Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
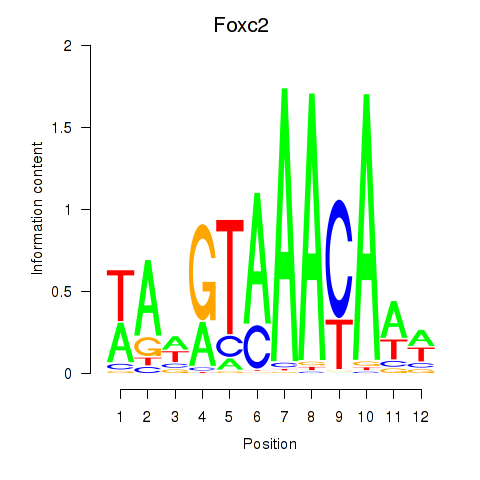
Results for Foxc2
Z-value: 1.35
Transcription factors associated with Foxc2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxc2
|
ENSMUSG00000046714.8 | Foxc2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxc2 | mm39_v1_chr8_+_121842902_121842916 | -0.46 | 4.5e-05 | Click! |
Activity profile of Foxc2 motif
Sorted Z-values of Foxc2 motif
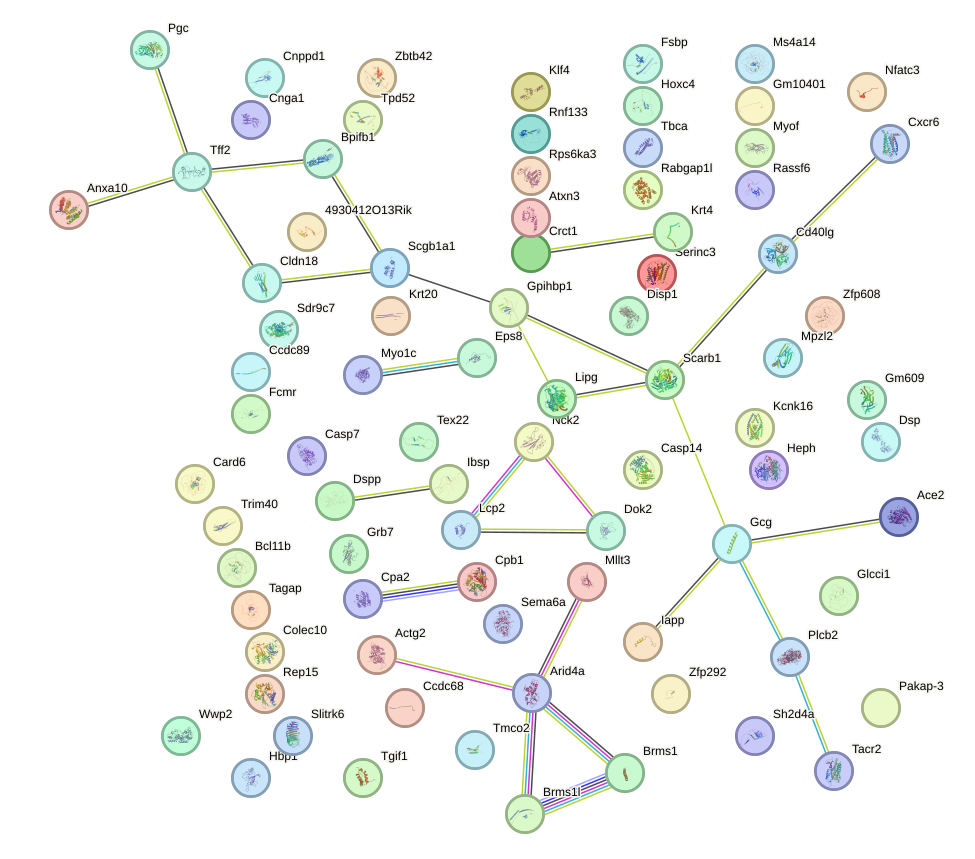
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxc2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_83513222 | 21.54 |
ENSMUST00000075161.12
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr6_-_83513184 | 19.73 |
ENSMUST00000205926.2
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr19_-_9065309 | 17.29 |
ENSMUST00000025554.3
|
Scgb1a1
|
secretoglobin, family 1A, member 1 (uteroglobin) |
| chr8_-_62576140 | 15.40 |
ENSMUST00000034052.14
ENSMUST00000034054.9 |
Anxa10
|
annexin A10 |
| chr4_-_55532453 | 14.52 |
ENSMUST00000132746.2
ENSMUST00000107619.3 |
Klf4
|
Kruppel-like factor 4 (gut) |
| chr16_-_48232770 | 11.23 |
ENSMUST00000212197.2
|
Gm5485
|
predicted gene 5485 |
| chr17_+_48037758 | 10.55 |
ENSMUST00000024782.12
ENSMUST00000144955.2 |
Pgc
|
progastricsin (pepsinogen C) |
| chr17_-_31363245 | 10.08 |
ENSMUST00000024826.8
|
Tff2
|
trefoil factor 2 (spasmolytic protein 1) |
| chr9_-_99599312 | 9.75 |
ENSMUST00000112882.9
ENSMUST00000131922.2 |
Cldn18
|
claudin 18 |
| chrX_+_162923474 | 9.30 |
ENSMUST00000073973.11
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr4_-_87724533 | 8.77 |
ENSMUST00000126353.8
ENSMUST00000149357.8 |
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr6_+_146934082 | 8.65 |
ENSMUST00000036194.6
|
Rep15
|
RAB15 effector protein |
| chr15_-_101833160 | 8.60 |
ENSMUST00000023797.8
|
Krt4
|
keratin 4 |
| chr12_+_112645237 | 8.30 |
ENSMUST00000174780.2
ENSMUST00000169593.2 ENSMUST00000173942.2 |
Zbtb42
|
zinc finger and BTB domain containing 42 |
| chr2_-_62313981 | 8.05 |
ENSMUST00000136686.2
ENSMUST00000102733.10 |
Gcg
|
glucagon |
| chr13_+_38335232 | 7.88 |
ENSMUST00000124830.3
|
Dsp
|
desmoplakin |
| chr1_-_160079007 | 7.84 |
ENSMUST00000191909.6
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chr3_-_92922976 | 7.69 |
ENSMUST00000107301.2
ENSMUST00000029521.5 |
Crct1
|
cysteine-rich C-terminal 1 |
| chr10_-_78554104 | 7.38 |
ENSMUST00000005488.9
|
Casp14
|
caspase 14 |
| chr11_-_99328969 | 7.28 |
ENSMUST00000017743.3
|
Krt20
|
keratin 20 |
| chr8_+_68729219 | 7.26 |
ENSMUST00000066594.4
|
Sh2d4a
|
SH2 domain containing 4A |
| chr9_+_44951075 | 6.91 |
ENSMUST00000217097.2
|
Mpzl2
|
myelin protein zero-like 2 |
| chr3_-_20329823 | 6.53 |
ENSMUST00000011607.6
|
Cpb1
|
carboxypeptidase B1 (tissue) |
| chr19_+_56414114 | 6.29 |
ENSMUST00000238892.2
|
Casp7
|
caspase 7 |
| chr9_-_99592116 | 5.79 |
ENSMUST00000035048.12
|
Cldn18
|
claudin 18 |
| chr19_-_38032006 | 5.68 |
ENSMUST00000172095.3
ENSMUST00000041475.16 |
Myof
|
myoferlin |
| chr9_-_99592058 | 5.21 |
ENSMUST00000136429.8
|
Cldn18
|
claudin 18 |
| chr18_+_70058533 | 5.04 |
ENSMUST00000043929.11
|
Ccdc68
|
coiled-coil domain containing 68 |
| chr19_-_38031774 | 4.94 |
ENSMUST00000226068.2
|
Myof
|
myoferlin |
| chr17_-_71158703 | 4.86 |
ENSMUST00000166395.9
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr3_-_9029097 | 4.73 |
ENSMUST00000091354.12
ENSMUST00000094381.11 |
Tpd52
|
tumor protein D52 |
| chr12_+_71063431 | 4.73 |
ENSMUST00000125125.2
|
Arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr14_+_71011744 | 4.66 |
ENSMUST00000022698.8
|
Dok2
|
docking protein 2 |
| chr17_+_29309942 | 4.44 |
ENSMUST00000119901.9
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A (P21) |
| chr2_+_154042291 | 4.43 |
ENSMUST00000028987.7
|
Bpifb1
|
BPI fold containing family B, member 1 |
| chr6_+_15185202 | 4.29 |
ENSMUST00000154448.2
|
Foxp2
|
forkhead box P2 |
| chr18_+_70058613 | 4.28 |
ENSMUST00000080050.6
|
Ccdc68
|
coiled-coil domain containing 68 |
| chr10_+_62088104 | 4.17 |
ENSMUST00000020278.6
|
Tacr2
|
tachykinin receptor 2 |
| chr18_-_75094323 | 3.83 |
ENSMUST00000066532.5
|
Lipg
|
lipase, endothelial |
| chr8_+_106785434 | 3.74 |
ENSMUST00000212742.2
ENSMUST00000211991.2 |
Nfatc3
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 3 |
| chr2_-_118558852 | 3.70 |
ENSMUST00000102524.8
|
Plcb2
|
phospholipase C, beta 2 |
| chr2_-_118558825 | 3.52 |
ENSMUST00000159756.2
|
Plcb2
|
phospholipase C, beta 2 |
| chr13_-_100753419 | 3.49 |
ENSMUST00000168772.2
ENSMUST00000163163.9 ENSMUST00000022137.14 |
Marveld2
|
MARVEL (membrane-associating) domain containing 2 |
| chrX_+_56257374 | 3.48 |
ENSMUST00000033466.2
|
Cd40lg
|
CD40 ligand |
| chr5_-_90788323 | 3.43 |
ENSMUST00000202784.4
ENSMUST00000031317.10 ENSMUST00000201370.2 |
Rassf6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr7_+_90075762 | 3.38 |
ENSMUST00000061391.9
|
Ccdc89
|
coiled-coil domain containing 89 |
| chr12_+_119291343 | 3.37 |
ENSMUST00000221917.2
|
Macc1
|
metastasis associated in colon cancer 1 |
| chr6_+_8520006 | 3.36 |
ENSMUST00000162567.8
ENSMUST00000161217.8 |
Glcci1
|
glucocorticoid induced transcript 1 |
| chr6_+_142244145 | 3.18 |
ENSMUST00000041993.3
|
Iapp
|
islet amyloid polypeptide |
| chr6_-_142268692 | 3.17 |
ENSMUST00000081380.10
|
Slco1a5
|
solute carrier organic anion transporter family, member 1a5 |
| chr15_+_54274151 | 3.16 |
ENSMUST00000036737.4
|
Colec10
|
collectin sub-family member 10 |
| chr19_-_11291805 | 3.12 |
ENSMUST00000187467.2
|
Ms4a14
|
membrane-spanning 4-domains, subfamily A, member 14 |
| chr1_-_75119277 | 3.09 |
ENSMUST00000168720.8
ENSMUST00000041213.12 ENSMUST00000189809.2 |
Cnppd1
|
cyclin Pas1/PHO80 domain containing 1 |
| chr5_-_115236354 | 3.05 |
ENSMUST00000100848.3
|
Gm10401
|
predicted gene 10401 |
| chr6_-_41535322 | 3.05 |
ENSMUST00000193003.2
|
Trbv31
|
T cell receptor beta, variable 31 |
| chr5_-_72800070 | 3.03 |
ENSMUST00000087213.12
|
Cnga1
|
cyclic nucleotide gated channel alpha 1 |
| chr11_+_98337655 | 3.03 |
ENSMUST00000019456.5
|
Grb7
|
growth factor receptor bound protein 7 |
| chr6_-_23650205 | 3.03 |
ENSMUST00000115354.2
|
Rnf133
|
ring finger protein 133 |
| chr10_+_127734384 | 3.00 |
ENSMUST00000047134.8
|
Sdr9c7
|
4short chain dehydrogenase/reductase family 9C, member 7 |
| chr11_+_33996920 | 2.97 |
ENSMUST00000052413.12
|
Lcp2
|
lymphocyte cytosolic protein 2 |
| chr18_+_36498826 | 2.96 |
ENSMUST00000144158.2
|
Cystm1
|
cysteine-rich transmembrane module containing 1 |
| chr3_+_3699205 | 2.93 |
ENSMUST00000108394.3
|
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr12_-_101924407 | 2.92 |
ENSMUST00000159883.2
ENSMUST00000160251.8 ENSMUST00000161011.8 ENSMUST00000021606.12 |
Atxn3
|
ataxin 3 |
| chr9_+_123635529 | 2.86 |
ENSMUST00000049810.9
|
Cxcr6
|
chemokine (C-X-C motif) receptor 6 |
| chr9_+_123635556 | 2.83 |
ENSMUST00000216072.2
|
Cxcr6
|
chemokine (C-X-C motif) receptor 6 |
| chr19_+_5091365 | 2.82 |
ENSMUST00000116567.4
ENSMUST00000235918.2 |
Brms1
|
breast cancer metastasis-suppressor 1 |
| chr14_-_110992533 | 2.82 |
ENSMUST00000078386.4
|
Slitrk6
|
SLIT and NTRK-like family, member 6 |
| chr15_+_102927366 | 2.81 |
ENSMUST00000165375.3
|
Hoxc4
|
homeobox C4 |
| chr12_-_32000534 | 2.80 |
ENSMUST00000172314.9
|
Hbp1
|
high mobility group box transcription factor 1 |
| chrX_+_158086253 | 2.76 |
ENSMUST00000112491.2
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr15_-_5137975 | 2.71 |
ENSMUST00000118365.3
|
Card6
|
caspase recruitment domain family, member 6 |
| chrX_+_95498965 | 2.59 |
ENSMUST00000033553.14
|
Heph
|
hephaestin |
| chr12_-_32000169 | 2.57 |
ENSMUST00000176520.8
|
Hbp1
|
high mobility group box transcription factor 1 |
| chr6_+_34686543 | 2.56 |
ENSMUST00000031775.13
|
Cald1
|
caldesmon 1 |
| chr6_-_142268667 | 2.56 |
ENSMUST00000128446.2
|
Slco1a5
|
solute carrier organic anion transporter family, member 1a5 |
| chr4_+_11579648 | 2.55 |
ENSMUST00000180239.2
|
Fsbp
|
fibrinogen silencer binding protein |
| chr8_+_108271643 | 2.52 |
ENSMUST00000212543.2
|
Wwp2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr11_+_33997114 | 2.52 |
ENSMUST00000109329.9
|
Lcp2
|
lymphocyte cytosolic protein 2 |
| chr4_+_57821050 | 2.51 |
ENSMUST00000238994.2
|
Pakap
|
paralemmin A kinase anchor protein |
| chr13_+_94954202 | 2.49 |
ENSMUST00000220825.2
|
Tbca
|
tubulin cofactor A |
| chr5_+_43673093 | 2.49 |
ENSMUST00000144558.3
|
C1qtnf7
|
C1q and tumor necrosis factor related protein 7 |
| chr1_-_182929025 | 2.49 |
ENSMUST00000171366.7
|
Disp1
|
dispatched RND transporter family member 1 |
| chr16_-_45313324 | 2.38 |
ENSMUST00000114585.3
|
Gm609
|
predicted gene 609 |
| chr1_+_130793406 | 2.38 |
ENSMUST00000038829.7
|
Fcmr
|
Fc fragment of IgM receptor |
| chr2_-_27365633 | 2.36 |
ENSMUST00000138693.8
ENSMUST00000113941.9 ENSMUST00000077737.13 |
Brd3
|
bromodomain containing 3 |
| chr4_-_58499398 | 2.33 |
ENSMUST00000107570.2
|
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr18_-_55123153 | 2.32 |
ENSMUST00000064763.7
|
Zfp608
|
zinc finger protein 608 |
| chr1_+_43484895 | 2.29 |
ENSMUST00000086421.9
|
Nck2
|
non-catalytic region of tyrosine kinase adaptor protein 2 |
| chr14_-_20319242 | 2.29 |
ENSMUST00000024155.9
|
Kcnk16
|
potassium channel, subfamily K, member 16 |
| chr2_+_9887427 | 2.27 |
ENSMUST00000114919.2
|
4930412O13Rik
|
RIKEN cDNA 4930412O13 gene |
| chr4_-_87724512 | 2.25 |
ENSMUST00000148059.2
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr17_+_8144822 | 2.25 |
ENSMUST00000036370.8
|
Tagap
|
T cell activation Rho GTPase activating protein |
| chr6_-_23650297 | 2.25 |
ENSMUST00000063548.4
|
Rnf133
|
ring finger protein 133 |
| chr5_+_43672856 | 2.24 |
ENSMUST00000076939.10
|
C1qtnf7
|
C1q and tumor necrosis factor related protein 7 |
| chr12_-_107969853 | 2.23 |
ENSMUST00000066060.11
|
Bcl11b
|
B cell leukemia/lymphoma 11B |
| chr12_+_113038376 | 2.22 |
ENSMUST00000109729.3
|
Tex22
|
testis expressed gene 22 |
| chr6_-_137548004 | 2.22 |
ENSMUST00000100841.9
|
Eps8
|
epidermal growth factor receptor pathway substrate 8 |
| chr6_+_30541581 | 2.16 |
ENSMUST00000096066.5
|
Cpa2
|
carboxypeptidase A2, pancreatic |
| chr6_+_14901343 | 2.16 |
ENSMUST00000115477.8
|
Foxp2
|
forkhead box P2 |
| chr4_-_120966396 | 2.13 |
ENSMUST00000106268.4
|
Tmco2
|
transmembrane and coiled-coil domains 2 |
| chr2_-_163486998 | 2.13 |
ENSMUST00000017851.4
|
Serinc3
|
serine incorporator 3 |
| chr17_-_37200927 | 2.10 |
ENSMUST00000087158.11
|
Trim40
|
tripartite motif-containing 40 |
| chr12_-_32000209 | 2.08 |
ENSMUST00000176084.2
ENSMUST00000176103.8 ENSMUST00000167458.9 |
Hbp1
|
high mobility group box transcription factor 1 |
| chr5_-_18093739 | 2.07 |
ENSMUST00000169095.6
ENSMUST00000197574.2 |
Cd36
|
CD36 molecule |
| chr6_-_41535292 | 2.06 |
ENSMUST00000103300.3
|
Trbv31
|
T cell receptor beta, variable 31 |
| chr2_-_51862941 | 2.01 |
ENSMUST00000145481.8
ENSMUST00000112705.9 |
Nmi
|
N-myc (and STAT) interactor |
| chr4_-_34882917 | 2.01 |
ENSMUST00000098163.9
ENSMUST00000047950.6 |
Zfp292
|
zinc finger protein 292 |
| chr11_+_54517164 | 1.96 |
ENSMUST00000239168.2
|
Rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr1_+_58841808 | 1.92 |
ENSMUST00000190213.2
|
Casp8
|
caspase 8 |
| chr18_-_47466378 | 1.90 |
ENSMUST00000126684.2
ENSMUST00000156422.8 |
Sema6a
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr17_+_47816042 | 1.89 |
ENSMUST00000183044.8
ENSMUST00000037333.17 |
Ccnd3
|
cyclin D3 |
| chr5_+_104447037 | 1.87 |
ENSMUST00000031246.9
|
Ibsp
|
integrin binding sialoprotein |
| chr5_-_90788460 | 1.81 |
ENSMUST00000202704.4
|
Rassf6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr7_-_34354924 | 1.80 |
ENSMUST00000032709.3
|
Kctd15
|
potassium channel tetramerisation domain containing 15 |
| chr13_-_104056803 | 1.79 |
ENSMUST00000091269.11
ENSMUST00000188997.7 ENSMUST00000169083.8 ENSMUST00000191275.7 |
Erbin
|
Erbb2 interacting protein |
| chr4_+_97665992 | 1.78 |
ENSMUST00000107062.9
ENSMUST00000052018.12 ENSMUST00000107057.8 |
Nfia
|
nuclear factor I/A |
| chr13_-_99121070 | 1.77 |
ENSMUST00000054425.7
|
H2bl1
|
H2B.L histone variant 1 |
| chr5_-_138169253 | 1.74 |
ENSMUST00000139983.8
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr16_-_89940652 | 1.74 |
ENSMUST00000114124.9
|
Tiam1
|
T cell lymphoma invasion and metastasis 1 |
| chr4_+_52607204 | 1.70 |
ENSMUST00000107671.2
|
Toporsl
|
topoisomerase I binding, arginine/serine-rich like |
| chr7_-_115459082 | 1.68 |
ENSMUST00000206123.2
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr14_+_61844899 | 1.64 |
ENSMUST00000225582.2
ENSMUST00000051184.10 |
Kcnrg
|
potassium channel regulator |
| chr15_-_66432938 | 1.63 |
ENSMUST00000048372.7
|
Tmem71
|
transmembrane protein 71 |
| chr10_-_61814852 | 1.59 |
ENSMUST00000105453.8
ENSMUST00000105452.9 ENSMUST00000105454.3 |
Col13a1
|
collagen, type XIII, alpha 1 |
| chr4_+_32238950 | 1.58 |
ENSMUST00000037416.13
|
Bach2
|
BTB and CNC homology, basic leucine zipper transcription factor 2 |
| chr12_-_11258973 | 1.58 |
ENSMUST00000049877.3
|
Msgn1
|
mesogenin 1 |
| chr17_+_47816074 | 1.57 |
ENSMUST00000183177.8
ENSMUST00000182848.8 |
Ccnd3
|
cyclin D3 |
| chr14_+_53478202 | 1.56 |
ENSMUST00000179583.3
|
Trav12n-3
|
T cell receptor alpha variable 12N-3 |
| chrX_-_159518743 | 1.54 |
ENSMUST00000135856.2
|
Ppef1
|
protein phosphatase with EF hand calcium-binding domain 1 |
| chr16_-_43484494 | 1.51 |
ENSMUST00000096065.6
|
Tigit
|
T cell immunoreceptor with Ig and ITIM domains |
| chr2_+_91376650 | 1.50 |
ENSMUST00000099716.11
ENSMUST00000046769.16 ENSMUST00000111337.3 |
Ckap5
|
cytoskeleton associated protein 5 |
| chr17_+_35188888 | 1.50 |
ENSMUST00000173680.2
|
Gm20481
|
predicted gene 20481 |
| chr1_+_156666485 | 1.45 |
ENSMUST00000111720.2
|
Angptl1
|
angiopoietin-like 1 |
| chr4_-_82803384 | 1.45 |
ENSMUST00000048430.4
|
Cer1
|
cerberus 1, DAN family BMP antagonist |
| chr2_-_60503998 | 1.43 |
ENSMUST00000059888.15
ENSMUST00000154764.2 |
Itgb6
|
integrin beta 6 |
| chr17_+_47815968 | 1.42 |
ENSMUST00000182129.8
ENSMUST00000171031.8 |
Ccnd3
|
cyclin D3 |
| chr4_-_102883905 | 1.42 |
ENSMUST00000084382.6
ENSMUST00000106869.3 |
Insl5
|
insulin-like 5 |
| chr12_-_32258469 | 1.42 |
ENSMUST00000085469.6
|
Pik3cg
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma |
| chr3_-_102690026 | 1.41 |
ENSMUST00000172026.8
ENSMUST00000170856.6 |
Tshb
|
thyroid stimulating hormone, beta subunit |
| chr2_-_51863203 | 1.39 |
ENSMUST00000028314.9
|
Nmi
|
N-myc (and STAT) interactor |
| chr11_+_70323452 | 1.38 |
ENSMUST00000084954.13
ENSMUST00000108568.10 ENSMUST00000079056.9 ENSMUST00000102564.11 ENSMUST00000124943.8 ENSMUST00000150076.8 ENSMUST00000102563.2 |
Arrb2
|
arrestin, beta 2 |
| chr11_-_68277799 | 1.36 |
ENSMUST00000135141.2
|
Ntn1
|
netrin 1 |
| chr16_-_45313244 | 1.35 |
ENSMUST00000232138.2
|
Gm609
|
predicted gene 609 |
| chr12_-_32258604 | 1.35 |
ENSMUST00000053215.14
|
Pik3cg
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma |
| chr5_-_123127148 | 1.28 |
ENSMUST00000046073.16
|
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr1_+_40554513 | 1.27 |
ENSMUST00000027237.12
|
Il18rap
|
interleukin 18 receptor accessory protein |
| chr12_-_32258331 | 1.27 |
ENSMUST00000220366.2
|
Pik3cg
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma |
| chr2_+_164328375 | 1.26 |
ENSMUST00000069385.15
ENSMUST00000143690.8 |
Dbndd2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr10_+_41179966 | 1.23 |
ENSMUST00000173494.4
|
Ak9
|
adenylate kinase 9 |
| chr17_+_28226319 | 1.21 |
ENSMUST00000139173.3
|
Anks1
|
ankyrin repeat and SAM domain containing 1 |
| chr5_+_3707171 | 1.21 |
ENSMUST00000198739.5
|
Tmbim7
|
transmembrane BAX inhibitor motif containing 7 |
| chr12_-_107969673 | 1.18 |
ENSMUST00000109887.8
ENSMUST00000109891.3 |
Bcl11b
|
B cell leukemia/lymphoma 11B |
| chr18_-_84104574 | 1.18 |
ENSMUST00000175783.3
|
Tshz1
|
teashirt zinc finger family member 1 |
| chr1_+_51328265 | 1.17 |
ENSMUST00000051572.8
|
Cavin2
|
caveolae associated 2 |
| chr19_+_56450062 | 1.13 |
ENSMUST00000178590.9
ENSMUST00000039666.8 |
Plekhs1
|
pleckstrin homology domain containing, family S member 1 |
| chr11_-_16958647 | 1.11 |
ENSMUST00000102881.10
|
Plek
|
pleckstrin |
| chr15_-_5137951 | 1.11 |
ENSMUST00000141020.2
|
Card6
|
caspase recruitment domain family, member 6 |
| chr18_-_84104507 | 1.10 |
ENSMUST00000060303.10
|
Tshz1
|
teashirt zinc finger family member 1 |
| chr2_-_73143045 | 1.09 |
ENSMUST00000058615.10
|
Cir1
|
corepressor interacting with RBPJ, 1 |
| chr9_+_57921954 | 1.03 |
ENSMUST00000034874.14
|
Cyp11a1
|
cytochrome P450, family 11, subfamily a, polypeptide 1 |
| chr6_+_14901439 | 1.02 |
ENSMUST00000128567.8
|
Foxp2
|
forkhead box P2 |
| chr10_+_38841511 | 1.01 |
ENSMUST00000019992.6
|
Lama4
|
laminin, alpha 4 |
| chr13_-_43632368 | 1.01 |
ENSMUST00000222651.2
|
Ranbp9
|
RAN binding protein 9 |
| chr17_+_47816137 | 1.00 |
ENSMUST00000182935.8
ENSMUST00000182506.8 |
Ccnd3
|
cyclin D3 |
| chr13_-_95615229 | 0.99 |
ENSMUST00000022186.5
|
S100z
|
S100 calcium binding protein, zeta |
| chr3_+_53396120 | 0.98 |
ENSMUST00000029307.4
|
Stoml3
|
stomatin (Epb7.2)-like 3 |
| chrX_+_162694397 | 0.98 |
ENSMUST00000140845.2
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chrX_+_132751729 | 0.92 |
ENSMUST00000033602.9
|
Tnmd
|
tenomodulin |
| chr11_-_68277631 | 0.91 |
ENSMUST00000021284.4
|
Ntn1
|
netrin 1 |
| chr6_-_141719536 | 0.87 |
ENSMUST00000148411.2
|
Gm5724
|
predicted gene 5724 |
| chr5_+_91175323 | 0.87 |
ENSMUST00000202724.4
ENSMUST00000041516.9 |
Epgn
|
epithelial mitogen |
| chr14_-_71004019 | 0.79 |
ENSMUST00000167242.8
|
Xpo7
|
exportin 7 |
| chr17_+_46471950 | 0.76 |
ENSMUST00000024748.14
ENSMUST00000172170.8 |
Gtpbp2
|
GTP binding protein 2 |
| chr5_-_104261556 | 0.69 |
ENSMUST00000031249.8
|
Sparcl1
|
SPARC-like 1 |
| chr6_-_136899167 | 0.66 |
ENSMUST00000032343.7
|
Erp27
|
endoplasmic reticulum protein 27 |
| chr4_-_127864744 | 0.65 |
ENSMUST00000030614.3
|
CK137956
|
cDNA sequence CK137956 |
| chr18_+_34891941 | 0.64 |
ENSMUST00000049281.12
|
Fam53c
|
family with sequence similarity 53, member C |
| chr14_+_48358331 | 0.63 |
ENSMUST00000226513.2
|
Peli2
|
pellino 2 |
| chr3_+_94745009 | 0.62 |
ENSMUST00000107266.8
ENSMUST00000042402.12 ENSMUST00000107269.2 |
Pogz
|
pogo transposable element with ZNF domain |
| chr5_+_16758777 | 0.59 |
ENSMUST00000030683.8
|
Hgf
|
hepatocyte growth factor |
| chr11_+_29413734 | 0.59 |
ENSMUST00000155854.8
|
Ccdc88a
|
coiled coil domain containing 88A |
| chr5_-_130284366 | 0.57 |
ENSMUST00000026387.11
|
Sbds
|
SBDS ribosome maturation factor |
| chr17_-_45047521 | 0.57 |
ENSMUST00000113572.9
|
Runx2
|
runt related transcription factor 2 |
| chr14_+_50618620 | 0.55 |
ENSMUST00000215263.2
ENSMUST00000213402.2 ENSMUST00000213755.2 ENSMUST00000215227.2 |
Olfr736
|
olfactory receptor 736 |
| chr11_+_77656414 | 0.53 |
ENSMUST00000164315.2
|
Myo18a
|
myosin XVIIIA |
| chr7_+_67305162 | 0.52 |
ENSMUST00000107470.2
|
Ttc23
|
tetratricopeptide repeat domain 23 |
| chr2_-_7400690 | 0.51 |
ENSMUST00000182404.8
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr11_-_86884507 | 0.51 |
ENSMUST00000018571.5
|
Ypel2
|
yippee like 2 |
| chr1_+_87332638 | 0.51 |
ENSMUST00000173152.2
ENSMUST00000173663.2 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr13_+_23879775 | 0.50 |
ENSMUST00000041052.5
|
H1f6
|
H1.6 linker histone, cluster member |
| chr9_+_40098375 | 0.48 |
ENSMUST00000062229.6
|
Olfr986
|
olfactory receptor 986 |
| chr10_-_41179167 | 0.47 |
ENSMUST00000043814.5
|
Fig4
|
FIG4 phosphoinositide 5-phosphatase |
| chr5_+_16758897 | 0.45 |
ENSMUST00000196645.2
|
Hgf
|
hepatocyte growth factor |
| chr6_-_134768288 | 0.45 |
ENSMUST00000149776.3
|
Dusp16
|
dual specificity phosphatase 16 |
| chr17_+_47815998 | 0.44 |
ENSMUST00000183210.2
|
Ccnd3
|
cyclin D3 |
| chr18_+_52958382 | 0.43 |
ENSMUST00000238707.2
|
Gm50457
|
predicted gene, 50457 |
| chr3_+_125197722 | 0.42 |
ENSMUST00000173932.8
|
Ndst4
|
N-deacetylase/N-sulfotransferase (heparin glucosaminyl) 4 |
| chr3_+_85946145 | 0.39 |
ENSMUST00000238331.2
|
Sh3d19
|
SH3 domain protein D19 |
| chr5_-_5714196 | 0.35 |
ENSMUST00000196165.5
ENSMUST00000061008.10 ENSMUST00000135252.3 ENSMUST00000054865.13 |
Cfap69
|
cilia and flagella associated protein 69 |
| chr12_-_111946560 | 0.30 |
ENSMUST00000190680.2
|
Rd3l
|
retinal degeneration 3-like |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 17.3 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 3.6 | 14.5 | GO:0045415 | negative regulation of interleukin-8 biosynthetic process(GO:0045415) |
| 3.0 | 20.7 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 2.3 | 9.3 | GO:0015827 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 2.1 | 10.5 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 1.4 | 10.1 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 1.3 | 6.3 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 1.2 | 3.5 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 1.1 | 3.4 | GO:0097534 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 1.1 | 3.4 | GO:1902524 | positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 1.0 | 8.0 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 1.0 | 3.8 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.9 | 7.4 | GO:0070268 | cornification(GO:0070268) |
| 0.9 | 7.9 | GO:0071896 | protein localization to adherens junction(GO:0071896) bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.8 | 4.2 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.8 | 2.5 | GO:0035638 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.8 | 4.0 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.6 | 10.6 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.6 | 2.3 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.6 | 1.7 | GO:1990751 | regulation of Schwann cell chemotaxis(GO:1904266) positive regulation of Schwann cell chemotaxis(GO:1904268) Schwann cell chemotaxis(GO:1990751) |
| 0.6 | 12.6 | GO:0007379 | segment specification(GO:0007379) |
| 0.6 | 2.2 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.5 | 8.7 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.5 | 2.5 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.5 | 7.7 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.5 | 1.4 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.5 | 4.7 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.5 | 2.8 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.5 | 2.3 | GO:1903898 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.4 | 2.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.4 | 2.1 | GO:0070543 | response to linoleic acid(GO:0070543) |
| 0.4 | 3.2 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.4 | 1.1 | GO:0010925 | regulation of inositol-polyphosphate 5-phosphatase activity(GO:0010924) positive regulation of inositol-polyphosphate 5-phosphatase activity(GO:0010925) phospholipase C-inhibiting G-protein coupled receptor signaling pathway(GO:0030845) regulation of cell diameter(GO:0060305) |
| 0.4 | 4.4 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.4 | 7.5 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.4 | 2.8 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.3 | 2.3 | GO:0060023 | soft palate development(GO:0060023) |
| 0.3 | 1.3 | GO:0021592 | fourth ventricle development(GO:0021592) initiation of neural tube closure(GO:0021993) |
| 0.3 | 1.2 | GO:0046066 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) purine deoxyribonucleoside triphosphate biosynthetic process(GO:0009216) dGDP metabolic process(GO:0046066) |
| 0.3 | 1.8 | GO:0070425 | negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 0.3 | 4.4 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.3 | 2.9 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.3 | 6.3 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.3 | 4.9 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.3 | 1.5 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.2 | 3.7 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.2 | 1.4 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.2 | 7.3 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.2 | 2.3 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.2 | 2.8 | GO:0021562 | vestibulocochlear nerve development(GO:0021562) |
| 0.2 | 7.8 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.2 | 1.7 | GO:0021779 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.2 | 0.5 | GO:1903028 | regulation of opsonization(GO:1903027) positive regulation of opsonization(GO:1903028) |
| 0.2 | 0.9 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.2 | 1.4 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.1 | 1.7 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 5.8 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 1.9 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.1 | 6.6 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 3.0 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 2.4 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.1 | 1.4 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 2.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 1.2 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 | 1.8 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 0.6 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.6 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.1 | 7.8 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 8.1 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 1.9 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 0.6 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 2.6 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 2.0 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.1 | 2.1 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 0.5 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.1 | 3.5 | GO:0061028 | establishment of endothelial barrier(GO:0061028) |
| 0.1 | 7.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 6.9 | GO:0033077 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.1 | 1.6 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.1 | 0.6 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 9.5 | GO:0050680 | negative regulation of epithelial cell proliferation(GO:0050680) |
| 0.1 | 0.9 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.5 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.2 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 1.2 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 1.6 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 3.5 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.3 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.0 | 0.5 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 2.6 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.0 | 2.8 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.0 | 0.2 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 2.1 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) |
| 0.0 | 1.0 | GO:0044705 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 | 3.0 | GO:0051899 | membrane depolarization(GO:0051899) |
| 0.0 | 0.5 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 | 1.0 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.0 | 2.7 | GO:0043122 | regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043122) |
| 0.0 | 0.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.5 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 2.2 | GO:0051607 | defense response to virus(GO:0051607) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.5 | GO:0036398 | TCR signalosome(GO:0036398) |
| 1.8 | 7.2 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 1.5 | 4.4 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.8 | 4.0 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.4 | 3.5 | GO:0033010 | paranodal junction(GO:0033010) tricellular tight junction(GO:0061689) |
| 0.4 | 1.9 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 0.4 | 3.0 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.4 | 15.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.4 | 7.9 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.3 | 1.4 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.3 | 2.6 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 10.1 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.2 | 11.0 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 2.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 2.9 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 1.7 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 17.3 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 1.7 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 2.8 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 5.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 29.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 19.7 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 2.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 9.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 13.9 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 1.0 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 6.9 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 1.8 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 1.0 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 3.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 10.3 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 5.0 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 1.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 4.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 5.3 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 4.8 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 3.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 2.3 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.1 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.3 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 1.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.0 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 2.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 4.8 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.2 | GO:0005922 | connexon complex(GO:0005922) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 14.5 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 1.9 | 17.3 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 1.6 | 9.3 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 1.5 | 4.4 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 1.2 | 3.5 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 1.1 | 15.6 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 1.0 | 4.2 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.8 | 5.7 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.7 | 10.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.7 | 5.7 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.6 | 3.8 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.5 | 3.0 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.5 | 1.4 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.5 | 1.4 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.4 | 2.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.4 | 2.9 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.4 | 10.5 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.3 | 2.3 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.3 | 4.7 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.3 | 8.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.3 | 4.0 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.3 | 1.2 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.3 | 7.2 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.3 | 1.0 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 0.2 | 15.4 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.2 | 2.6 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 4.9 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.2 | 2.1 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.2 | 2.5 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.1 | 1.8 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 3.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 6.3 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 2.3 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 1.3 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.1 | 10.2 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 0.5 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 1.6 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 1.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 2.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 2.0 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 2.8 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 1.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 1.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 16.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 2.8 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.6 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 3.3 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 2.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 2.8 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 7.0 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 2.5 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 35.5 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 1.7 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 1.2 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 2.5 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 1.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 1.0 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.1 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.0 | 0.9 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.6 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.9 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 1.3 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 5.8 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 0.6 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 2.6 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.4 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.9 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 1.7 | GO:0042826 | histone deacetylase binding(GO:0042826) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 41.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.4 | 4.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.4 | 27.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.3 | 7.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.3 | 5.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 8.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.2 | 7.5 | PID MYC PATHWAY | C-MYC pathway |
| 0.2 | 5.4 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 7.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 4.7 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 2.8 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 4.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 24.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 6.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 3.0 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 10.6 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 10.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 2.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 1.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 2.9 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 5.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 2.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 2.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 1.5 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.7 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 2.2 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.1 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.6 | PID FGF PATHWAY | FGF signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 22.6 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.8 | 43.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.6 | 7.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.5 | 20.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.4 | 7.9 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.3 | 7.9 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.3 | 6.1 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 8.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 9.5 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.2 | 10.2 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.2 | 2.3 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.2 | 1.7 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.2 | 2.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 2.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 4.9 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 2.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.7 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 5.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 5.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 1.9 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 2.8 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 1.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 3.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 1.0 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 2.1 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.6 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 3.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 4.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 2.1 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 2.3 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 0.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.0 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |