9
|
Zfp385b
|
zinc finger protein 385B |
| chr10_-_95678786 | 2.78 |
ENSMUST00000211096.2
|
Gm33543
|
predicted gene, 33543 |
| chr3_+_20011201 | 2.74 |
ENSMUST00000091309.12
ENSMUST00000108329.8 ENSMUST00000003714.13 |
Cp
|
ceruloplasmin |
| chr3_-_106126794 | 2.70 |
ENSMUST00000082219.6
|
Chil4
|
chitinase-like 4 |
| chr15_-_99717956 | 2.64 |
ENSMUST00000109024.9
|
Lima1
|
LIM domain and actin binding 1 |
| chr11_-_69553451 | 2.52 |
ENSMUST00000018905.12
|
Mpdu1
|
mannose-P-dolichol utilization defect 1 |
| chr7_-_84328553 | 2.51 |
ENSMUST00000069537.3
ENSMUST00000207865.2 ENSMUST00000178385.9 ENSMUST00000208782.2 |
Zfand6
|
zinc finger, AN1-type domain 6 |
| chrX_+_141011173 | 2.51 |
ENSMUST00000112914.8
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr6_+_34686373 | 2.45 |
ENSMUST00000115021.8
|
Cald1
|
caldesmon 1 |
| chr15_+_9071655 | 2.40 |
ENSMUST00000227682.3
|
Nadk2
|
NAD kinase 2, mitochondrial |
| chr11_+_78356523 | 2.40 |
ENSMUST00000001126.4
|
Slc46a1
|
solute carrier family 46, member 1 |
| chr13_+_23922783 | 2.32 |
ENSMUST00000040914.3
|
H1f2
|
H1.2 linker histone, cluster member |
| chr9_+_66065488 | 2.28 |
ENSMUST00000034944.9
ENSMUST00000238682.2 |
Dapk2
|
death-associated protein kinase 2 |
| chr15_+_9071331 | 2.26 |
ENSMUST00000190591.10
|
Nadk2
|
NAD kinase 2, mitochondrial |
| chr4_+_101574601 | 2.26 |
ENSMUST00000102777.10
ENSMUST00000106921.9 ENSMUST00000037552.10 ENSMUST00000145024.2 |
Lepr
|
leptin receptor |
| chr7_+_18962252 | 2.25 |
ENSMUST00000063976.9
|
Opa3
|
optic atrophy 3 |
| chr4_-_127247864 | 2.21 |
ENSMUST00000106090.8
ENSMUST00000060419.2 |
Gjb4
|
gap junction protein, beta 4 |
| chr6_-_86742789 | 2.19 |
ENSMUST00000123732.4
|
Anxa4
|
annexin A4 |
| chr10_-_128755127 | 2.17 |
ENSMUST00000149961.2
ENSMUST00000026406.14 |
Rdh5
|
retinol dehydrogenase 5 |
| chr7_+_130633776 | 2.07 |
ENSMUST00000084509.7
ENSMUST00000213064.3 ENSMUST00000208311.4 |
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr3_-_148696155 | 2.01 |
ENSMUST00000196526.5
ENSMUST00000200543.5 ENSMUST00000200154.5 |
Adgrl2
|
adhesion G protein-coupled receptor L2 |
| chrX_+_141010919 | 1.93 |
ENSMUST00000042329.12
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr14_-_20319242 | 1.89 |
ENSMUST00000024155.9
|
Kcnk16
|
potassium channel, subfamily K, member 16 |
| chr1_-_54233207 | 1.89 |
ENSMUST00000120904.8
|
Hecw2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr11_+_108811626 | 1.87 |
ENSMUST00000140821.2
|
Axin2
|
axin 2 |
| chrX_+_108138965 | 1.86 |
ENSMUST00000033598.9
|
Sh3bgrl
|
SH3-binding domain glutamic acid-rich protein like |
| chr11_+_94455865 | 1.85 |
ENSMUST00000040418.9
|
Chad
|
chondroadherin |
| chr2_+_24235300 | 1.84 |
ENSMUST00000114485.9
ENSMUST00000114482.3 |
Il1rn
|
interleukin 1 receptor antagonist |
| chr18_+_36498826 | 1.83 |
ENSMUST00000144158.2
|
Cystm1
|
cysteine-rich transmembrane module containing 1 |
| chr2_-_62313981 | 1.79 |
ENSMUST00000136686.2
ENSMUST00000102733.10 |
Gcg
|
glucagon |
| chr19_-_12206908 | 1.78 |
ENSMUST00000180978.3
|
Olfr1432
|
olfactory receptor 1432 |
| chr11_-_99328969 | 1.77 |
ENSMUST00000017743.3
|
Krt20
|
keratin 20 |
| chrX_-_144131890 | 1.73 |
ENSMUST00000040084.10
ENSMUST00000123443.2 |
Lhfpl1
|
lipoma HMGIC fusion partner-like 1 |
| chr12_+_112645237 | 1.63 |
ENSMUST00000174780.2
ENSMUST00000169593.2 ENSMUST00000173942.2 |
Zbtb42
|
zinc finger and BTB domain containing 42 |
| chr17_-_80203457 | 1.63 |
ENSMUST00000068282.7
ENSMUST00000112437.8 |
Atl2
|
atlastin GTPase 2 |
| chr15_-_3333003 | 1.62 |
ENSMUST00000165386.2
|
Ccdc152
|
coiled-coil domain containing 152 |
| chrX_-_140508177 | 1.60 |
ENSMUST00000067841.8
|
Irs4
|
insulin receptor substrate 4 |
| chr2_+_153003212 | 1.55 |
ENSMUST00000089027.3
|
Tm9sf4
|
transmembrane 9 superfamily member 4 |
| chr14_+_32321824 | 1.44 |
ENSMUST00000068938.7
ENSMUST00000228878.2 |
Prrxl1
|
paired related homeobox protein-like 1 |
| chr18_-_39051695 | 1.44 |
ENSMUST00000040647.11
|
Fgf1
|
fibroblast growth factor 1 |
| chr6_-_86742847 | 1.41 |
ENSMUST00000113675.8
|
Anxa4
|
annexin A4 |
| chr2_-_51039112 | 1.38 |
ENSMUST00000154545.2
ENSMUST00000017288.9 |
Rnd3
|
Rho family GTPase 3 |
| chr17_+_29077385 | 1.37 |
ENSMUST00000056866.8
|
Pnpla1
|
patatin-like phospholipase domain containing 1 |
| chr2_+_164675697 | 1.36 |
ENSMUST00000143780.9
|
Ctsa
|
cathepsin A |
| chr14_-_40907106 | 1.36 |
ENSMUST00000077136.5
|
Sftpd
|
surfactant associated protein D |
| chr11_+_46701619 | 1.35 |
ENSMUST00000068877.7
|
Timd4
|
T cell immunoglobulin and mucin domain containing 4 |
| chr11_-_69563133 | 1.31 |
ENSMUST00000163666.3
|
Eif4a1
|
eukaryotic translation initiation factor 4A1 |
| chr17_-_43003135 | 1.31 |
ENSMUST00000170723.8
ENSMUST00000164524.2 ENSMUST00000024711.11 ENSMUST00000167993.8 |
Adgrf4
|
adhesion G protein-coupled receptor F4 |
| chr2_-_34261121 | 1.30 |
ENSMUST00000127353.3
ENSMUST00000141653.3 |
Pbx3
|
pre B cell leukemia homeobox 3 |
| chr1_+_75119419 | 1.30 |
ENSMUST00000097694.11
ENSMUST00000190240.7 |
Retreg2
|
reticulophagy regulator family member 2 |
| chr8_-_26275182 | 1.27 |
ENSMUST00000038498.10
|
Bag4
|
BCL2-associated athanogene 4 |
| chr3_-_92346078 | 1.20 |
ENSMUST00000062160.4
|
Sprr1b
|
small proline-rich protein 1B |
| chr16_-_35891739 | 1.17 |
ENSMUST00000231351.2
ENSMUST00000004057.9 |
Fam162a
|
family with sequence similarity 162, member A |
| chr6_+_42377172 | 1.16 |
ENSMUST00000057398.4
|
Tas2r143
|
taste receptor, type 2, member 143 |
| chr14_+_32321341 | 1.15 |
ENSMUST00000187377.7
ENSMUST00000189022.8 ENSMUST00000186452.7 |
Prrxl1
|
paired related homeobox protein-like 1 |
| chrX_-_55643429 | 1.15 |
ENSMUST00000059899.3
|
Mmgt1
|
membrane magnesium transporter 1 |
| chr14_-_48900192 | 1.15 |
ENSMUST00000122009.8
|
Otx2
|
orthodenticle homeobox 2 |
| chr1_+_75119472 | 1.11 |
ENSMUST00000189650.7
|
Retreg2
|
reticulophagy regulator family member 2 |
| chr13_+_36301331 | 1.08 |
ENSMUST00000021857.13
|
Fars2
|
phenylalanine-tRNA synthetase 2 (mitochondrial) |
| chr15_-_54783357 | 1.03 |
ENSMUST00000167541.3
ENSMUST00000171545.9 ENSMUST00000041591.16 ENSMUST00000173516.8 |
Enpp2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr13_+_89687915 | 1.00 |
ENSMUST00000022108.9
|
Hapln1
|
hyaluronan and proteoglycan link protein 1 |
| chr16_-_48232770 | 1.00 |
ENSMUST00000212197.2
|
Gm5485
|
predicted gene 5485 |
| chr6_-_40590244 | 0.99 |
ENSMUST00000076565.3
|
Tas2r138
|
taste receptor, type 2, member 138 |
| chr1_-_82933734 | 0.98 |
ENSMUST00000222426.2
|
Gm7544
|
predicted gene 7544 |
| chr10_-_129627483 | 0.95 |
ENSMUST00000091986.4
|
Olfr810
|
olfactory receptor 810 |
| chr2_-_111062182 | 0.94 |
ENSMUST00000099620.5
|
Olfr1275
|
olfactory receptor 1275 |
| chr12_-_108241597 | 0.91 |
ENSMUST00000222310.2
|
Ccdc85c
|
coiled-coil domain containing 85C |
| chr3_-_27764522 | 0.88 |
ENSMUST00000195008.6
|
Fndc3b
|
fibronectin type III domain containing 3B |
| chr14_-_50519728 | 0.88 |
ENSMUST00000071208.3
|
Olfr732
|
olfactory receptor 732 |
| chr4_-_55532453 | 0.83 |
ENSMUST00000132746.2
ENSMUST00000107619.3 |
Klf4
|
Kruppel-like factor 4 (gut) |
| chrX_-_107877909 | 0.83 |
ENSMUST00000101283.4
ENSMUST00000150434.8 |
Brwd3
|
bromodomain and WD repeat domain containing 3 |
| chr10_-_52071340 | 0.81 |
ENSMUST00000020045.10
|
Ros1
|
Ros1 proto-oncogene |
| chr6_+_41945074 | 0.81 |
ENSMUST00000080742.3
|
Sval3
|
seminal vesicle antigen-like 3 |
| chr5_+_88635834 | 0.80 |
ENSMUST00000199104.5
ENSMUST00000031222.9 |
Enam
|
enamelin |
| chr12_-_32000169 | 0.78 |
ENSMUST00000176520.8
|
Hbp1
|
high mobility group box transcription factor 1 |
| chr10_+_97400990 | 0.77 |
ENSMUST00000038160.6
|
Lum
|
lumican |
| chr7_+_18962301 | 0.76 |
ENSMUST00000161711.2
|
Opa3
|
optic atrophy 3 |
| chr10_-_57408585 | 0.75 |
ENSMUST00000020027.11
|
Serinc1
|
serine incorporator 1 |
| chr3_-_102871440 | 0.72 |
ENSMUST00000058899.13
|
Nr1h5
|
nuclear receptor subfamily 1, group H, member 5 |
| chr17_+_38485977 | 0.71 |
ENSMUST00000074883.2
|
Olfr134
|
olfactory receptor 134 |
| chr10_-_57408512 | 0.69 |
ENSMUST00000169122.8
|
Serinc1
|
serine incorporator 1 |
| chr17_+_38106337 | 0.68 |
ENSMUST00000054748.6
|
Olfr123
|
olfactory receptor 123 |
| chr2_+_86338805 | 0.66 |
ENSMUST00000076263.2
|
Olfr1076
|
olfactory receptor 1076 |
| chr4_-_43823866 | 0.65 |
ENSMUST00000215406.2
ENSMUST00000079234.6 ENSMUST00000214843.2 |
Olfr156
|
olfactory receptor 156 |
| chr17_-_42922286 | 0.64 |
ENSMUST00000068355.8
|
Opn5
|
opsin 5 |
| chr9_+_107457316 | 0.63 |
ENSMUST00000093785.6
|
Naa80
|
N(alpha)-acetyltransferase 80, NatH catalytic subunit |
| chr10_+_21868114 | 0.62 |
ENSMUST00000150089.8
ENSMUST00000100036.10 |
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr11_+_59503792 | 0.62 |
ENSMUST00000055276.6
|
Olfr225
|
olfactory receptor 225 |
| chr19_-_12773472 | 0.61 |
ENSMUST00000038627.9
|
Zfp91
|
zinc finger protein 91 |
| chr2_+_153984800 | 0.59 |
ENSMUST00000028985.8
|
Bpifa1
|
BPI fold containing family A, member 1 |
| chr6_+_29348068 | 0.57 |
ENSMUST00000173216.8
ENSMUST00000173694.5 ENSMUST00000172974.8 ENSMUST00000031779.17 ENSMUST00000090481.14 |
Calu
|
calumenin |
| chr12_-_32000209 | 0.57 |
ENSMUST00000176084.2
ENSMUST00000176103.8 ENSMUST00000167458.9 |
Hbp1
|
high mobility group box transcription factor 1 |
| chr3_+_107137924 | 0.55 |
ENSMUST00000179399.3
|
A630076J17Rik
|
RIKEN cDNA A630076J17 gene |
| chr2_-_84255602 | 0.53 |
ENSMUST00000074262.9
|
Calcrl
|
calcitonin receptor-like |
| chr13_+_23398297 | 0.50 |
ENSMUST00000236177.2
|
Vmn1r221
|
vomeronasal 1 receptor 221 |
| chr17_+_33410276 | 0.49 |
ENSMUST00000214406.2
ENSMUST00000213731.2 |
Olfr239
|
olfactory receptor 239 |
| chr3_+_96127174 | 0.49 |
ENSMUST00000073115.5
|
H2ac21
|
H2A clustered histone 21 |
| chr13_+_22454692 | 0.47 |
ENSMUST00000228711.2
|
Vmn1r195
|
vomeronasal 1 receptor 195 |
| chr11_+_108811168 | 0.46 |
ENSMUST00000052915.14
|
Axin2
|
axin 2 |
| chr13_-_115226666 | 0.46 |
ENSMUST00000109226.5
|
Pelo
|
pelota mRNA surveillance and ribosome rescue factor |
| chr14_-_110992533 | 0.45 |
ENSMUST00000078386.4
|
Slitrk6
|
SLIT and NTRK-like family, member 6 |
| chr12_+_59178072 | 0.44 |
ENSMUST00000176464.8
ENSMUST00000170992.9 ENSMUST00000176322.8 |
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr2_-_39116457 | 0.42 |
ENSMUST00000028087.6
|
Ppp6c
|
protein phosphatase 6, catalytic subunit |
| chr5_+_104447037 | 0.40 |
ENSMUST00000031246.9
|
Ibsp
|
integrin binding sialoprotein |
| chr9_-_79920131 | 0.37 |
ENSMUST00000217264.2
|
Filip1
|
filamin A interacting protein 1 |
| chr13_+_24118417 | 0.37 |
ENSMUST00000072391.2
|
H2ac1
|
H2A clustered histone 1 |
| chr11_-_99884818 | 0.36 |
ENSMUST00000105049.2
|
Krtap17-1
|
keratin associated protein 17-1 |
| chr12_+_59178258 | 0.36 |
ENSMUST00000177162.8
|
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr2_+_85597442 | 0.35 |
ENSMUST00000216397.3
|
Olfr1013
|
olfactory receptor 1013 |
| chr18_+_67266784 | 0.34 |
ENSMUST00000236918.2
|
Gnal
|
guanine nucleotide binding protein, alpha stimulating, olfactory type |
| chr11_-_70560110 | 0.30 |
ENSMUST00000129434.2
ENSMUST00000018431.13 |
Spag7
|
sperm associated antigen 7 |
| chr1_+_173093568 | 0.29 |
ENSMUST00000213420.2
|
Olfr418
|
olfactory receptor 418 |
| chr2_-_39116284 | 0.29 |
ENSMUST00000204701.3
|
Ppp6c
|
protein phosphatase 6, catalytic subunit |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.9 | 47.4 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 5.6 | 22.3 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 3.7 | 52.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 3.3 | 9.8 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 3.2 | 12.9 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 2.7 | 18.9 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 2.4 | 14.2 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 2.2 | 9.0 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 2.1 | 14.9 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 2.1 | 10.6 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 2.0 | 10.0 | GO:0070859 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 1.8 | 12.5 | GO:0015886 | heme transport(GO:0015886) |
| 1.8 | 16.0 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 1.7 | 25.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 1.5 | 7.4 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 1.5 | 13.3 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 1.4 | 11.0 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 1.3 | 5.2 | GO:0015855 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 1.3 | 16.6 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 1.2 | 3.5 | GO:1903028 | regulation of opsonization(GO:1903027) positive regulation of opsonization(GO:1903028) |
| 1.0 | 9.4 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.9 | 12.8 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.8 | 3.3 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.8 | 11.7 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.8 | 2.3 | GO:2000424 | regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 0.7 | 22.2 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.7 | 5.6 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.7 | 8.9 | GO:0097421 | smooth muscle cell-matrix adhesion(GO:0061302) liver regeneration(GO:0097421) |
| 0.6 | 6.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.6 | 4.8 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.6 | 4.7 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.6 | 9.3 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.6 | 3.5 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.6 | 2.3 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 0.6 | 3.9 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.6 | 5.0 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.5 | 6.5 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.5 | 3.7 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.5 | 4.7 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.5 | 2.8 | GO:0010616 | negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.5 | 3.3 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.5 | 5.5 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.4 | 1.3 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 0.4 | 2.3 | GO:0032423 | regulation of mismatch repair(GO:0032423) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.4 | 1.8 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.4 | 5.7 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.3 | 5.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.3 | 1.4 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.3 | 5.3 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.3 | 6.3 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.3 | 16.5 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.3 | 8.6 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.3 | 1.4 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.3 | 0.8 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.3 | 4.4 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.2 | 9.7 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.2 | 1.8 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.2 | 1.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.2 | 1.4 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.2 | 1.4 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.2 | 17.9 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.2 | 4.2 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.2 | 1.1 | GO:0042706 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.2 | 0.8 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.2 | 1.8 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 0.2 | 4.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.6 | GO:1900191 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.1 | 2.2 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.1 | 2.6 | GO:0021559 | trigeminal nerve development(GO:0021559) |
| 0.1 | 0.8 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.1 | 0.5 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.1 | 1.0 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 3.3 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.2 | GO:0034136 | negative regulation of toll-like receptor 2 signaling pathway(GO:0034136) |
| 0.1 | 5.3 | GO:0060706 | cell differentiation involved in embryonic placenta development(GO:0060706) |
| 0.1 | 0.8 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.1 | 1.1 | GO:0015684 | ferrous iron transport(GO:0015684) |
| 0.1 | 1.9 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.5 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 2.5 | GO:0072662 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.1 | 3.0 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 4.5 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.1 | 8.0 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.1 | 1.3 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.1 | 0.9 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.1 | 2.1 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 12.6 | GO:0009166 | nucleotide catabolic process(GO:0009166) |
| 0.1 | 8.2 | GO:0007586 | digestion(GO:0007586) |
| 0.1 | 0.4 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 1.8 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.1 | 1.4 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 17.2 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.3 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 1.7 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.6 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 1.2 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 1.2 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.4 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.0 | 4.4 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.3 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 1.9 | GO:0030071 | regulation of mitotic metaphase/anaphase transition(GO:0030071) |
| 0.0 | 0.6 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 1.3 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.8 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 2.4 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 4.0 | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation(GO:0050731) |
| 0.0 | 16.7 | GO:0055114 | oxidation-reduction process(GO:0055114) |
| 0.0 | 0.2 | GO:0042509 | regulation of tyrosine phosphorylation of STAT protein(GO:0042509) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.4 | 52.1 | GO:0034359 | mature chylomicron(GO:0034359) |
| 9.5 | 47.4 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 4.2 | 37.9 | GO:0005579 | membrane attack complex(GO:0005579) |
| 2.1 | 10.6 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 2.0 | 6.1 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 1.3 | 8.9 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 1.3 | 14.0 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 1.0 | 9.6 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.8 | 8.0 | GO:0030478 | actin cap(GO:0030478) |
| 0.7 | 7.4 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.6 | 5.3 | GO:1990357 | terminal web(GO:1990357) |
| 0.3 | 35.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.2 | 2.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 38.6 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 19.8 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 44.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 9.8 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 1.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 2.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 2.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 7.4 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 2.6 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 8.9 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 72.6 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 3.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 2.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 3.8 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.8 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.8 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 1.2 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.8 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 23.4 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 1.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 44.3 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 4.2 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 2.6 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.2 | GO:0031985 | Golgi cisterna(GO:0031985) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.8 | 47.4 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 7.4 | 22.3 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 4.7 | 14.2 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) norepinephrine transmembrane transporter activity(GO:0005333) acetate ester transmembrane transporter activity(GO:1901375) |
| 4.2 | 21.2 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 4.0 | 52.1 | GO:0035473 | lipase binding(GO:0035473) |
| 3.0 | 12.2 | GO:0019862 | IgA binding(GO:0019862) |
| 2.8 | 22.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 2.7 | 13.3 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 2.3 | 14.0 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 2.1 | 12.5 | GO:0015232 | heme transporter activity(GO:0015232) |
| 2.0 | 9.8 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 1.8 | 7.4 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 1.8 | 16.0 | GO:0004568 | chitinase activity(GO:0004568) |
| 1.6 | 12.6 | GO:0005534 | galactose binding(GO:0005534) |
| 1.3 | 27.6 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 1.3 | 6.3 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 1.2 | 5.0 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 1.1 | 14.9 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 1.1 | 7.4 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 1.0 | 5.2 | GO:0005350 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 0.9 | 20.9 | GO:0005537 | mannose binding(GO:0005537) |
| 0.7 | 8.6 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.7 | 5.6 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.7 | 6.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.6 | 1.9 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.5 | 3.3 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.5 | 20.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.5 | 7.6 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.5 | 1.8 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.5 | 3.6 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.4 | 9.6 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.4 | 4.0 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.3 | 4.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.3 | 0.6 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.3 | 8.9 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.3 | 4.7 | GO:0019841 | retinol binding(GO:0019841) |
| 0.3 | 4.8 | GO:0008430 | selenium binding(GO:0008430) |
| 0.3 | 0.8 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.3 | 4.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.2 | 6.2 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.2 | 35.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 0.8 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.2 | 10.0 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.2 | 1.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.2 | 0.5 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.2 | 13.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.2 | 9.0 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.1 | 3.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 1.1 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.1 | 2.3 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 3.5 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 2.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 14.7 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.1 | 1.9 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 0.6 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 1.7 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 1.4 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 5.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 3.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 1.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 2.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 3.5 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 1.4 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 1.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.1 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.9 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 2.5 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 5.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 5.3 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 1.6 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 2.1 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.5 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 13.3 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 1.3 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 3.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 5.1 | GO:0048037 | cofactor binding(GO:0048037) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 1.3 | GO:0051087 | chaperone binding(GO:0051087) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 70.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.7 | 47.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.3 | 11.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.3 | 56.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.2 | 10.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.2 | 4.7 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 37.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 8.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 2.3 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 4.6 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 2.2 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 3.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 2.4 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.8 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 2.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 2.3 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 1.3 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 1.4 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.2 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 1.9 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.2 | PID AURORA A PATHWAY | Aurora A signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 99.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 2.8 | 22.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 2.0 | 22.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 1.9 | 21.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 1.6 | 37.9 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 1.0 | 13.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 1.0 | 20.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.9 | 14.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.7 | 14.0 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.6 | 6.5 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.6 | 10.0 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.5 | 18.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.4 | 9.8 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.4 | 5.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.4 | 9.0 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 13.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.2 | 8.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.2 | 8.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 6.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 2.6 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 1.9 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 7.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 4.7 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 2.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 3.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 1.4 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 16.6 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 6.6 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 0.6 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.3 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 1.1 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 2.2 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 1.8 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 3.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 4.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 3.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |