Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Foxc1
Z-value: 1.44
Transcription factors associated with Foxc1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxc1
|
ENSMUSG00000050295.5 | Foxc1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxc1 | mm39_v1_chr13_+_31990604_31990633 | -0.34 | 3.7e-03 | Click! |
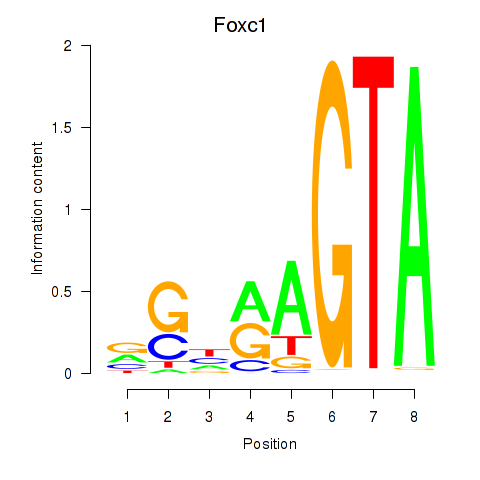
Activity profile of Foxc1 motif
Sorted Z-values of Foxc1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxc1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_103099262 | 14.61 |
ENSMUST00000170904.2
|
Trf
|
transferrin |
| chr11_+_108286114 | 13.22 |
ENSMUST00000000049.6
|
Apoh
|
apolipoprotein H |
| chr19_+_40078132 | 13.20 |
ENSMUST00000068094.13
ENSMUST00000080171.3 |
Cyp2c50
|
cytochrome P450, family 2, subfamily c, polypeptide 50 |
| chr9_+_123603605 | 11.58 |
ENSMUST00000180093.2
|
Ccr9
|
chemokine (C-C motif) receptor 9 |
| chr8_+_105775224 | 11.26 |
ENSMUST00000093222.13
ENSMUST00000093223.5 |
Ces3a
|
carboxylesterase 3A |
| chr14_-_31362909 | 10.26 |
ENSMUST00000022437.16
|
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr6_+_41520150 | 10.20 |
ENSMUST00000103295.2
|
Trbj2-3
|
T cell receptor beta joining 2-3 |
| chr1_-_172722589 | 10.14 |
ENSMUST00000027824.7
|
Apcs
|
serum amyloid P-component |
| chr14_-_31362835 | 9.79 |
ENSMUST00000167066.8
ENSMUST00000127204.9 |
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr8_+_105810380 | 9.55 |
ENSMUST00000093221.13
ENSMUST00000074403.13 |
Ces3b
|
carboxylesterase 3B |
| chr11_+_69945157 | 8.23 |
ENSMUST00000108585.9
ENSMUST00000018699.13 |
Asgr1
|
asialoglycoprotein receptor 1 |
| chr14_+_54043954 | 8.00 |
ENSMUST00000103672.9
ENSMUST00000186545.2 |
Trav17
|
T cell receptor alpha variable 17 |
| chr15_+_80507671 | 7.91 |
ENSMUST00000043149.9
|
Grap2
|
GRB2-related adaptor protein 2 |
| chr14_+_53913598 | 7.39 |
ENSMUST00000103662.6
|
Trav9-4
|
T cell receptor alpha variable 9-4 |
| chr7_+_30193047 | 7.36 |
ENSMUST00000058280.13
ENSMUST00000133318.8 ENSMUST00000142575.8 ENSMUST00000131040.2 |
Prodh2
|
proline dehydrogenase (oxidase) 2 |
| chr5_+_146016064 | 7.33 |
ENSMUST00000035571.10
|
Cyp3a59
|
cytochrome P450, family 3, subfamily a, polypeptide 59 |
| chr9_-_44920899 | 6.97 |
ENSMUST00000102832.3
|
Cd3e
|
CD3 antigen, epsilon polypeptide |
| chr6_-_55152002 | 6.72 |
ENSMUST00000003569.6
|
Inmt
|
indolethylamine N-methyltransferase |
| chr16_-_48592319 | 6.64 |
ENSMUST00000239408.2
|
Trat1
|
T cell receptor associated transmembrane adaptor 1 |
| chr9_+_123596276 | 6.47 |
ENSMUST00000166236.9
ENSMUST00000111454.4 ENSMUST00000168910.2 |
Ccr9
|
chemokine (C-C motif) receptor 9 |
| chr16_+_17149235 | 6.17 |
ENSMUST00000023450.15
ENSMUST00000231884.2 |
Serpind1
|
serine (or cysteine) peptidase inhibitor, clade D, member 1 |
| chr14_+_53129643 | 6.08 |
ENSMUST00000199746.5
ENSMUST00000178824.2 |
Trav9d-2
|
T cell receptor alpha variable 9D-2 |
| chr16_-_48592372 | 6.06 |
ENSMUST00000231701.3
|
Trat1
|
T cell receptor associated transmembrane adaptor 1 |
| chr6_+_41118120 | 5.89 |
ENSMUST00000103273.3
|
Trbv15
|
T cell receptor beta, variable 15 |
| chr12_-_73093953 | 5.87 |
ENSMUST00000050029.8
|
Six1
|
sine oculis-related homeobox 1 |
| chr2_-_10135449 | 5.75 |
ENSMUST00000042290.14
|
Itih2
|
inter-alpha trypsin inhibitor, heavy chain 2 |
| chr7_+_119360141 | 5.34 |
ENSMUST00000106528.8
ENSMUST00000106527.8 ENSMUST00000063770.10 ENSMUST00000106529.8 |
Acsm3
|
acyl-CoA synthetase medium-chain family member 3 |
| chr16_-_19703014 | 5.32 |
ENSMUST00000100083.5
ENSMUST00000231564.2 |
A930003A15Rik
|
RIKEN cDNA A930003A15 gene |
| chr2_+_58644922 | 5.20 |
ENSMUST00000059102.13
|
Upp2
|
uridine phosphorylase 2 |
| chr2_+_58645189 | 5.19 |
ENSMUST00000102755.4
ENSMUST00000230627.2 ENSMUST00000229923.2 |
Upp2
|
uridine phosphorylase 2 |
| chr10_+_127157784 | 5.04 |
ENSMUST00000219511.2
|
Arhgap9
|
Rho GTPase activating protein 9 |
| chr11_+_70410009 | 4.82 |
ENSMUST00000057685.3
|
Gltpd2
|
glycolipid transfer protein domain containing 2 |
| chr12_-_113386312 | 4.80 |
ENSMUST00000177715.8
ENSMUST00000103426.3 |
Ighm
|
immunoglobulin heavy constant mu |
| chr14_+_53980561 | 4.78 |
ENSMUST00000103667.6
|
Trav16
|
T cell receptor alpha variable 16 |
| chr8_-_25066313 | 4.72 |
ENSMUST00000121992.2
|
Ido2
|
indoleamine 2,3-dioxygenase 2 |
| chr3_+_94284739 | 4.63 |
ENSMUST00000197040.5
|
Rorc
|
RAR-related orphan receptor gamma |
| chr9_+_110248815 | 4.59 |
ENSMUST00000035061.9
|
Ngp
|
neutrophilic granule protein |
| chr2_-_103315483 | 4.41 |
ENSMUST00000028610.10
|
Cat
|
catalase |
| chr8_-_84769170 | 4.38 |
ENSMUST00000005601.9
|
Il27ra
|
interleukin 27 receptor, alpha |
| chr14_-_52252318 | 4.29 |
ENSMUST00000228051.2
|
Zfp219
|
zinc finger protein 219 |
| chr3_+_94284812 | 4.22 |
ENSMUST00000200009.2
|
Rorc
|
RAR-related orphan receptor gamma |
| chr14_+_54453748 | 4.18 |
ENSMUST00000103736.2
|
Traj4
|
T cell receptor alpha joining 4 |
| chr14_+_31363004 | 4.17 |
ENSMUST00000090147.7
|
Btd
|
biotinidase |
| chr10_+_127159609 | 4.10 |
ENSMUST00000069548.7
|
Arhgap9
|
Rho GTPase activating protein 9 |
| chr10_+_127159568 | 4.08 |
ENSMUST00000219026.2
|
Arhgap9
|
Rho GTPase activating protein 9 |
| chr10_-_89369432 | 4.02 |
ENSMUST00000105297.2
|
Nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr14_+_53153256 | 3.98 |
ENSMUST00000197007.2
ENSMUST00000103593.3 |
Trav12d-2
|
T cell receptor alpha variable 12D-2 |
| chr3_-_14873406 | 3.98 |
ENSMUST00000181860.8
ENSMUST00000144327.3 |
Car1
|
carbonic anhydrase 1 |
| chr14_+_54439232 | 3.89 |
ENSMUST00000103724.2
|
Traj17
|
T cell receptor alpha joining 17 |
| chr3_+_40904253 | 3.84 |
ENSMUST00000048490.13
|
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr8_+_118428643 | 3.83 |
ENSMUST00000034304.9
|
Hsd17b2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr13_-_19495474 | 3.59 |
ENSMUST00000197113.2
|
Trgj2
|
T cell receptor gamma joining 2 |
| chr14_+_54429757 | 3.48 |
ENSMUST00000103714.2
|
Traj27
|
T cell receptor alpha joining 27 |
| chr14_+_53245069 | 3.44 |
ENSMUST00000178650.3
|
Trav6d-7
|
T cell receptor alpha variable 6D-7 |
| chr9_+_123635556 | 3.30 |
ENSMUST00000216072.2
|
Cxcr6
|
chemokine (C-X-C motif) receptor 6 |
| chr5_-_135601887 | 3.29 |
ENSMUST00000004936.10
ENSMUST00000201401.2 |
Ccl24
|
chemokine (C-C motif) ligand 24 |
| chr5_-_24732200 | 3.23 |
ENSMUST00000088311.6
|
Gbx1
|
gastrulation brain homeobox 1 |
| chr9_+_7558449 | 3.22 |
ENSMUST00000018765.4
|
Mmp8
|
matrix metallopeptidase 8 |
| chr17_-_52117894 | 3.21 |
ENSMUST00000176669.8
|
Satb1
|
special AT-rich sequence binding protein 1 |
| chr11_+_100973391 | 3.20 |
ENSMUST00000001806.10
ENSMUST00000107308.4 |
Coasy
|
Coenzyme A synthase |
| chr7_+_79048884 | 3.15 |
ENSMUST00000137667.3
|
Fanci
|
Fanconi anemia, complementation group I |
| chr1_+_75213171 | 3.09 |
ENSMUST00000187058.7
|
Dnajb2
|
DnaJ heat shock protein family (Hsp40) member B2 |
| chr7_+_114344920 | 3.09 |
ENSMUST00000136645.8
ENSMUST00000169913.8 |
Insc
|
INSC spindle orientation adaptor protein |
| chr14_+_53743184 | 3.01 |
ENSMUST00000103583.5
|
Trav10
|
T cell receptor alpha variable 10 |
| chr14_-_52252432 | 3.01 |
ENSMUST00000226527.2
|
Zfp219
|
zinc finger protein 219 |
| chr14_+_53167246 | 2.95 |
ENSMUST00000177703.3
|
Trav12d-3
|
T cell receptor alpha variable 12D-3 |
| chr3_-_102871440 | 2.89 |
ENSMUST00000058899.13
|
Nr1h5
|
nuclear receptor subfamily 1, group H, member 5 |
| chr14_+_78141679 | 2.81 |
ENSMUST00000022591.16
ENSMUST00000169978.2 ENSMUST00000227903.2 |
Epsti1
|
epithelial stromal interaction 1 (breast) |
| chr17_+_34457868 | 2.80 |
ENSMUST00000095342.11
ENSMUST00000167280.8 ENSMUST00000236838.2 |
H2-Ob
|
histocompatibility 2, O region beta locus |
| chr11_+_61017573 | 2.79 |
ENSMUST00000010286.8
ENSMUST00000146033.8 ENSMUST00000139422.8 |
Tnfrsf13b
|
tumor necrosis factor receptor superfamily, member 13b |
| chr1_+_75213082 | 2.79 |
ENSMUST00000055223.14
ENSMUST00000082158.13 ENSMUST00000188346.7 |
Dnajb2
|
DnaJ heat shock protein family (Hsp40) member B2 |
| chr11_-_53509485 | 2.76 |
ENSMUST00000000889.7
|
Il4
|
interleukin 4 |
| chr9_+_54771064 | 2.73 |
ENSMUST00000034843.9
|
Ireb2
|
iron responsive element binding protein 2 |
| chr11_-_84058292 | 2.54 |
ENSMUST00000050771.8
|
Gm11437
|
predicted gene 11437 |
| chr9_+_44245981 | 2.50 |
ENSMUST00000052686.4
|
H2ax
|
H2A.X variant histone |
| chr14_-_50425655 | 2.48 |
ENSMUST00000205837.3
|
Olfr730
|
olfactory receptor 730 |
| chr1_+_75213114 | 2.45 |
ENSMUST00000188290.7
|
Dnajb2
|
DnaJ heat shock protein family (Hsp40) member B2 |
| chr8_-_46605196 | 2.44 |
ENSMUST00000110378.9
|
Snx25
|
sorting nexin 25 |
| chrX_-_108056995 | 2.40 |
ENSMUST00000033597.9
|
Hmgn5
|
high-mobility group nucleosome binding domain 5 |
| chr14_+_53059379 | 2.39 |
ENSMUST00000197754.2
|
Trav6d-6
|
T cell receptor alpha variable 6D-6 |
| chr4_+_100336003 | 2.35 |
ENSMUST00000133493.9
ENSMUST00000092730.5 |
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr14_+_53029773 | 2.34 |
ENSMUST00000178426.4
|
Trav9d-1
|
T cell receptor alpha variable 9D-1 |
| chr1_+_75213044 | 2.23 |
ENSMUST00000188931.7
|
Dnajb2
|
DnaJ heat shock protein family (Hsp40) member B2 |
| chr13_+_19526322 | 2.22 |
ENSMUST00000184430.2
|
Trgj4
|
T cell receptor gamma joining 4 |
| chr17_+_57318469 | 2.15 |
ENSMUST00000210548.2
|
Gm17949
|
predicted gene, 17949 |
| chr14_+_53220913 | 2.15 |
ENSMUST00000167409.2
|
Trav9d-4
|
T cell receptor alpha variable 9D-4 |
| chr1_+_135980508 | 2.06 |
ENSMUST00000112068.10
|
Cacna1s
|
calcium channel, voltage-dependent, L type, alpha 1S subunit |
| chr13_+_38010203 | 2.02 |
ENSMUST00000128570.9
|
Rreb1
|
ras responsive element binding protein 1 |
| chr6_-_129637519 | 1.98 |
ENSMUST00000119533.2
ENSMUST00000145984.8 ENSMUST00000118401.8 ENSMUST00000112057.9 ENSMUST00000071920.11 |
Klrc2
|
killer cell lectin-like receptor subfamily C, member 2 |
| chr17_+_48666919 | 1.96 |
ENSMUST00000224001.2
ENSMUST00000024792.8 ENSMUST00000225849.2 |
Treml1
|
triggering receptor expressed on myeloid cells-like 1 |
| chr1_+_135980488 | 1.95 |
ENSMUST00000160641.8
|
Cacna1s
|
calcium channel, voltage-dependent, L type, alpha 1S subunit |
| chr14_-_48900192 | 1.94 |
ENSMUST00000122009.8
|
Otx2
|
orthodenticle homeobox 2 |
| chr1_-_136888118 | 1.91 |
ENSMUST00000192357.6
ENSMUST00000027649.14 |
Nr5a2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr15_-_78290038 | 1.87 |
ENSMUST00000058659.9
|
Tst
|
thiosulfate sulfurtransferase, mitochondrial |
| chr5_-_33591320 | 1.86 |
ENSMUST00000173348.2
|
Nkx1-1
|
NK1 homeobox 1 |
| chr8_-_120301883 | 1.83 |
ENSMUST00000212198.2
ENSMUST00000212065.2 ENSMUST00000036049.6 |
Hsdl1
|
hydroxysteroid dehydrogenase like 1 |
| chr10_-_78448514 | 1.81 |
ENSMUST00000205085.2
|
Olfr1357
|
olfactory receptor 1357 |
| chr1_+_75213258 | 1.81 |
ENSMUST00000185654.3
|
Dnajb2
|
DnaJ heat shock protein family (Hsp40) member B2 |
| chrY_+_1010543 | 1.81 |
ENSMUST00000091197.4
|
Eif2s3y
|
eukaryotic translation initiation factor 2, subunit 3, structural gene Y-linked |
| chr10_+_115405891 | 1.75 |
ENSMUST00000173620.2
|
A930009A15Rik
|
RIKEN cDNA A930009A15 gene |
| chr11_-_78441584 | 1.74 |
ENSMUST00000103242.5
|
Tmem97
|
transmembrane protein 97 |
| chr12_+_76417040 | 1.74 |
ENSMUST00000042779.4
|
Zbtb1
|
zinc finger and BTB domain containing 1 |
| chr14_+_53239170 | 1.73 |
ENSMUST00000179701.2
|
Trav5d-4
|
T cell receptor alpha variable 5D-4 |
| chr4_-_116484675 | 1.72 |
ENSMUST00000081182.5
ENSMUST00000030457.12 |
Nasp
|
nuclear autoantigenic sperm protein (histone-binding) |
| chr2_-_172212426 | 1.72 |
ENSMUST00000109139.8
ENSMUST00000028997.8 ENSMUST00000109140.10 |
Aurka
|
aurora kinase A |
| chr18_-_3280999 | 1.63 |
ENSMUST00000049942.13
|
Crem
|
cAMP responsive element modulator |
| chr2_+_174485327 | 1.63 |
ENSMUST00000059452.6
|
Zfp831
|
zinc finger protein 831 |
| chr17_+_35056219 | 1.60 |
ENSMUST00000173995.2
ENSMUST00000174092.8 |
Dxo
|
decapping exoribonuclease |
| chr6_-_42442130 | 1.59 |
ENSMUST00000216650.2
|
Olfr458
|
olfactory receptor 458 |
| chr5_-_66308666 | 1.58 |
ENSMUST00000201561.4
|
Rbm47
|
RNA binding motif protein 47 |
| chr5_-_66308421 | 1.57 |
ENSMUST00000200775.4
ENSMUST00000094756.11 |
Rbm47
|
RNA binding motif protein 47 |
| chr5_+_16758777 | 1.54 |
ENSMUST00000030683.8
|
Hgf
|
hepatocyte growth factor |
| chr14_-_52733146 | 1.53 |
ENSMUST00000206931.2
ENSMUST00000206062.4 |
Olfr1507
|
olfactory receptor 1507 |
| chr13_-_40441487 | 1.52 |
ENSMUST00000054635.8
|
Ofcc1
|
orofacial cleft 1 candidate 1 |
| chr5_+_147125488 | 1.51 |
ENSMUST00000065382.6
|
Gsx1
|
GS homeobox 1 |
| chr7_-_110682204 | 1.51 |
ENSMUST00000161051.8
ENSMUST00000160132.8 ENSMUST00000162415.9 |
Eif4g2
|
eukaryotic translation initiation factor 4, gamma 2 |
| chr1_+_88093726 | 1.51 |
ENSMUST00000097659.5
|
Ugt1a5
|
UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr18_-_3281089 | 1.50 |
ENSMUST00000139537.2
ENSMUST00000124747.8 |
Crem
|
cAMP responsive element modulator |
| chr13_-_49473695 | 1.46 |
ENSMUST00000110086.2
|
Fgd3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr15_+_10177709 | 1.45 |
ENSMUST00000124470.8
|
Prlr
|
prolactin receptor |
| chr1_+_107439145 | 1.42 |
ENSMUST00000009356.11
ENSMUST00000064916.9 |
Serpinb2
|
serine (or cysteine) peptidase inhibitor, clade B, member 2 |
| chr6_-_30680502 | 1.41 |
ENSMUST00000133373.8
|
Cep41
|
centrosomal protein 41 |
| chr6_-_68746087 | 1.41 |
ENSMUST00000103333.4
|
Igkv4-91
|
immunoglobulin kappa chain variable 4-91 |
| chr9_-_51240201 | 1.40 |
ENSMUST00000039959.11
ENSMUST00000238450.3 |
1810046K07Rik
|
RIKEN cDNA 1810046K07 gene |
| chr14_+_63235512 | 1.35 |
ENSMUST00000100492.5
|
Defb47
|
defensin beta 47 |
| chr10_+_127927443 | 1.33 |
ENSMUST00000238829.2
ENSMUST00000217851.2 ENSMUST00000220049.2 |
Baz2a
|
bromodomain adjacent to zinc finger domain, 2A |
| chr19_+_12839106 | 1.32 |
ENSMUST00000059675.4
|
Olfr1444
|
olfactory receptor 1444 |
| chr11_+_3152683 | 1.31 |
ENSMUST00000125637.8
|
Eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chrX_-_94521712 | 1.30 |
ENSMUST00000033549.3
|
Asb12
|
ankyrin repeat and SOCS box-containing 12 |
| chr14_+_53370136 | 1.29 |
ENSMUST00000181793.3
|
Trav6n-6
|
T cell receptor alpha variable 6N-6 |
| chr17_-_36582721 | 1.28 |
ENSMUST00000042717.13
|
Trim39
|
tripartite motif-containing 39 |
| chr10_+_128540049 | 1.27 |
ENSMUST00000217836.2
|
Pmel
|
premelanosome protein |
| chr4_-_94817025 | 1.25 |
ENSMUST00000030309.6
|
Eqtn
|
equatorin, sperm acrosome associated |
| chr5_+_88635834 | 1.22 |
ENSMUST00000199104.5
ENSMUST00000031222.9 |
Enam
|
enamelin |
| chr13_-_18556626 | 1.21 |
ENSMUST00000139064.10
ENSMUST00000175703.9 |
Pou6f2
|
POU domain, class 6, transcription factor 2 |
| chr10_+_39245746 | 1.20 |
ENSMUST00000063091.13
ENSMUST00000099967.10 ENSMUST00000126486.8 |
Fyn
|
Fyn proto-oncogene |
| chr9_-_106768601 | 1.19 |
ENSMUST00000069036.14
|
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr11_+_3152874 | 1.18 |
ENSMUST00000179770.8
ENSMUST00000110048.8 |
Eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr18_-_78166595 | 1.16 |
ENSMUST00000091813.12
|
Slc14a1
|
solute carrier family 14 (urea transporter), member 1 |
| chr19_-_46314945 | 1.15 |
ENSMUST00000225781.2
ENSMUST00000223903.2 |
Psd
|
pleckstrin and Sec7 domain containing |
| chr8_+_120301974 | 1.13 |
ENSMUST00000093100.3
|
Dnaaf1
|
dynein, axonemal assembly factor 1 |
| chr14_+_53370308 | 1.12 |
ENSMUST00000103613.3
|
Trav6n-6
|
T cell receptor alpha variable 6N-6 |
| chr11_-_4544751 | 1.12 |
ENSMUST00000109943.10
|
Mtmr3
|
myotubularin related protein 3 |
| chr3_+_94744844 | 1.09 |
ENSMUST00000107270.9
|
Pogz
|
pogo transposable element with ZNF domain |
| chr6_-_42453259 | 1.07 |
ENSMUST00000204324.3
ENSMUST00000203396.3 |
Olfr457
|
olfactory receptor 457 |
| chr3_-_37180093 | 1.06 |
ENSMUST00000029275.6
|
Il2
|
interleukin 2 |
| chr8_-_69541852 | 1.06 |
ENSMUST00000037478.13
ENSMUST00000148856.2 |
Slc18a1
|
solute carrier family 18 (vesicular monoamine), member 1 |
| chr15_-_100497863 | 1.04 |
ENSMUST00000073837.13
|
Pou6f1
|
POU domain, class 6, transcription factor 1 |
| chr12_-_69245191 | 1.03 |
ENSMUST00000021356.6
|
Dnaaf2
|
dynein, axonemal assembly factor 2 |
| chr6_+_42382368 | 1.03 |
ENSMUST00000070178.5
|
Tas2r135
|
taste receptor, type 2, member 135 |
| chr3_-_89905547 | 1.02 |
ENSMUST00000199740.2
ENSMUST00000198782.2 |
Hax1
|
HCLS1 associated X-1 |
| chr9_-_56759229 | 1.00 |
ENSMUST00000055036.7
|
Odf3l1
|
outer dense fiber of sperm tails 3-like 1 |
| chr18_+_65021974 | 1.00 |
ENSMUST00000225261.3
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like |
| chr1_-_16689660 | 1.00 |
ENSMUST00000117146.9
|
Ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr10_-_23968192 | 0.99 |
ENSMUST00000092654.4
|
Taar8b
|
trace amine-associated receptor 8B |
| chr7_-_5128936 | 0.99 |
ENSMUST00000147835.4
|
Rasl2-9
|
RAS-like, family 2, locus 9 |
| chrX_+_100492684 | 0.98 |
ENSMUST00000033674.6
|
Itgb1bp2
|
integrin beta 1 binding protein 2 |
| chr16_+_35803794 | 0.97 |
ENSMUST00000173555.8
|
Kpna1
|
karyopherin (importin) alpha 1 |
| chr15_+_78481247 | 0.96 |
ENSMUST00000043069.6
ENSMUST00000231180.2 ENSMUST00000229796.2 ENSMUST00000229295.2 |
Cyth4
|
cytohesin 4 |
| chr4_+_49521176 | 0.96 |
ENSMUST00000042964.13
ENSMUST00000107696.2 |
Zfp189
|
zinc finger protein 189 |
| chr5_-_100186728 | 0.94 |
ENSMUST00000153442.8
|
Hnrnpdl
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr18_+_58689296 | 0.93 |
ENSMUST00000025500.7
|
Slc27a6
|
solute carrier family 27 (fatty acid transporter), member 6 |
| chr3_+_68598757 | 0.93 |
ENSMUST00000107816.4
|
Il12a
|
interleukin 12a |
| chr11_-_54158102 | 0.93 |
ENSMUST00000019058.6
|
Il3
|
interleukin 3 |
| chr1_-_131025562 | 0.92 |
ENSMUST00000016672.11
|
Mapkapk2
|
MAP kinase-activated protein kinase 2 |
| chr7_+_79674562 | 0.92 |
ENSMUST00000164056.9
ENSMUST00000166250.8 |
Zfp710
|
zinc finger protein 710 |
| chr1_+_131890679 | 0.92 |
ENSMUST00000191034.2
ENSMUST00000177943.8 |
Gm29103
Slc45a3
|
predicted gene 29103 solute carrier family 45, member 3 |
| chr1_+_170060318 | 0.92 |
ENSMUST00000162752.2
|
Sh2d1b2
|
SH2 domain containing 1B2 |
| chr16_+_43067641 | 0.91 |
ENSMUST00000079441.13
ENSMUST00000114691.8 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr19_-_53932867 | 0.89 |
ENSMUST00000235688.2
ENSMUST00000235348.2 |
Bbip1
|
BBSome interacting protein 1 |
| chr17_-_33904386 | 0.87 |
ENSMUST00000087582.13
|
Hnrnpm
|
heterogeneous nuclear ribonucleoprotein M |
| chr2_+_172392678 | 0.87 |
ENSMUST00000099058.10
|
Tfap2c
|
transcription factor AP-2, gamma |
| chr5_-_30401416 | 0.82 |
ENSMUST00000125367.4
|
Adgrf3
|
adhesion G protein-coupled receptor F3 |
| chr18_-_37771772 | 0.82 |
ENSMUST00000058635.5
|
Slc25a2
|
solute carrier family 25 (mitochondrial carrier, ornithine transporter) member 2 |
| chr5_-_149559667 | 0.82 |
ENSMUST00000074846.14
|
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr1_-_4430481 | 0.82 |
ENSMUST00000027032.6
|
Rp1
|
retinitis pigmentosa 1 (human) |
| chr6_-_93889483 | 0.81 |
ENSMUST00000205116.3
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr18_-_78166539 | 0.79 |
ENSMUST00000160292.8
|
Slc14a1
|
solute carrier family 14 (urea transporter), member 1 |
| chr5_+_16758897 | 0.79 |
ENSMUST00000196645.2
|
Hgf
|
hepatocyte growth factor |
| chr11_+_104983022 | 0.78 |
ENSMUST00000021029.6
|
Efcab3
|
EF-hand calcium binding domain 3 |
| chr17_-_37574640 | 0.77 |
ENSMUST00000080759.5
|
Olfr98
|
olfactory receptor 98 |
| chr7_+_141996067 | 0.76 |
ENSMUST00000149529.8
|
Tnni2
|
troponin I, skeletal, fast 2 |
| chr8_+_46604786 | 0.76 |
ENSMUST00000154040.2
|
Cfap97
|
cilia and flagella associated protein 97 |
| chr12_-_85421467 | 0.76 |
ENSMUST00000040766.9
|
Tmed10
|
transmembrane p24 trafficking protein 10 |
| chr1_-_132953068 | 0.76 |
ENSMUST00000186617.7
ENSMUST00000067429.10 ENSMUST00000067398.13 ENSMUST00000188090.7 |
Mdm4
|
transformed mouse 3T3 cell double minute 4 |
| chr18_-_6490808 | 0.75 |
ENSMUST00000028100.13
ENSMUST00000050542.6 |
Epc1
|
enhancer of polycomb homolog 1 |
| chr5_+_45611093 | 0.72 |
ENSMUST00000053250.5
|
Clrn2
|
clarin 2 |
| chr9_+_27308087 | 0.71 |
ENSMUST00000214158.2
ENSMUST00000034473.7 |
Spata19
|
spermatogenesis associated 19 |
| chr13_+_78173013 | 0.71 |
ENSMUST00000175955.4
|
Pou5f2
|
POU domain class 5, transcription factor 2 |
| chr12_-_115031622 | 0.69 |
ENSMUST00000194257.2
|
Ighv8-5
|
immunoglobulin heavy variable V8-5 |
| chr19_-_11301919 | 0.69 |
ENSMUST00000159269.2
|
Ms4a7
|
membrane-spanning 4-domains, subfamily A, member 7 |
| chr1_+_163822438 | 0.68 |
ENSMUST00000045694.14
ENSMUST00000111490.2 |
Mettl18
|
methyltransferase like 18 |
| chrX_-_56384089 | 0.66 |
ENSMUST00000033468.11
|
Arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr13_-_74918745 | 0.64 |
ENSMUST00000223033.2
ENSMUST00000222588.2 |
Cast
|
calpastatin |
| chrX_-_74460137 | 0.64 |
ENSMUST00000033542.11
|
Mtcp1
|
mature T cell proliferation 1 |
| chr17_-_33904345 | 0.62 |
ENSMUST00000234474.2
ENSMUST00000139302.8 ENSMUST00000114385.9 |
Hnrnpm
|
heterogeneous nuclear ribonucleoprotein M |
| chr3_+_89986925 | 0.61 |
ENSMUST00000118566.8
ENSMUST00000119158.8 |
Tpm3
|
tropomyosin 3, gamma |
| chr16_-_88571089 | 0.60 |
ENSMUST00000054223.4
|
2310057N15Rik
|
RIKEN cDNA 2310057N15 gene |
| chrX_-_74460168 | 0.59 |
ENSMUST00000033543.14
ENSMUST00000149863.3 ENSMUST00000114081.2 |
Cmc4
Mtcp1
|
C-x(9)-C motif containing 4 mature T cell proliferation 1 |
| chr4_-_117786690 | 0.59 |
ENSMUST00000097913.9
|
Artn
|
artemin |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.0 | 18.0 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 3.4 | 10.1 | GO:0052203 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 2.9 | 20.0 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 2.5 | 7.4 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 2.1 | 12.7 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 2.1 | 14.6 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 2.0 | 5.9 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 1.8 | 12.4 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 1.7 | 13.2 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 1.6 | 4.8 | GO:0002343 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 1.2 | 6.0 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 1.2 | 12.9 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 1.1 | 4.6 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 1.1 | 13.2 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 1.1 | 3.2 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.9 | 2.8 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.9 | 2.7 | GO:0090650 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.7 | 7.0 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.6 | 1.9 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.6 | 1.7 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) |
| 0.4 | 4.0 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.4 | 1.7 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.4 | 4.7 | GO:0006569 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) |
| 0.4 | 3.8 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.4 | 1.2 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.4 | 1.2 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.3 | 1.9 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.3 | 4.4 | GO:0002829 | negative regulation of type 2 immune response(GO:0002829) |
| 0.3 | 4.4 | GO:0009650 | UV protection(GO:0009650) |
| 0.3 | 2.0 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.3 | 1.9 | GO:0042706 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.3 | 0.8 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.3 | 3.5 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.3 | 1.1 | GO:2000320 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.2 | 1.5 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.2 | 0.9 | GO:0038156 | interleukin-3-mediated signaling pathway(GO:0038156) |
| 0.2 | 0.9 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.2 | 0.2 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.2 | 0.9 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.2 | 1.2 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.2 | 2.6 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.2 | 0.6 | GO:0097021 | lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.2 | 0.6 | GO:0009629 | response to gravity(GO:0009629) |
| 0.2 | 0.8 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.2 | 0.6 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.2 | 3.2 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.2 | 3.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.2 | 1.0 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.2 | 1.1 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.2 | 1.9 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.2 | 1.1 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.1 | 1.2 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.1 | 1.5 | GO:0051135 | positive regulation of NK T cell activation(GO:0051135) |
| 0.1 | 1.5 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 0.5 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.1 | 5.8 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 1.3 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.1 | 2.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 3.2 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 2.8 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.1 | 0.6 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 | 3.2 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 0.6 | GO:0070384 | growth plate cartilage chondrocyte growth(GO:0003430) Harderian gland development(GO:0070384) |
| 0.1 | 4.2 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.1 | 1.6 | GO:0071028 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.1 | 1.1 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 21.7 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 0.9 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.1 | 1.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 1.3 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.1 | 4.0 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 1.5 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.1 | 0.9 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 3.1 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.1 | 0.8 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 1.1 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 2.1 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 6.6 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 0.2 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) |
| 0.1 | 0.9 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.1 | 1.0 | GO:0000054 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) |
| 0.1 | 1.3 | GO:2000059 | negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.1 | 5.3 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 1.5 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 1.0 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 3.1 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 6.7 | GO:0009308 | amine metabolic process(GO:0009308) |
| 0.0 | 9.4 | GO:0031668 | cellular response to extracellular stimulus(GO:0031668) |
| 0.0 | 2.4 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 1.0 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.0 | 1.9 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 7.1 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 0.8 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 1.0 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.5 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.3 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.0 | 1.7 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.9 | GO:0045954 | positive regulation of natural killer cell mediated immunity(GO:0002717) positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.0 | 1.2 | GO:0043467 | regulation of generation of precursor metabolites and energy(GO:0043467) |
| 0.0 | 0.9 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.2 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.8 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 6.9 | GO:0009116 | nucleoside metabolic process(GO:0009116) |
| 0.0 | 0.2 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 1.5 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.2 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 1.8 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 3.8 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.0 | 0.3 | GO:0014823 | response to activity(GO:0014823) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 14.6 | GO:0097433 | dense body(GO:0097433) |
| 0.9 | 10.4 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.7 | 4.8 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.6 | 19.7 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.6 | 13.2 | GO:0042627 | chylomicron(GO:0042627) |
| 0.3 | 2.4 | GO:0033503 | HULC complex(GO:0033503) |
| 0.3 | 1.7 | GO:0042585 | chromosome passenger complex(GO:0032133) germinal vesicle(GO:0042585) |
| 0.3 | 1.3 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.2 | 2.8 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 1.3 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.2 | 3.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 1.5 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 1.9 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 1.3 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.1 | 0.8 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 12.4 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.1 | 24.5 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 0.5 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.1 | 0.9 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.8 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 4.0 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 0.8 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.5 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.6 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 1.8 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.6 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 3.2 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 1.0 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.0 | 0.6 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 5.2 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 2.5 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.2 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 7.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.8 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 6.6 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 2.4 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 1.2 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 4.0 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.3 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 1.1 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.1 | GO:0005687 | U4 snRNP(GO:0005687) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 13.2 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 2.2 | 13.2 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 2.1 | 14.6 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 1.7 | 10.4 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 1.7 | 20.0 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 1.6 | 4.7 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 1.5 | 8.8 | GO:0008142 | oxysterol binding(GO:0008142) |
| 1.5 | 7.3 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 1.3 | 10.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 1.1 | 4.4 | GO:0004096 | catalase activity(GO:0004096) |
| 1.1 | 3.3 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 1.1 | 3.2 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.9 | 18.0 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.9 | 2.7 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.8 | 6.7 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.8 | 4.0 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.7 | 5.3 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.6 | 2.4 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.5 | 3.8 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.5 | 12.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.5 | 3.3 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.4 | 1.2 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.4 | 4.0 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.3 | 1.9 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.3 | 0.9 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.3 | 0.9 | GO:0005135 | interleukin-3 receptor binding(GO:0005135) |
| 0.3 | 2.8 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.3 | 1.5 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.3 | 1.9 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.3 | 1.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.2 | 1.2 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.2 | 0.9 | GO:0015154 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.2 | 7.4 | GO:0071949 | FAD binding(GO:0071949) |
| 0.2 | 0.6 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.2 | 1.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.2 | 13.2 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.2 | 5.9 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.2 | 4.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 1.5 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 1.1 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.8 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.1 | 3.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 0.6 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.1 | 1.5 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.1 | 2.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.1 | 0.8 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 0.6 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 20.8 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.1 | 2.8 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 1.2 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.1 | 1.3 | GO:0001164 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.1 | 0.9 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 1.6 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.1 | 13.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.7 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.1 | 0.8 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.1 | 6.6 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 1.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 2.6 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.9 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 1.7 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 2.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.9 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 0.6 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 2.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 3.7 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 4.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 5.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.6 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 4.0 | GO:0038024 | cargo receptor activity(GO:0038024) |
| 0.0 | 0.9 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 1.0 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 4.2 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 1.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.3 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 1.1 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 2.5 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 11.7 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.6 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 3.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.1 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 4.9 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 1.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 2.9 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 0.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 1.0 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.7 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.8 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 7.9 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 8.0 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.2 | 4.5 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.2 | 6.8 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 11.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 4.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 3.2 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 1.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 2.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 13.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.7 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 3.2 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.0 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.9 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 3.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.5 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.9 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.8 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.1 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.9 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.5 | PID IL4 2PATHWAY | IL4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 7.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.5 | 20.0 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.5 | 10.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.4 | 12.7 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.4 | 21.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.3 | 4.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.3 | 10.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.3 | 7.9 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.3 | 4.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.2 | 3.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 2.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.5 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 11.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 3.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 1.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 3.0 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 1.0 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 1.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 1.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.9 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.4 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 1.7 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 4.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 1.5 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.9 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |