Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Foxd2
Z-value: 0.63
Transcription factors associated with Foxd2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxd2
|
ENSMUSG00000055210.5 | Foxd2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxd2 | mm39_v1_chr4_-_114766070_114766111 | 0.13 | 2.7e-01 | Click! |
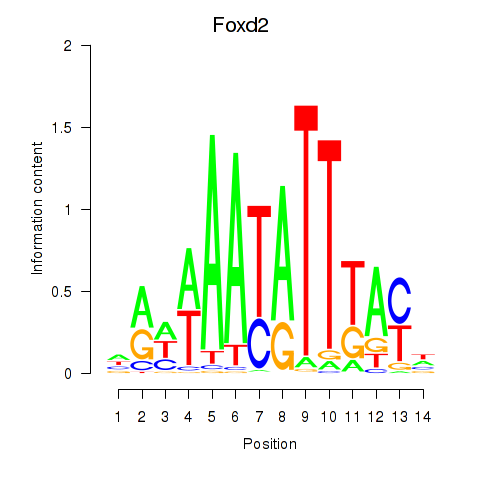
Activity profile of Foxd2 motif
Sorted Z-values of Foxd2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxd2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_173105344 | 8.57 |
ENSMUST00000111224.5
|
Mptx2
|
mucosal pentraxin 2 |
| chr7_+_130633776 | 6.92 |
ENSMUST00000084509.7
ENSMUST00000213064.3 ENSMUST00000208311.4 |
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr1_+_174158083 | 6.87 |
ENSMUST00000027816.5
|
Mptx1
|
mucosal pentraxin 1 |
| chr3_-_144738526 | 4.75 |
ENSMUST00000029919.7
|
Clca1
|
chloride channel accessory 1 |
| chr2_-_62313981 | 4.22 |
ENSMUST00000136686.2
ENSMUST00000102733.10 |
Gcg
|
glucagon |
| chr7_+_97480125 | 4.16 |
ENSMUST00000206351.2
|
Pak1
|
p21 (RAC1) activated kinase 1 |
| chr3_-_72875187 | 3.71 |
ENSMUST00000167334.8
|
Sis
|
sucrase isomaltase (alpha-glucosidase) |
| chr18_+_37637317 | 3.46 |
ENSMUST00000052179.8
|
Pcdhb20
|
protocadherin beta 20 |
| chr2_+_14393127 | 3.42 |
ENSMUST00000114731.8
ENSMUST00000082290.8 |
Slc39a12
|
solute carrier family 39 (zinc transporter), member 12 |
| chr16_+_13804461 | 3.21 |
ENSMUST00000056521.12
ENSMUST00000118412.8 ENSMUST00000131608.2 |
Bmerb1
|
bMERB domain containing 1 |
| chr17_-_37334091 | 3.21 |
ENSMUST00000167275.3
|
Mog
|
myelin oligodendrocyte glycoprotein |
| chr3_-_144511566 | 3.14 |
ENSMUST00000199029.2
|
Clca3a2
|
chloride channel accessory 3A2 |
| chr9_+_50466127 | 2.83 |
ENSMUST00000213916.2
|
Il18
|
interleukin 18 |
| chr17_+_29309942 | 2.78 |
ENSMUST00000119901.9
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A (P21) |
| chr10_+_42736771 | 2.64 |
ENSMUST00000105494.8
|
Scml4
|
Scm polycomb group protein like 4 |
| chr3_+_75982890 | 2.61 |
ENSMUST00000160261.8
|
Fstl5
|
follistatin-like 5 |
| chr17_+_34311314 | 2.61 |
ENSMUST00000025192.8
|
H2-Oa
|
histocompatibility 2, O region alpha locus |
| chr10_-_18887701 | 2.60 |
ENSMUST00000105527.2
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr2_-_110591909 | 2.51 |
ENSMUST00000140777.2
|
Ano3
|
anoctamin 3 |
| chr7_+_91321500 | 2.39 |
ENSMUST00000238619.2
ENSMUST00000238467.2 |
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr12_-_41536430 | 2.33 |
ENSMUST00000043884.6
|
Lrrn3
|
leucine rich repeat protein 3, neuronal |
| chr15_-_98505508 | 2.20 |
ENSMUST00000096224.6
|
Adcy6
|
adenylate cyclase 6 |
| chr1_-_37535202 | 2.17 |
ENSMUST00000143636.8
|
Mgat4a
|
mannoside acetylglucosaminyltransferase 4, isoenzyme A |
| chr14_+_80237691 | 2.15 |
ENSMUST00000228749.2
ENSMUST00000088735.4 |
Olfm4
|
olfactomedin 4 |
| chr7_+_91321694 | 2.05 |
ENSMUST00000238608.2
|
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr13_-_95170755 | 1.95 |
ENSMUST00000162670.8
|
Pde8b
|
phosphodiesterase 8B |
| chr12_+_38833501 | 1.87 |
ENSMUST00000159334.8
|
Etv1
|
ets variant 1 |
| chr18_-_6516089 | 1.85 |
ENSMUST00000115870.9
|
Epc1
|
enhancer of polycomb homolog 1 |
| chrX_+_162922317 | 1.80 |
ENSMUST00000112271.10
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr14_-_70867588 | 1.75 |
ENSMUST00000228009.2
|
Dmtn
|
dematin actin binding protein |
| chr12_+_38833454 | 1.73 |
ENSMUST00000161980.8
ENSMUST00000160701.8 |
Etv1
|
ets variant 1 |
| chr10_-_128579879 | 1.73 |
ENSMUST00000026414.9
|
Dgka
|
diacylglycerol kinase, alpha |
| chr5_+_107645626 | 1.72 |
ENSMUST00000152474.8
ENSMUST00000060553.8 |
Btbd8
|
BTB (POZ) domain containing 8 |
| chr4_-_136626073 | 1.71 |
ENSMUST00000046285.6
|
C1qa
|
complement component 1, q subcomponent, alpha polypeptide |
| chr2_+_14393245 | 1.69 |
ENSMUST00000133258.2
|
Slc39a12
|
solute carrier family 39 (zinc transporter), member 12 |
| chr14_+_102077937 | 1.69 |
ENSMUST00000159026.8
|
Lmo7
|
LIM domain only 7 |
| chr18_+_73996743 | 1.68 |
ENSMUST00000134847.2
|
Mro
|
maestro |
| chr9_+_36744016 | 1.66 |
ENSMUST00000214772.2
|
Fez1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr15_+_100052260 | 1.65 |
ENSMUST00000023768.14
|
Dip2b
|
disco interacting protein 2 homolog B |
| chr1_-_37535170 | 1.63 |
ENSMUST00000148047.2
|
Mgat4a
|
mannoside acetylglucosaminyltransferase 4, isoenzyme A |
| chrX_+_158480304 | 1.60 |
ENSMUST00000123433.8
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr16_-_16345197 | 1.56 |
ENSMUST00000069284.14
|
Fgd4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr2_-_104241358 | 1.54 |
ENSMUST00000230671.2
|
D430041D05Rik
|
RIKEN cDNA D430041D05 gene |
| chr1_+_107456731 | 1.51 |
ENSMUST00000182198.8
|
Serpinb10
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 10 |
| chrX_-_104973003 | 1.50 |
ENSMUST00000130980.2
ENSMUST00000113573.8 |
Atrx
|
ATRX, chromatin remodeler |
| chr14_+_102078038 | 1.50 |
ENSMUST00000159314.8
|
Lmo7
|
LIM domain only 7 |
| chrX_-_104972938 | 1.48 |
ENSMUST00000198448.5
ENSMUST00000199233.5 ENSMUST00000134507.8 ENSMUST00000154866.8 ENSMUST00000128968.8 ENSMUST00000134381.8 ENSMUST00000150914.8 |
Atrx
|
ATRX, chromatin remodeler |
| chr14_+_54113415 | 1.47 |
ENSMUST00000180938.3
|
Trav21-dv12
|
T cell receptor alpha variable 21-DV12 |
| chr11_+_78811613 | 1.47 |
ENSMUST00000018610.7
|
Nos2
|
nitric oxide synthase 2, inducible |
| chr11_+_97692738 | 1.46 |
ENSMUST00000127033.9
|
Lasp1
|
LIM and SH3 protein 1 |
| chr6_+_77219627 | 1.41 |
ENSMUST00000159616.2
|
Lrrtm1
|
leucine rich repeat transmembrane neuronal 1 |
| chr13_-_97235745 | 1.40 |
ENSMUST00000071118.7
|
Gm6169
|
predicted gene 6169 |
| chr6_+_77219698 | 1.39 |
ENSMUST00000161677.2
|
Lrrtm1
|
leucine rich repeat transmembrane neuronal 1 |
| chr1_-_80687213 | 1.39 |
ENSMUST00000186087.7
|
Dock10
|
dedicator of cytokinesis 10 |
| chr6_+_38895902 | 1.36 |
ENSMUST00000003017.13
|
Tbxas1
|
thromboxane A synthase 1, platelet |
| chr5_-_23889607 | 1.33 |
ENSMUST00000197985.5
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr5_+_148202011 | 1.32 |
ENSMUST00000110515.9
|
Mtus2
|
microtubule associated tumor suppressor candidate 2 |
| chr16_-_45313244 | 1.30 |
ENSMUST00000232138.2
|
Gm609
|
predicted gene 609 |
| chr3_+_7494108 | 1.30 |
ENSMUST00000193330.2
|
Pkia
|
protein kinase inhibitor, alpha |
| chr16_-_45313324 | 1.30 |
ENSMUST00000114585.3
|
Gm609
|
predicted gene 609 |
| chrX_+_136552469 | 1.26 |
ENSMUST00000075471.4
|
Il1rapl2
|
interleukin 1 receptor accessory protein-like 2 |
| chr5_-_120750623 | 1.25 |
ENSMUST00000140554.2
ENSMUST00000031599.9 ENSMUST00000177800.8 |
Rita1
|
RBPJ interacting and tubulin associated 1 |
| chr18_+_37646674 | 1.24 |
ENSMUST00000061405.6
|
Pcdhb21
|
protocadherin beta 21 |
| chr5_-_103247920 | 1.23 |
ENSMUST00000112848.8
|
Mapk10
|
mitogen-activated protein kinase 10 |
| chr2_+_24235300 | 1.21 |
ENSMUST00000114485.9
ENSMUST00000114482.3 |
Il1rn
|
interleukin 1 receptor antagonist |
| chr15_-_101833160 | 1.19 |
ENSMUST00000023797.8
|
Krt4
|
keratin 4 |
| chr16_+_48637219 | 1.19 |
ENSMUST00000023328.8
|
Retnlb
|
resistin like beta |
| chr2_+_124978518 | 1.18 |
ENSMUST00000238754.2
|
Ctxn2
|
cortexin 2 |
| chr7_+_43284131 | 1.17 |
ENSMUST00000032663.10
|
Ceacam18
|
carcinoembryonic antigen-related cell adhesion molecule 18 |
| chrX_-_104972844 | 1.16 |
ENSMUST00000198441.5
ENSMUST00000137453.8 |
Atrx
|
ATRX, chromatin remodeler |
| chr5_-_110220379 | 1.13 |
ENSMUST00000210275.2
|
Gm17655
|
predicted gene, 17655 |
| chr18_+_37858753 | 1.12 |
ENSMUST00000066149.9
|
Pcdhga8
|
protocadherin gamma subfamily A, 8 |
| chr3_-_75177378 | 1.09 |
ENSMUST00000039047.5
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr3_-_123483772 | 1.08 |
ENSMUST00000172537.3
|
Ndst3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr2_-_25498459 | 1.08 |
ENSMUST00000058137.9
|
Rabl6
|
RAB, member RAS oncogene family-like 6 |
| chr2_+_124978612 | 1.07 |
ENSMUST00000099452.3
ENSMUST00000238377.2 |
Ctxn2
|
cortexin 2 |
| chr10_-_116732813 | 1.06 |
ENSMUST00000048229.9
|
Myrfl
|
myelin regulatory factor-like |
| chr5_+_148202117 | 0.98 |
ENSMUST00000110514.8
|
Mtus2
|
microtubule associated tumor suppressor candidate 2 |
| chr2_+_132532040 | 0.97 |
ENSMUST00000148271.8
ENSMUST00000110132.3 |
Shld1
|
shieldin complex subunit 1 |
| chr1_+_171668173 | 0.94 |
ENSMUST00000136479.8
|
Cd84
|
CD84 antigen |
| chr9_+_94551929 | 0.94 |
ENSMUST00000033463.10
|
Slc9a9
|
solute carrier family 9 (sodium/hydrogen exchanger), member 9 |
| chr5_+_32404695 | 0.92 |
ENSMUST00000101376.3
|
Plb1
|
phospholipase B1 |
| chr3_+_114874614 | 0.88 |
ENSMUST00000051309.9
|
Olfm3
|
olfactomedin 3 |
| chr5_+_20433169 | 0.87 |
ENSMUST00000197553.5
ENSMUST00000208219.2 |
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr14_+_75368532 | 0.87 |
ENSMUST00000143539.8
ENSMUST00000134114.8 |
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr18_+_89224219 | 0.85 |
ENSMUST00000236835.2
|
Cd226
|
CD226 antigen |
| chr10_-_10956700 | 0.83 |
ENSMUST00000105560.2
|
Grm1
|
glutamate receptor, metabotropic 1 |
| chr7_+_105290259 | 0.83 |
ENSMUST00000209445.2
|
Timm10b
|
translocase of inner mitochondrial membrane 10B |
| chr11_+_98689479 | 0.81 |
ENSMUST00000037930.13
|
Msl1
|
male specific lethal 1 |
| chr11_-_107080150 | 0.76 |
ENSMUST00000106757.8
ENSMUST00000018577.8 |
Nol11
|
nucleolar protein 11 |
| chr10_-_88440869 | 0.76 |
ENSMUST00000119185.8
ENSMUST00000238199.2 |
Mybpc1
|
myosin binding protein C, slow-type |
| chr17_-_35265702 | 0.76 |
ENSMUST00000097338.11
|
Msh5
|
mutS homolog 5 |
| chr18_+_68470616 | 0.74 |
ENSMUST00000172148.4
ENSMUST00000239469.2 |
Mc5r
|
melanocortin 5 receptor |
| chr17_+_8144822 | 0.74 |
ENSMUST00000036370.8
|
Tagap
|
T cell activation Rho GTPase activating protein |
| chr9_+_96140781 | 0.73 |
ENSMUST00000190104.7
ENSMUST00000179416.8 ENSMUST00000189606.7 |
Tfdp2
|
transcription factor Dp 2 |
| chr5_-_87845916 | 0.73 |
ENSMUST00000196163.2
|
Csn2
|
casein beta |
| chr10_+_57521958 | 0.72 |
ENSMUST00000177473.8
|
Pkib
|
protein kinase inhibitor beta, cAMP dependent, testis specific |
| chr19_-_53577499 | 0.69 |
ENSMUST00000095978.5
|
Nutf2-ps1
|
nuclear transport factor 2, pseudogene 1 |
| chr6_-_129600798 | 0.67 |
ENSMUST00000095412.10
|
Klrk1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chr5_+_18167547 | 0.64 |
ENSMUST00000030561.9
|
Gnat3
|
guanine nucleotide binding protein, alpha transducing 3 |
| chr5_+_148202075 | 0.60 |
ENSMUST00000071878.12
|
Mtus2
|
microtubule associated tumor suppressor candidate 2 |
| chr7_-_108769719 | 0.59 |
ENSMUST00000208136.2
ENSMUST00000036992.9 |
Lmo1
|
LIM domain only 1 |
| chr6_-_57827328 | 0.59 |
ENSMUST00000203310.3
ENSMUST00000203488.3 |
Vmn1r21
|
vomeronasal 1 receptor 21 |
| chr13_-_98152768 | 0.56 |
ENSMUST00000238746.2
|
Arhgef28
|
Rho guanine nucleotide exchange factor (GEF) 28 |
| chrX_-_100312629 | 0.56 |
ENSMUST00000117736.2
|
Gm20489
|
predicted gene 20489 |
| chr6_+_17636979 | 0.56 |
ENSMUST00000015877.14
ENSMUST00000152005.3 |
Capza2
|
capping protein (actin filament) muscle Z-line, alpha 2 |
| chr7_-_126795096 | 0.55 |
ENSMUST00000206026.2
ENSMUST00000205321.2 ENSMUST00000206587.2 ENSMUST00000205316.2 ENSMUST00000166791.8 |
Cd2bp2
|
CD2 cytoplasmic tail binding protein 2 |
| chr7_-_126795040 | 0.55 |
ENSMUST00000035771.5
|
Cd2bp2
|
CD2 cytoplasmic tail binding protein 2 |
| chr4_-_45108038 | 0.55 |
ENSMUST00000107809.9
ENSMUST00000107808.3 ENSMUST00000107807.2 ENSMUST00000107810.3 |
Tomm5
|
translocase of outer mitochondrial membrane 5 |
| chr5_+_149601688 | 0.54 |
ENSMUST00000100404.6
|
B3glct
|
beta-3-glucosyltransferase |
| chr2_-_87881473 | 0.54 |
ENSMUST00000183862.3
|
Olfr1162
|
olfactory receptor 1162 |
| chr14_+_75368939 | 0.52 |
ENSMUST00000125833.8
ENSMUST00000124499.8 |
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr4_+_115594918 | 0.52 |
ENSMUST00000106525.9
|
Efcab14
|
EF-hand calcium binding domain 14 |
| chr3_+_89427458 | 0.51 |
ENSMUST00000000811.8
|
Kcnn3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
| chr6_+_11926757 | 0.50 |
ENSMUST00000133776.2
|
Phf14
|
PHD finger protein 14 |
| chr6_+_142244145 | 0.50 |
ENSMUST00000041993.3
|
Iapp
|
islet amyloid polypeptide |
| chr1_+_171238873 | 0.49 |
ENSMUST00000159207.8
ENSMUST00000161241.8 |
Usf1
|
upstream transcription factor 1 |
| chr4_+_115594951 | 0.49 |
ENSMUST00000106522.9
|
Efcab14
|
EF-hand calcium binding domain 14 |
| chr6_-_129600812 | 0.48 |
ENSMUST00000168919.8
|
Klrk1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chr1_-_173018204 | 0.48 |
ENSMUST00000215878.2
ENSMUST00000201132.3 |
Olfr1406
|
olfactory receptor 1406 |
| chr19_-_8691460 | 0.47 |
ENSMUST00000206560.2
ENSMUST00000205538.2 |
Slc3a2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 |
| chr4_+_128621378 | 0.47 |
ENSMUST00000106079.10
ENSMUST00000133439.8 |
Phc2
|
polyhomeotic 2 |
| chr13_-_20008397 | 0.45 |
ENSMUST00000222664.2
ENSMUST00000065335.3 |
Gpr141
|
G protein-coupled receptor 141 |
| chr2_-_17465410 | 0.45 |
ENSMUST00000145492.2
|
Nebl
|
nebulette |
| chr6_+_136495784 | 0.45 |
ENSMUST00000032335.13
ENSMUST00000185724.7 |
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr2_-_87868043 | 0.44 |
ENSMUST00000129056.3
|
Olfr73
|
olfactory receptor 73 |
| chr2_-_125624754 | 0.44 |
ENSMUST00000053699.13
|
Secisbp2l
|
SECIS binding protein 2-like |
| chr14_+_54436898 | 0.42 |
ENSMUST00000103721.3
|
Traj20
|
T cell receptor alpha joining 20 |
| chr1_-_58009191 | 0.42 |
ENSMUST00000159826.2
ENSMUST00000164963.8 |
Kctd18
|
potassium channel tetramerisation domain containing 18 |
| chr2_-_111779785 | 0.41 |
ENSMUST00000099604.6
|
Olfr1307
|
olfactory receptor 1307 |
| chr2_+_87853118 | 0.41 |
ENSMUST00000214438.2
|
Olfr1161
|
olfactory receptor 1161 |
| chr7_+_6346723 | 0.41 |
ENSMUST00000207173.3
|
Gm3854
|
predicted gene 3854 |
| chr3_-_14676261 | 0.40 |
ENSMUST00000108365.4
|
Rbis
|
ribosomal biogenesis factor |
| chr9_+_37995368 | 0.38 |
ENSMUST00000212502.4
|
Olfr887
|
olfactory receptor 887 |
| chr4_+_108576846 | 0.38 |
ENSMUST00000178992.2
|
3110021N24Rik
|
RIKEN cDNA 3110021N24 gene |
| chr9_+_78016468 | 0.38 |
ENSMUST00000118869.8
ENSMUST00000125615.2 |
Cilk1
|
ciliogenesis associated kinase 1 |
| chr6_-_3494587 | 0.37 |
ENSMUST00000049985.15
|
Hepacam2
|
HEPACAM family member 2 |
| chrX_+_106192510 | 0.37 |
ENSMUST00000147521.8
ENSMUST00000167673.2 |
P2ry10b
|
purinergic receptor P2Y, G-protein coupled 10B |
| chr7_+_28834276 | 0.37 |
ENSMUST00000161522.8
ENSMUST00000204845.3 ENSMUST00000205027.3 ENSMUST00000204194.3 ENSMUST00000203070.3 ENSMUST00000203380.3 |
Rasgrp4
|
RAS guanyl releasing protein 4 |
| chrX_-_50031587 | 0.36 |
ENSMUST00000060650.7
|
Frmd7
|
FERM domain containing 7 |
| chr7_+_140226365 | 0.36 |
ENSMUST00000084456.6
ENSMUST00000211057.2 ENSMUST00000211399.2 |
Olfr53
|
olfactory receptor 53 |
| chr11_+_29413734 | 0.35 |
ENSMUST00000155854.8
|
Ccdc88a
|
coiled coil domain containing 88A |
| chr12_-_101924407 | 0.35 |
ENSMUST00000159883.2
ENSMUST00000160251.8 ENSMUST00000161011.8 ENSMUST00000021606.12 |
Atxn3
|
ataxin 3 |
| chr10_+_66932235 | 0.34 |
ENSMUST00000174317.8
|
Jmjd1c
|
jumonji domain containing 1C |
| chr2_-_86061745 | 0.33 |
ENSMUST00000216056.2
|
Olfr1047
|
olfactory receptor 1047 |
| chr10_-_88440996 | 0.32 |
ENSMUST00000121629.8
|
Mybpc1
|
myosin binding protein C, slow-type |
| chr12_+_65122355 | 0.32 |
ENSMUST00000058889.5
|
Fancm
|
Fanconi anemia, complementation group M |
| chr6_-_57992144 | 0.32 |
ENSMUST00000228070.2
ENSMUST00000228040.2 |
Vmn1r26
|
vomeronasal 1 receptor 26 |
| chr6_+_132824105 | 0.32 |
ENSMUST00000071696.2
|
Tas2r123
|
taste receptor, type 2, member 123 |
| chr9_+_96140750 | 0.31 |
ENSMUST00000186609.7
|
Tfdp2
|
transcription factor Dp 2 |
| chr1_-_58009263 | 0.29 |
ENSMUST00000114410.10
|
Kctd18
|
potassium channel tetramerisation domain containing 18 |
| chr17_-_37625328 | 0.29 |
ENSMUST00000169373.2
|
Olfr102
|
olfactory receptor 102 |
| chr9_-_20791012 | 0.29 |
ENSMUST00000043726.8
|
Angptl6
|
angiopoietin-like 6 |
| chr2_+_111198936 | 0.29 |
ENSMUST00000090328.5
|
Olfr1283
|
olfactory receptor 1283 |
| chr9_+_38340751 | 0.29 |
ENSMUST00000216502.3
ENSMUST00000216644.2 |
Olfr901
|
olfactory receptor 901 |
| chr2_-_11608003 | 0.29 |
ENSMUST00000040314.12
|
Rbm17
|
RNA binding motif protein 17 |
| chr5_+_87955941 | 0.29 |
ENSMUST00000072539.12
ENSMUST00000113279.8 ENSMUST00000101057.8 ENSMUST00000197301.2 |
Csn1s2b
|
casein alpha s2-like B |
| chr7_-_102804952 | 0.28 |
ENSMUST00000061055.2
|
Olfr589
|
olfactory receptor 589 |
| chrX_+_82898974 | 0.28 |
ENSMUST00000239269.2
|
Dmd
|
dystrophin, muscular dystrophy |
| chr11_+_58549642 | 0.26 |
ENSMUST00000214392.2
|
Olfr322
|
olfactory receptor 322 |
| chr1_-_157084252 | 0.24 |
ENSMUST00000134543.8
|
Rasal2
|
RAS protein activator like 2 |
| chr14_-_50380137 | 0.23 |
ENSMUST00000213685.2
|
Olfr728
|
olfactory receptor 728 |
| chr2_+_120807498 | 0.23 |
ENSMUST00000067582.14
|
Tmem62
|
transmembrane protein 62 |
| chrX_+_108240356 | 0.21 |
ENSMUST00000139259.2
ENSMUST00000060013.4 |
Gm6377
|
predicted gene 6377 |
| chrX_+_132751729 | 0.21 |
ENSMUST00000033602.9
|
Tnmd
|
tenomodulin |
| chr16_-_19132814 | 0.21 |
ENSMUST00000216157.2
|
Olfr164
|
olfactory receptor 164 |
| chr14_+_53270305 | 0.20 |
ENSMUST00000179512.3
|
Trav13d-3
|
T cell receptor alpha variable 13D-3 |
| chr5_-_87847268 | 0.20 |
ENSMUST00000196869.5
ENSMUST00000199624.5 ENSMUST00000198057.5 ENSMUST00000082370.10 |
Csn2
|
casein beta |
| chr7_-_102143980 | 0.17 |
ENSMUST00000058750.4
|
Olfr545
|
olfactory receptor 545 |
| chr10_+_57521930 | 0.17 |
ENSMUST00000177325.8
|
Pkib
|
protein kinase inhibitor beta, cAMP dependent, testis specific |
| chr7_-_13723513 | 0.17 |
ENSMUST00000165167.8
ENSMUST00000108520.4 |
Sult2a4
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 4 |
| chr2_-_87504008 | 0.16 |
ENSMUST00000213835.2
|
Olfr1135
|
olfactory receptor 1135 |
| chr5_-_86475595 | 0.16 |
ENSMUST00000122377.2
|
Tmprss11d
|
transmembrane protease, serine 11d |
| chr7_-_103113358 | 0.15 |
ENSMUST00000214347.2
|
Olfr607
|
olfactory receptor 607 |
| chr4_-_130033152 | 0.15 |
ENSMUST00000119423.8
|
Hcrtr1
|
hypocretin (orexin) receptor 1 |
| chr17_-_56583715 | 0.14 |
ENSMUST00000058136.9
|
Ticam1
|
toll-like receptor adaptor molecule 1 |
| chr6_-_54949587 | 0.13 |
ENSMUST00000060655.15
|
Nod1
|
nucleotide-binding oligomerization domain containing 1 |
| chr1_-_118239146 | 0.13 |
ENSMUST00000027623.9
|
Tsn
|
translin |
| chr9_+_37683153 | 0.11 |
ENSMUST00000215128.2
|
Olfr875
|
olfactory receptor 875 |
| chr2_-_86008164 | 0.11 |
ENSMUST00000215171.2
|
Olfr1044
|
olfactory receptor 1044 |
| chr18_-_62729391 | 0.10 |
ENSMUST00000076194.6
|
Spink7
|
serine peptidase inhibitor, Kazal type 7 (putative) |
| chr1_-_170695328 | 0.10 |
ENSMUST00000027974.7
|
Atf6
|
activating transcription factor 6 |
| chr4_-_82768958 | 0.09 |
ENSMUST00000139401.2
|
Zdhhc21
|
zinc finger, DHHC domain containing 21 |
| chr9_-_95632387 | 0.08 |
ENSMUST00000189137.7
ENSMUST00000053785.10 |
Trpc1
|
transient receptor potential cation channel, subfamily C, member 1 |
| chr17_+_38110779 | 0.07 |
ENSMUST00000215168.2
ENSMUST00000216478.2 |
Olfr124
|
olfactory receptor 124 |
| chr2_-_109111064 | 0.05 |
ENSMUST00000147770.2
|
Mettl15
|
methyltransferase like 15 |
| chr2_+_164674782 | 0.04 |
ENSMUST00000103093.10
|
Ctsa
|
cathepsin A |
| chr10_-_89457115 | 0.03 |
ENSMUST00000020102.14
|
Slc17a8
|
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 8 |
| chr9_+_7628201 | 0.03 |
ENSMUST00000034487.4
|
Mmp20
|
matrix metallopeptidase 20 (enamelysin) |
| chr2_+_36886739 | 0.03 |
ENSMUST00000069578.5
|
Olfr357
|
olfactory receptor 357 |
| chr12_-_111946560 | 0.02 |
ENSMUST00000190680.2
|
Rd3l
|
retinal degeneration 3-like |
| chr16_+_84590434 | 0.02 |
ENSMUST00000231910.2
|
Jam2
|
junction adhesion molecule 2 |
| chr11_+_95227836 | 0.01 |
ENSMUST00000037502.7
|
Fam117a
|
family with sequence similarity 117, member A |
| chr2_+_164674801 | 0.01 |
ENSMUST00000103092.9
ENSMUST00000151493.3 ENSMUST00000127650.8 |
Ctsa
|
cathepsin A |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.2 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.9 | 2.6 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.9 | 2.6 | GO:0002632 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) |
| 0.7 | 2.2 | GO:1904117 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.7 | 2.8 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.7 | 4.1 | GO:0097394 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.5 | 4.2 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.4 | 1.8 | GO:0015827 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 0.4 | 1.2 | GO:0030887 | positive regulation of myeloid dendritic cell activation(GO:0030887) |
| 0.3 | 0.9 | GO:0032685 | negative regulation of granulocyte macrophage colony-stimulating factor production(GO:0032685) |
| 0.3 | 2.5 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.3 | 1.7 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.3 | 5.1 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.3 | 6.9 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.2 | 1.9 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.2 | 1.2 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.2 | 0.9 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.2 | 1.5 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.2 | 4.4 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.2 | 3.6 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.2 | 1.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.2 | 2.8 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.2 | 1.6 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.2 | 0.8 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.1 | 0.9 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.1 | 2.8 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.1 | 1.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 0.6 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.1 | 1.4 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 2.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 7.9 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 0.9 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 1.7 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.1 | 0.5 | GO:2000584 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.1 | 0.5 | GO:0019086 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) late viral transcription(GO:0019086) |
| 0.1 | 0.5 | GO:0060356 | leucine import(GO:0060356) |
| 0.1 | 1.5 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 0.3 | GO:0021629 | olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 0.1 | 1.4 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.1 | 0.6 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 0.9 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.9 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.1 | 1.4 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 1.8 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 1.1 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.8 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.1 | 0.5 | GO:0038042 | dimeric G-protein coupled receptor signaling pathway(GO:0038042) |
| 0.1 | 0.5 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 0.4 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.8 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.1 | GO:0035666 | TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.0 | 0.4 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.6 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 2.2 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 7.0 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 2.6 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.3 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 3.8 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.3 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.8 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 1.1 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.3 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.4 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.1 | GO:0002606 | regulation of dendritic cell antigen processing and presentation(GO:0002604) positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.9 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.4 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.6 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.7 | 4.1 | GO:1990707 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 0.5 | 6.9 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.3 | 1.8 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.3 | 0.8 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.2 | 1.7 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.2 | 2.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 4.7 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.2 | 4.2 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 0.8 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 4.4 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 2.2 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 0.8 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 2.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 0.3 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.1 | 0.8 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 2.3 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.9 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.5 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 1.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 2.9 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 1.1 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 1.1 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 6.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.5 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.6 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 1.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 3.9 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 2.4 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 0.1 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.6 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.3 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 3.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.9 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 1.6 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 4.1 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.0 | 1.0 | GO:0043197 | dendritic spine(GO:0043197) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.2 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.9 | 3.7 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.9 | 2.8 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.6 | 4.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.4 | 1.2 | GO:0032394 | MHC class Ib receptor activity(GO:0032394) |
| 0.4 | 3.0 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.4 | 1.5 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.3 | 3.8 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.3 | 10.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.3 | 2.2 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.3 | 1.3 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.3 | 0.9 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.3 | 1.2 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.3 | 1.8 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.3 | 0.8 | GO:0099530 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.2 | 1.2 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.2 | 4.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.2 | 1.1 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.1 | 0.9 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.1 | 5.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.7 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.1 | 1.7 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 2.6 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 0.9 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 2.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 0.8 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 0.5 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 1.1 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 1.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 4.2 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.4 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 5.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 2.6 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.0 | 3.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 1.8 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 1.4 | GO:0016860 | intramolecular oxidoreductase activity(GO:0016860) |
| 0.0 | 2.2 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 1.7 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.4 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.6 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 2.7 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 7.5 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.8 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 2.8 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 4.2 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 3.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 2.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 2.6 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 1.2 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 1.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 2.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.2 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 6.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 2.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.6 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.9 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 1.2 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.2 | 4.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 3.8 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 2.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 1.2 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.1 | 2.8 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 2.7 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 1.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.8 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 1.6 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 1.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 1.2 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 2.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.5 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 1.1 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 2.8 | REACTOME SIGNALING BY ILS | Genes involved in Signaling by Interleukins |
| 0.0 | 0.6 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.9 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.8 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.0 | 0.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 2.4 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 1.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.5 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |