Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
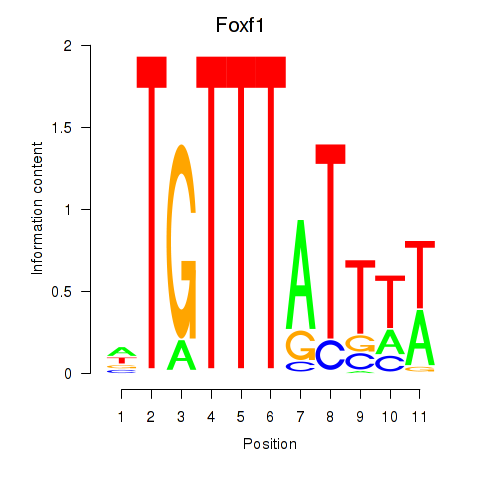
Results for Foxf1
Z-value: 1.35
Transcription factors associated with Foxf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxf1
|
ENSMUSG00000042812.6 | Foxf1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxf1 | mm39_v1_chr8_+_121811091_121811125 | 0.09 | 4.3e-01 | Click! |
Activity profile of Foxf1 motif
Sorted Z-values of Foxf1 motif
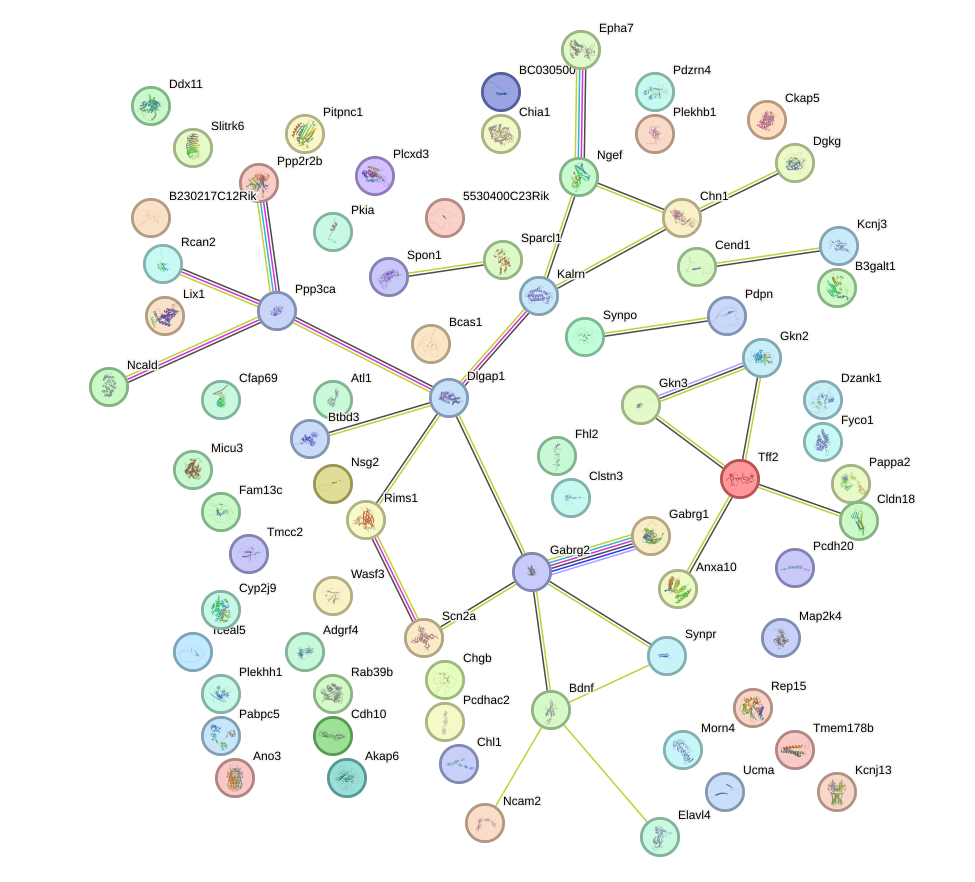
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxf1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_106020545 | 15.62 |
ENSMUST00000079132.12
ENSMUST00000139086.2 |
Chia1
|
chitinase, acidic 1 |
| chr5_-_104261285 | 13.82 |
ENSMUST00000199947.2
|
Sparcl1
|
SPARC-like 1 |
| chr1_+_66426127 | 11.90 |
ENSMUST00000145419.8
|
Map2
|
microtubule-associated protein 2 |
| chr14_-_9015639 | 11.52 |
ENSMUST00000112656.4
|
Synpr
|
synaptoporin |
| chr2_-_144369261 | 10.96 |
ENSMUST00000163701.2
ENSMUST00000081982.12 |
Dzank1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr5_+_88731386 | 10.13 |
ENSMUST00000031229.11
|
Rufy3
|
RUN and FYVE domain containing 3 |
| chr2_-_79738773 | 9.20 |
ENSMUST00000102652.10
ENSMUST00000102651.10 |
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr5_+_88731366 | 9.18 |
ENSMUST00000199312.5
|
Rufy3
|
RUN and FYVE domain containing 3 |
| chr7_-_4847673 | 8.83 |
ENSMUST00000066041.12
ENSMUST00000119433.4 |
Shisa7
|
shisa family member 7 |
| chr12_+_52746158 | 8.43 |
ENSMUST00000095737.5
|
Akap6
|
A kinase (PRKA) anchor protein 6 |
| chr6_+_87350292 | 8.17 |
ENSMUST00000032128.6
|
Gkn2
|
gastrokine 2 |
| chr14_-_88708782 | 8.16 |
ENSMUST00000192557.2
ENSMUST00000061628.7 |
Pcdh20
|
protocadherin 20 |
| chr2_-_79738734 | 8.04 |
ENSMUST00000090756.11
|
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr2_+_138120401 | 7.91 |
ENSMUST00000075410.5
|
Btbd3
|
BTB (POZ) domain containing 3 |
| chr2_+_65499097 | 7.56 |
ENSMUST00000200829.4
|
Scn2a
|
sodium channel, voltage-gated, type II, alpha |
| chr11_+_31950452 | 7.27 |
ENSMUST00000109409.8
ENSMUST00000020537.9 |
Nsg2
|
neuron specific gene family member 2 |
| chr14_-_9015757 | 7.06 |
ENSMUST00000153954.8
|
Synpr
|
synaptoporin |
| chr17_+_70829050 | 7.05 |
ENSMUST00000133717.9
ENSMUST00000148486.8 |
Dlgap1
|
DLG associated protein 1 |
| chr3_+_7431717 | 6.88 |
ENSMUST00000192468.2
ENSMUST00000028999.12 |
Pkia
|
protein kinase inhibitor, alpha |
| chr16_-_34083549 | 6.75 |
ENSMUST00000114949.8
ENSMUST00000114954.8 |
Kalrn
|
kalirin, RhoGEF kinase |
| chr2_+_109522781 | 6.71 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr2_+_67948057 | 6.68 |
ENSMUST00000112346.3
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr7_-_100306160 | 6.63 |
ENSMUST00000107046.8
ENSMUST00000107045.9 ENSMUST00000139708.9 |
Plekhb1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr15_-_37734579 | 6.50 |
ENSMUST00000145909.9
ENSMUST00000153775.9 |
Gm49397
Ncald
|
predicted gene, 49397 neurocalcin delta |
| chr2_+_55327110 | 6.40 |
ENSMUST00000112633.3
ENSMUST00000112632.2 |
Kcnj3
|
potassium inwardly-rectifying channel, subfamily J, member 3 |
| chr19_-_42074777 | 6.36 |
ENSMUST00000051772.10
|
Morn4
|
MORN repeat containing 4 |
| chr2_+_65451100 | 6.35 |
ENSMUST00000144254.6
ENSMUST00000028377.14 |
Scn2a
|
sodium channel, voltage-gated, type II, alpha |
| chr18_-_60781365 | 6.30 |
ENSMUST00000143275.3
|
Synpo
|
synaptopodin |
| chr6_+_39848367 | 6.27 |
ENSMUST00000239008.2
|
Tmem178b
|
transmembrane protein 178B |
| chr18_+_66005891 | 6.25 |
ENSMUST00000173985.10
|
Grp
|
gastrin releasing peptide |
| chr17_-_31363245 | 6.21 |
ENSMUST00000024826.8
|
Tff2
|
trefoil factor 2 (spasmolytic protein 1) |
| chr7_-_141009264 | 6.19 |
ENSMUST00000164387.2
ENSMUST00000137488.2 ENSMUST00000084436.10 |
Cend1
|
cell cycle exit and neuronal differentiation 1 |
| chr17_+_70829144 | 6.18 |
ENSMUST00000140728.8
|
Dlgap1
|
DLG associated protein 1 |
| chr2_-_170248421 | 6.05 |
ENSMUST00000154650.8
|
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr4_-_110148081 | 5.89 |
ENSMUST00000142722.2
|
Elavl4
|
ELAV like RNA binding protein 4 |
| chr5_-_103359117 | 5.86 |
ENSMUST00000112846.8
ENSMUST00000170792.9 ENSMUST00000112847.9 ENSMUST00000238446.3 ENSMUST00000133069.8 |
Mapk10
|
mitogen-activated protein kinase 10 |
| chr19_+_38252984 | 5.84 |
ENSMUST00000198518.5
ENSMUST00000199812.5 |
Lgi1
|
leucine-rich repeat LGI family, member 1 |
| chr5_+_146321757 | 5.79 |
ENSMUST00000016143.9
|
Wasf3
|
WASP family, member 3 |
| chr1_-_22551594 | 5.78 |
ENSMUST00000239255.2
|
Rims1
|
regulating synaptic membrane exocytosis 1 |
| chr6_+_146934082 | 5.69 |
ENSMUST00000036194.6
|
Rep15
|
RAB15 effector protein |
| chr6_-_113696809 | 5.65 |
ENSMUST00000203770.3
ENSMUST00000064993.8 |
Ghrl
|
ghrelin |
| chr16_-_34083315 | 5.65 |
ENSMUST00000114953.8
|
Kalrn
|
kalirin, RhoGEF kinase |
| chr8_+_94537910 | 5.63 |
ENSMUST00000138659.9
|
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chr8_-_65582206 | 5.59 |
ENSMUST00000098713.5
|
Smim31
|
small integral membrane protein 31 |
| chr1_-_43235914 | 5.54 |
ENSMUST00000187357.2
|
Fhl2
|
four and a half LIM domains 2 |
| chr1_-_87501548 | 5.51 |
ENSMUST00000068681.12
|
Ngef
|
neuronal guanine nucleotide exchange factor |
| chr5_-_70999547 | 5.47 |
ENSMUST00000199705.2
|
Gabrg1
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 1 |
| chr17_+_17669082 | 5.34 |
ENSMUST00000140134.2
|
Lix1
|
limb and CNS expressed 1 |
| chr6_-_87365859 | 5.32 |
ENSMUST00000032127.6
|
Gkn3
|
gastrokine 3 |
| chr6_-_113696390 | 5.17 |
ENSMUST00000203588.2
ENSMUST00000204163.3 ENSMUST00000203363.3 |
Ghrl
|
ghrelin |
| chr19_+_38253077 | 5.15 |
ENSMUST00000198045.5
|
Lgi1
|
leucine-rich repeat LGI family, member 1 |
| chr7_-_141009346 | 5.15 |
ENSMUST00000124444.2
|
Cend1
|
cell cycle exit and neuronal differentiation 1 |
| chr19_+_38253105 | 5.11 |
ENSMUST00000196090.2
|
Lgi1
|
leucine-rich repeat LGI family, member 1 |
| chr1_-_132318039 | 5.09 |
ENSMUST00000132435.2
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr2_-_110591909 | 5.07 |
ENSMUST00000140777.2
|
Ano3
|
anoctamin 3 |
| chr1_-_87322443 | 5.06 |
ENSMUST00000113212.4
|
Kcnj13
|
potassium inwardly-rectifying channel, subfamily J, member 13 |
| chr18_+_66006119 | 5.05 |
ENSMUST00000025395.10
|
Grp
|
gastrin releasing peptide |
| chrX_-_74621828 | 5.02 |
ENSMUST00000033545.6
|
Rab39b
|
RAB39B, member RAS oncogene family |
| chr11_-_41891111 | 5.00 |
ENSMUST00000109290.2
|
Gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 2 |
| chr2_+_132623198 | 4.98 |
ENSMUST00000028826.4
|
Chgb
|
chromogranin B |
| chr8_-_62576140 | 4.91 |
ENSMUST00000034052.14
ENSMUST00000034054.9 |
Anxa10
|
annexin A10 |
| chr5_-_5714196 | 4.91 |
ENSMUST00000196165.5
ENSMUST00000061008.10 ENSMUST00000135252.3 ENSMUST00000054865.13 |
Cfap69
|
cilia and flagella associated protein 69 |
| chr18_-_43032359 | 4.89 |
ENSMUST00000117687.8
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr2_+_85715984 | 4.89 |
ENSMUST00000213441.3
|
Olfr1023
|
olfactory receptor 1023 |
| chr16_+_80997580 | 4.87 |
ENSMUST00000037785.14
ENSMUST00000067602.5 |
Ncam2
|
neural cell adhesion molecule 2 |
| chr10_+_90412827 | 4.85 |
ENSMUST00000182550.8
ENSMUST00000099364.12 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chrX_-_135104386 | 4.75 |
ENSMUST00000151592.8
ENSMUST00000131510.2 |
Tceal5
|
transcription elongation factor A (SII)-like 5 |
| chr18_-_43032535 | 4.69 |
ENSMUST00000120632.2
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr9_-_99599312 | 4.67 |
ENSMUST00000112882.9
ENSMUST00000131922.2 |
Cldn18
|
claudin 18 |
| chr6_-_124410452 | 4.63 |
ENSMUST00000124998.2
ENSMUST00000238807.2 |
Clstn3
|
calsyntenin 3 |
| chr17_+_44337566 | 4.56 |
ENSMUST00000229939.2
|
Rcan2
|
regulator of calcineurin 2 |
| chr2_+_71884943 | 4.51 |
ENSMUST00000028525.6
|
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr11_+_31950496 | 4.38 |
ENSMUST00000093219.4
|
Nsg2
|
neuron specific gene family member 2 |
| chr15_+_18819033 | 4.32 |
ENSMUST00000166873.9
|
Cdh10
|
cadherin 10 |
| chr4_-_96479793 | 4.30 |
ENSMUST00000055693.9
|
Cyp2j9
|
cytochrome P450, family 2, subfamily j, polypeptide 9 |
| chr17_+_29309942 | 4.21 |
ENSMUST00000119901.9
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A (P21) |
| chr10_+_70276604 | 4.21 |
ENSMUST00000173042.9
ENSMUST00000062883.7 |
Fam13c
|
family with sequence similarity 13, member C |
| chr8_+_40807344 | 4.20 |
ENSMUST00000136835.2
|
Micu3
|
mitochondrial calcium uptake family, member 3 |
| chr12_+_69954506 | 4.09 |
ENSMUST00000223456.2
|
Atl1
|
atlastin GTPase 1 |
| chr11_+_97732108 | 4.09 |
ENSMUST00000155954.3
ENSMUST00000164364.2 ENSMUST00000170806.2 |
B230217C12Rik
|
RIKEN cDNA B230217C12 gene |
| chr18_+_37072232 | 4.06 |
ENSMUST00000115662.9
ENSMUST00000195590.2 |
Pcdha2
|
protocadherin alpha 2 |
| chr18_-_22983794 | 4.05 |
ENSMUST00000092015.11
ENSMUST00000069215.13 |
Nol4
|
nucleolar protein 4 |
| chr8_+_59365291 | 3.91 |
ENSMUST00000160055.2
|
BC030500
|
cDNA sequence BC030500 |
| chr15_+_4404965 | 3.90 |
ENSMUST00000061925.5
|
Plcxd3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr12_+_79075924 | 3.89 |
ENSMUST00000039928.7
|
Plekhh1
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1 |
| chr2_-_73605387 | 3.88 |
ENSMUST00000166199.9
|
Chn1
|
chimerin 1 |
| chr17_-_43003135 | 3.84 |
ENSMUST00000170723.8
ENSMUST00000164524.2 ENSMUST00000024711.11 ENSMUST00000167993.8 |
Adgrf4
|
adhesion G protein-coupled receptor F4 |
| chr4_-_36056726 | 3.80 |
ENSMUST00000108124.4
|
Lingo2
|
leucine rich repeat and Ig domain containing 2 |
| chr7_+_113365235 | 3.75 |
ENSMUST00000046687.16
|
Spon1
|
spondin 1, (f-spondin) extracellular matrix protein |
| chrX_+_100419965 | 3.75 |
ENSMUST00000119080.8
|
Gjb1
|
gap junction protein, beta 1 |
| chr6_+_103487541 | 3.74 |
ENSMUST00000203912.3
|
Chl1
|
cell adhesion molecule L1-like |
| chr11_+_100902572 | 3.70 |
ENSMUST00000092663.4
|
Atp6v0a1
|
ATPase, H+ transporting, lysosomal V0 subunit A1 |
| chr16_-_22475960 | 3.69 |
ENSMUST00000023578.14
|
Dgkg
|
diacylglycerol kinase, gamma |
| chr4_-_143026068 | 3.69 |
ENSMUST00000030317.14
|
Pdpn
|
podoplanin |
| chr15_+_92495007 | 3.57 |
ENSMUST00000035399.10
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chr18_+_37822865 | 3.55 |
ENSMUST00000195112.2
|
Pcdhgb2
|
protocadherin gamma subfamily B, 2 |
| chr18_+_37827413 | 3.54 |
ENSMUST00000193414.2
|
Pcdhga5
|
protocadherin gamma subfamily A, 5 |
| chr11_-_65636651 | 3.47 |
ENSMUST00000138093.2
|
Map2k4
|
mitogen-activated protein kinase kinase 4 |
| chr16_-_22475915 | 3.44 |
ENSMUST00000089925.10
|
Dgkg
|
diacylglycerol kinase, gamma |
| chr8_+_46111703 | 3.35 |
ENSMUST00000134675.8
ENSMUST00000139869.8 ENSMUST00000126067.8 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr18_+_37100672 | 3.32 |
ENSMUST00000193777.6
ENSMUST00000193389.2 |
Pcdha6
|
protocadherin alpha 6 |
| chr11_-_107238956 | 3.27 |
ENSMUST00000134763.2
|
Pitpnc1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr4_+_28813152 | 3.27 |
ENSMUST00000108194.9
ENSMUST00000108191.2 |
Epha7
|
Eph receptor A7 |
| chr16_+_5703134 | 3.24 |
ENSMUST00000230658.2
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr14_-_110992533 | 3.24 |
ENSMUST00000078386.4
|
Slitrk6
|
SLIT and NTRK-like family, member 6 |
| chr9_+_78020554 | 3.22 |
ENSMUST00000009972.6
ENSMUST00000117330.8 ENSMUST00000044551.8 |
Cilk1
|
ciliogenesis associated kinase 1 |
| chr3_+_136376440 | 3.14 |
ENSMUST00000056758.9
|
Ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isoform |
| chr11_-_87805829 | 3.10 |
ENSMUST00000074874.5
|
Olfr464
|
olfactory receptor 464 |
| chr4_+_28813125 | 3.04 |
ENSMUST00000080934.11
ENSMUST00000029964.12 |
Epha7
|
Eph receptor A7 |
| chrX_+_118836893 | 3.03 |
ENSMUST00000040961.3
ENSMUST00000113366.2 |
Pabpc5
|
poly(A) binding protein, cytoplasmic 5 |
| chr2_+_91376650 | 3.00 |
ENSMUST00000099716.11
ENSMUST00000046769.16 ENSMUST00000111337.3 |
Ckap5
|
cytoskeleton associated protein 5 |
| chr10_+_70276473 | 3.00 |
ENSMUST00000105436.9
|
Fam13c
|
family with sequence similarity 13, member C |
| chr18_-_43032514 | 3.00 |
ENSMUST00000236238.2
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr1_+_37338964 | 2.94 |
ENSMUST00000027287.11
ENSMUST00000140264.8 |
Inpp4a
|
inositol polyphosphate-4-phosphatase, type I |
| chr2_+_67935015 | 2.91 |
ENSMUST00000042456.4
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr4_-_143026033 | 2.91 |
ENSMUST00000119654.2
|
Pdpn
|
podoplanin |
| chr4_-_102883905 | 2.90 |
ENSMUST00000084382.6
ENSMUST00000106869.3 |
Insl5
|
insulin-like 5 |
| chr1_-_158642039 | 2.89 |
ENSMUST00000161589.3
|
Pappa2
|
pappalysin 2 |
| chr1_+_34116308 | 2.89 |
ENSMUST00000239001.2
|
Dst
|
dystonin |
| chr17_+_38485977 | 2.88 |
ENSMUST00000074883.2
|
Olfr134
|
olfactory receptor 134 |
| chr1_+_179788675 | 2.87 |
ENSMUST00000076687.12
ENSMUST00000097450.10 ENSMUST00000212756.2 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr18_+_37580692 | 2.87 |
ENSMUST00000052387.5
|
Pcdhb14
|
protocadherin beta 14 |
| chr6_+_129510117 | 2.84 |
ENSMUST00000032264.9
|
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr1_+_179938904 | 2.83 |
ENSMUST00000145181.2
|
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr6_+_129510145 | 2.78 |
ENSMUST00000204487.3
|
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr7_-_86044743 | 2.72 |
ENSMUST00000053958.6
|
Olfr303
|
olfactory receptor 303 |
| chr18_+_37622518 | 2.67 |
ENSMUST00000055949.4
|
Pcdhb18
|
protocadherin beta 18 |
| chr9_+_53212871 | 2.64 |
ENSMUST00000051014.2
|
Exph5
|
exophilin 5 |
| chrX_-_50770733 | 2.63 |
ENSMUST00000114871.2
|
Hs6st2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr18_-_43071297 | 2.62 |
ENSMUST00000153737.2
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chrM_+_9870 | 2.61 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr14_-_45715308 | 2.59 |
ENSMUST00000141424.2
|
Fermt2
|
fermitin family member 2 |
| chr5_-_148931957 | 2.58 |
ENSMUST00000147473.6
|
Gm42791
|
predicted gene 42791 |
| chrX_+_94942639 | 2.57 |
ENSMUST00000082183.8
|
Zc3h12b
|
zinc finger CCCH-type containing 12B |
| chrM_+_10167 | 2.57 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr9_-_64160899 | 2.56 |
ENSMUST00000005066.9
|
Map2k1
|
mitogen-activated protein kinase kinase 1 |
| chr15_+_22549108 | 2.54 |
ENSMUST00000163361.8
|
Cdh18
|
cadherin 18 |
| chr19_-_37184692 | 2.53 |
ENSMUST00000132580.8
ENSMUST00000079754.11 ENSMUST00000136286.8 ENSMUST00000126188.8 ENSMUST00000126781.2 |
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr10_+_94412116 | 2.50 |
ENSMUST00000117929.2
|
Tmcc3
|
transmembrane and coiled coil domains 3 |
| chr17_-_16050913 | 2.48 |
ENSMUST00000231281.2
|
Rgmb
|
repulsive guidance molecule family member B |
| chr2_-_71198091 | 2.42 |
ENSMUST00000151937.8
|
Slc25a12
|
solute carrier family 25 (mitochondrial carrier, Aralar), member 12 |
| chr4_-_87724533 | 2.42 |
ENSMUST00000126353.8
ENSMUST00000149357.8 |
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr4_+_102427247 | 2.41 |
ENSMUST00000097950.9
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr19_-_6886965 | 2.41 |
ENSMUST00000173091.2
|
Prdx5
|
peroxiredoxin 5 |
| chr18_-_39062201 | 2.40 |
ENSMUST00000134864.2
|
Fgf1
|
fibroblast growth factor 1 |
| chr3_-_148696155 | 2.37 |
ENSMUST00000196526.5
ENSMUST00000200543.5 ENSMUST00000200154.5 |
Adgrl2
|
adhesion G protein-coupled receptor L2 |
| chr7_+_108266625 | 2.34 |
ENSMUST00000076289.2
|
Olfr510
|
olfactory receptor 510 |
| chr19_-_19088543 | 2.31 |
ENSMUST00000112832.8
|
Rorb
|
RAR-related orphan receptor beta |
| chrX_+_141010919 | 2.27 |
ENSMUST00000042329.12
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr13_-_21722197 | 2.25 |
ENSMUST00000168629.2
ENSMUST00000218154.2 |
Olfr1366
|
olfactory receptor 1366 |
| chr11_-_100244866 | 2.23 |
ENSMUST00000173630.8
|
Hap1
|
huntingtin-associated protein 1 |
| chr13_-_36301466 | 2.20 |
ENSMUST00000053265.8
|
Lyrm4
|
LYR motif containing 4 |
| chr7_+_113365356 | 2.16 |
ENSMUST00000084696.6
|
Spon1
|
spondin 1, (f-spondin) extracellular matrix protein |
| chr6_+_129510331 | 2.13 |
ENSMUST00000204956.2
ENSMUST00000204639.2 |
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr14_+_120513106 | 2.12 |
ENSMUST00000227012.2
ENSMUST00000167459.3 |
Mbnl2
|
muscleblind like splicing factor 2 |
| chr2_+_69553141 | 2.10 |
ENSMUST00000090858.10
|
Ppig
|
peptidyl-prolyl isomerase G (cyclophilin G) |
| chr2_+_3115250 | 2.09 |
ENSMUST00000072955.12
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr18_+_69652837 | 2.07 |
ENSMUST00000201410.4
ENSMUST00000202937.4 |
Tcf4
|
transcription factor 4 |
| chr19_-_58444336 | 2.07 |
ENSMUST00000131877.2
|
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr5_-_148329615 | 2.06 |
ENSMUST00000138257.8
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr4_+_102287244 | 2.04 |
ENSMUST00000172616.2
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr3_+_133942244 | 2.00 |
ENSMUST00000181904.3
|
Cxxc4
|
CXXC finger 4 |
| chr19_-_6886898 | 2.00 |
ENSMUST00000238095.2
|
Prdx5
|
peroxiredoxin 5 |
| chr4_+_43384320 | 1.99 |
ENSMUST00000136360.2
|
Rusc2
|
RUN and SH3 domain containing 2 |
| chr13_+_83672708 | 1.98 |
ENSMUST00000199105.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr19_-_6887361 | 1.98 |
ENSMUST00000025904.12
|
Prdx5
|
peroxiredoxin 5 |
| chr16_+_38562616 | 1.96 |
ENSMUST00000023482.13
ENSMUST00000114712.8 ENSMUST00000231655.2 |
B4galt4
|
UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 4 |
| chr2_-_90309509 | 1.95 |
ENSMUST00000111495.9
|
Ptprj
|
protein tyrosine phosphatase, receptor type, J |
| chr9_+_98372575 | 1.93 |
ENSMUST00000035029.3
|
Rbp2
|
retinol binding protein 2, cellular |
| chr6_-_66656668 | 1.93 |
ENSMUST00000071414.2
|
Vmn1r35
|
vomeronasal 1 receptor 35 |
| chr7_-_83381610 | 1.92 |
ENSMUST00000131916.2
|
Il16
|
interleukin 16 |
| chr9_-_105398346 | 1.89 |
ENSMUST00000176770.8
ENSMUST00000085133.13 |
Atp2c1
|
ATPase, Ca++-sequestering |
| chr1_-_168259264 | 1.85 |
ENSMUST00000176790.8
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr12_+_119291343 | 1.85 |
ENSMUST00000221917.2
|
Macc1
|
metastasis associated in colon cancer 1 |
| chrX_+_82898974 | 1.84 |
ENSMUST00000239269.2
|
Dmd
|
dystrophin, muscular dystrophy |
| chr1_-_169938298 | 1.82 |
ENSMUST00000192312.6
|
Ddr2
|
discoidin domain receptor family, member 2 |
| chr2_+_173579285 | 1.78 |
ENSMUST00000067530.6
|
Vapb
|
vesicle-associated membrane protein, associated protein B and C |
| chr5_-_73789764 | 1.78 |
ENSMUST00000087177.4
|
Lrrc66
|
leucine rich repeat containing 66 |
| chr1_+_34275665 | 1.76 |
ENSMUST00000194192.3
|
Dst
|
dystonin |
| chrX_+_162694397 | 1.76 |
ENSMUST00000140845.2
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr6_-_59001325 | 1.76 |
ENSMUST00000173193.2
|
Fam13a
|
family with sequence similarity 13, member A |
| chr8_-_26275182 | 1.74 |
ENSMUST00000038498.10
|
Bag4
|
BCL2-associated athanogene 4 |
| chr6_-_98319684 | 1.73 |
ENSMUST00000164491.3
|
Mdfic2
|
MyoD family inhibitor domain containing 2 |
| chr11_+_87938128 | 1.69 |
ENSMUST00000139129.9
|
Srsf1
|
serine and arginine-rich splicing factor 1 |
| chr19_+_12655487 | 1.69 |
ENSMUST00000215134.2
ENSMUST00000049724.8 |
Olfr1443
|
olfactory receptor 1443 |
| chr9_+_53757448 | 1.66 |
ENSMUST00000048485.7
|
Sln
|
sarcolipin |
| chr18_+_67266784 | 1.65 |
ENSMUST00000236918.2
|
Gnal
|
guanine nucleotide binding protein, alpha stimulating, olfactory type |
| chr6_+_41279199 | 1.64 |
ENSMUST00000031913.5
|
Try4
|
trypsin 4 |
| chr7_-_133966588 | 1.64 |
ENSMUST00000172947.8
|
D7Ertd443e
|
DNA segment, Chr 7, ERATO Doi 443, expressed |
| chr12_-_108241597 | 1.61 |
ENSMUST00000222310.2
|
Ccdc85c
|
coiled-coil domain containing 85C |
| chr9_+_65583826 | 1.59 |
ENSMUST00000153700.9
ENSMUST00000046490.14 |
Oaz2
|
ornithine decarboxylase antizyme 2 |
| chr6_-_57821483 | 1.57 |
ENSMUST00000226191.2
|
Vmn1r21
|
vomeronasal 1 receptor 21 |
| chr6_-_59001455 | 1.57 |
ENSMUST00000089860.12
|
Fam13a
|
family with sequence similarity 13, member A |
| chr1_-_168259070 | 1.56 |
ENSMUST00000064438.11
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr3_+_33853941 | 1.56 |
ENSMUST00000099153.10
|
Ttc14
|
tetratricopeptide repeat domain 14 |
| chr6_+_48604157 | 1.51 |
ENSMUST00000203088.3
ENSMUST00000204958.2 |
AI854703
|
expressed sequence AI854703 |
| chr19_-_18978463 | 1.51 |
ENSMUST00000040153.15
ENSMUST00000112828.8 |
Rorb
|
RAR-related orphan receptor beta |
| chr11_+_4936824 | 1.49 |
ENSMUST00000109897.8
ENSMUST00000009234.16 |
Ap1b1
|
adaptor protein complex AP-1, beta 1 subunit |
| chr8_+_26401698 | 1.48 |
ENSMUST00000120653.8
ENSMUST00000126226.2 |
Kcnu1
|
potassium channel, subfamily U, member 1 |
| chr19_+_58717319 | 1.46 |
ENSMUST00000048644.6
ENSMUST00000236445.2 |
Pnliprp1
|
pancreatic lipase related protein 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 6.6 | GO:1904328 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 2.8 | 11.3 | GO:0021933 | radial glia guided migration of cerebellar granule cell(GO:0021933) negative regulation of cerebellar granule cell precursor proliferation(GO:0021941) |
| 2.7 | 10.8 | GO:2000506 | regulation of gastric mucosal blood circulation(GO:1904344) positive regulation of gastric mucosal blood circulation(GO:1904346) gastric mucosal blood circulation(GO:1990768) negative regulation of energy homeostasis(GO:2000506) |
| 2.3 | 11.3 | GO:1900738 | psychomotor behavior(GO:0036343) positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 2.2 | 6.7 | GO:0061193 | taste bud development(GO:0061193) |
| 1.6 | 6.4 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 1.5 | 12.4 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 1.4 | 8.4 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 1.4 | 5.5 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 1.3 | 13.9 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 1.3 | 6.3 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 1.2 | 10.9 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 1.2 | 5.8 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 1.1 | 9.6 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 1.0 | 3.1 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 1.0 | 6.2 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 1.0 | 13.2 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.9 | 4.5 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.8 | 15.2 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.8 | 2.5 | GO:1900247 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.8 | 5.9 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.8 | 17.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.7 | 2.7 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.7 | 4.7 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.6 | 6.4 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.6 | 5.1 | GO:0061590 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.6 | 6.3 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.6 | 1.8 | GO:0021629 | olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 0.6 | 19.3 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.6 | 1.7 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 0.5 | 3.0 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.5 | 5.5 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.5 | 7.7 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.5 | 2.4 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.5 | 1.9 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.5 | 0.9 | GO:0035483 | gastric emptying(GO:0035483) |
| 0.4 | 2.2 | GO:0032901 | positive regulation of neurotrophin production(GO:0032901) |
| 0.4 | 1.8 | GO:0046725 | modulation by host of viral RNA genome replication(GO:0044830) negative regulation by virus of viral protein levels in host cell(GO:0046725) |
| 0.4 | 4.9 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.4 | 2.6 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.4 | 8.8 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.4 | 4.5 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.4 | 13.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.4 | 4.6 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.4 | 5.7 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.3 | 3.5 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.3 | 4.4 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.3 | 6.9 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.3 | 3.7 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.3 | 1.2 | GO:0046881 | sperm ejaculation(GO:0042713) positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.3 | 3.8 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.3 | 1.1 | GO:0009609 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.3 | 2.6 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.3 | 3.9 | GO:0035768 | cell chemotaxis to fibroblast growth factor(GO:0035766) endothelial cell chemotaxis to fibroblast growth factor(GO:0035768) regulation of cell chemotaxis to fibroblast growth factor(GO:1904847) regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000544) |
| 0.3 | 3.7 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.3 | 4.2 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.2 | 3.2 | GO:0021562 | vestibulocochlear nerve development(GO:0021562) |
| 0.2 | 1.6 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.2 | 10.5 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.2 | 3.5 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.2 | 10.4 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.2 | 3.2 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.2 | 4.6 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.2 | 0.7 | GO:0019389 | glucuronoside metabolic process(GO:0019389) |
| 0.2 | 1.9 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.2 | 5.9 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.2 | 1.7 | GO:1901894 | regulation of calcium-transporting ATPase activity(GO:1901894) |
| 0.2 | 2.9 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.2 | 1.4 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.2 | 4.6 | GO:0031987 | short-term memory(GO:0007614) locomotion involved in locomotory behavior(GO:0031987) |
| 0.2 | 1.7 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.2 | 1.4 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.2 | 5.8 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.2 | 1.4 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 6.0 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 1.0 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 1.9 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.1 | 20.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.8 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.1 | 4.4 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.1 | 5.3 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 2.6 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 1.5 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.1 | 2.1 | GO:0015809 | arginine transport(GO:0015809) |
| 0.1 | 3.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 4.9 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 1.0 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 3.2 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 3.4 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.1 | 5.1 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.1 | 1.9 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.1 | 0.4 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.1 | 0.7 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 | 3.9 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 6.3 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.8 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 | 16.1 | GO:0030307 | positive regulation of cell growth(GO:0030307) |
| 0.1 | 5.4 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 1.0 | GO:0051608 | histamine transport(GO:0051608) |
| 0.1 | 0.7 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 13.5 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.1 | 0.4 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 1.8 | GO:0033622 | integrin activation(GO:0033622) |
| 0.1 | 0.9 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 | 2.5 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 1.0 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 4.4 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.6 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 3.1 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.7 | GO:0098780 | response to mitochondrial depolarisation(GO:0098780) |
| 0.0 | 0.6 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 5.3 | GO:0050680 | negative regulation of epithelial cell proliferation(GO:0050680) |
| 0.0 | 0.7 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 0.4 | GO:0071073 | positive regulation of phospholipid biosynthetic process(GO:0071073) |
| 0.0 | 1.3 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 1.1 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 4.4 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 1.1 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 1.8 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.0 | 0.2 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 0.6 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 0.9 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 1.9 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 3.5 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.9 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.5 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.7 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 3.8 | GO:0050808 | synapse organization(GO:0050808) |
| 0.0 | 3.1 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 2.0 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 1.3 | GO:0050684 | regulation of mRNA processing(GO:0050684) |
| 0.0 | 14.9 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 1.7 | GO:0030509 | BMP signaling pathway(GO:0030509) |
| 0.0 | 1.3 | GO:0021987 | cerebral cortex development(GO:0021987) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 8.4 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 1.4 | 4.2 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 1.3 | 11.9 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 1.1 | 4.5 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 1.0 | 25.9 | GO:0071437 | invadopodium(GO:0071437) |
| 1.0 | 5.8 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.9 | 6.3 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.8 | 22.3 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.6 | 4.6 | GO:0031673 | H zone(GO:0031673) |
| 0.5 | 13.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.5 | 15.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.4 | 3.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.4 | 1.2 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.4 | 5.9 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.4 | 6.7 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.3 | 13.8 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.3 | 4.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.2 | 7.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.2 | 6.4 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.2 | 8.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.2 | 1.4 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.2 | 4.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.2 | 4.3 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 1.4 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.1 | 3.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 5.5 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.7 | GO:0036396 | MIS complex(GO:0036396) |
| 0.1 | 7.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 5.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 2.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 43.2 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.1 | 1.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 7.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 0.9 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 6.4 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 3.9 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 7.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 8.2 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.1 | 1.7 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 1.5 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 0.9 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 5.1 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 7.8 | GO:0042641 | actomyosin(GO:0042641) |
| 0.1 | 1.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 2.4 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 3.5 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 18.9 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.1 | 2.4 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 3.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.5 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 1.7 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 15.4 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 18.1 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 7.1 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 19.0 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 1.1 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 0.9 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 28.5 | GO:0031410 | cytoplasmic vesicle(GO:0031410) |
| 0.0 | 1.8 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 3.7 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 15.1 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 5.1 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 9.6 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.2 | GO:0016012 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.8 | GO:0031768 | ghrelin receptor binding(GO:0031768) |
| 2.4 | 9.6 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 2.2 | 6.7 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 1.6 | 6.4 | GO:0072541 | peroxynitrite reductase activity(GO:0072541) |
| 1.6 | 17.2 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 1.4 | 4.2 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 1.3 | 11.9 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 1.2 | 10.9 | GO:0004568 | chitinase activity(GO:0004568) |
| 1.2 | 5.9 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 1.0 | 11.6 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.8 | 13.9 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.7 | 2.9 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.7 | 6.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.7 | 5.6 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.7 | 3.4 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.7 | 2.7 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.7 | 2.6 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.6 | 6.3 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) axon guidance receptor activity(GO:0008046) |
| 0.6 | 1.8 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.6 | 3.5 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.5 | 3.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.5 | 4.6 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.5 | 2.4 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.5 | 1.9 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.5 | 8.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.4 | 7.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.4 | 2.0 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.4 | 7.7 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.3 | 12.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.3 | 11.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.3 | 6.6 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.3 | 9.8 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.3 | 3.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.3 | 6.9 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.3 | 4.4 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.3 | 3.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.3 | 9.0 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.2 | 1.7 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.2 | 1.4 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.2 | 2.1 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.2 | 3.7 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.2 | 1.4 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.2 | 15.3 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.2 | 8.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.2 | 6.2 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.2 | 5.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.2 | 1.5 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.2 | 0.7 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.2 | 4.5 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.2 | 3.9 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.2 | 1.9 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.2 | 2.2 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.2 | 5.1 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.2 | 3.0 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 2.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.2 | 1.2 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.1 | 1.0 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 2.6 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 1.9 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 10.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 1.8 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 6.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.7 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 3.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 3.7 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 1.9 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 1.0 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 1.6 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 1.4 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 1.0 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 0.7 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 3.3 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 1.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 1.3 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.1 | 0.5 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.2 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.0 | 1.0 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 3.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.9 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.0 | 1.1 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 2.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 9.2 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 2.6 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 18.4 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.6 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.8 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 1.1 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 2.8 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 0.5 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.8 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 3.8 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 6.1 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 2.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.9 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.1 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 3.5 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 7.6 | GO:0005509 | calcium ion binding(GO:0005509) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.4 | 5.6 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.4 | 12.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.3 | 15.2 | PID ATR PATHWAY | ATR signaling pathway |
| 0.2 | 9.3 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.2 | 6.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.2 | 11.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 5.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 6.0 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 28.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 7.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 5.2 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 1.6 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 3.0 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 6.8 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 3.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 2.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 1.9 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 3.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 2.0 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 2.0 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 2.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.2 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.5 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.7 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.8 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.7 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 11.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.5 | 17.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.5 | 10.8 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.5 | 6.8 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.4 | 9.3 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.3 | 6.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.3 | 13.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 6.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.2 | 1.6 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.2 | 3.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.2 | 5.0 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 4.5 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.2 | 6.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 17.8 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.2 | 3.9 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.2 | 2.6 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.2 | 7.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.2 | 7.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 3.7 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 4.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 5.6 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.1 | 5.6 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 7.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 2.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 0.7 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 2.0 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 2.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 1.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 3.8 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 1.3 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 7.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 2.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.9 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 1.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.0 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.7 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 3.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 1.4 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 1.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 1.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.9 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 1.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.5 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |