Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Foxk1_Foxj1
Z-value: 1.00
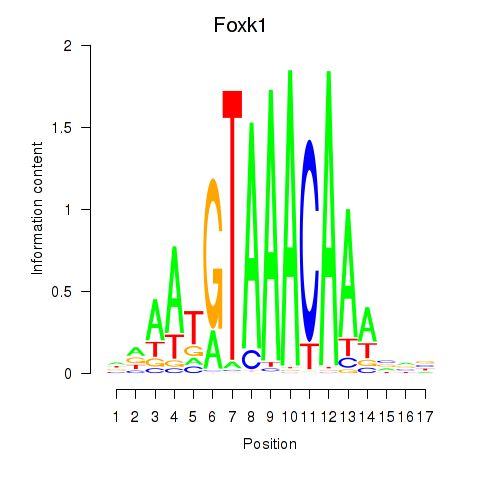
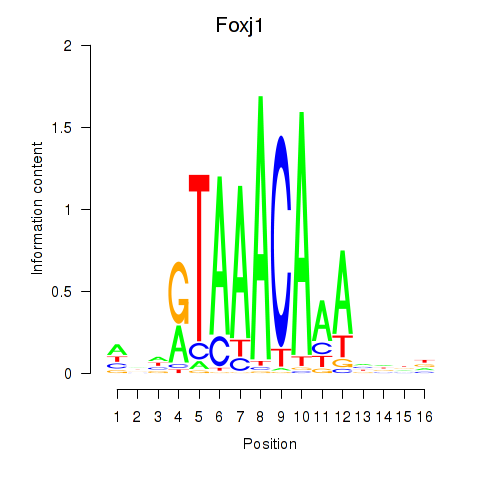
Transcription factors associated with Foxk1_Foxj1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxk1
|
ENSMUSG00000056493.10 | Foxk1 |
|
Foxj1
|
ENSMUSG00000034227.8 | Foxj1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxj1 | mm39_v1_chr11_-_116226175_116226225 | 0.11 | 3.5e-01 | Click! |
| Foxk1 | mm39_v1_chr5_+_142387226_142387284 | 0.07 | 5.4e-01 | Click! |
Activity profile of Foxk1_Foxj1 motif
Sorted Z-values of Foxk1_Foxj1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxk1_Foxj1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_11291805 | 6.90 |
ENSMUST00000187467.2
|
Ms4a14
|
membrane-spanning 4-domains, subfamily A, member 14 |
| chr6_-_83513222 | 5.65 |
ENSMUST00000075161.12
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr4_-_117035922 | 5.44 |
ENSMUST00000153953.2
ENSMUST00000106436.8 |
Kif2c
|
kinesin family member 2C |
| chr6_-_83513184 | 5.23 |
ENSMUST00000205926.2
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chrX_-_161747552 | 4.47 |
ENSMUST00000038769.3
|
S100g
|
S100 calcium binding protein G |
| chr6_-_23650297 | 4.25 |
ENSMUST00000063548.4
|
Rnf133
|
ring finger protein 133 |
| chr6_-_23650205 | 4.19 |
ENSMUST00000115354.2
|
Rnf133
|
ring finger protein 133 |
| chr19_+_53317844 | 4.19 |
ENSMUST00000111737.3
ENSMUST00000025998.15 ENSMUST00000237837.2 |
Mxi1
|
MAX interactor 1, dimerization protein |
| chr3_-_88455556 | 4.11 |
ENSMUST00000131775.2
ENSMUST00000008745.13 |
Rab25
|
RAB25, member RAS oncogene family |
| chr4_-_44710408 | 4.09 |
ENSMUST00000134968.9
ENSMUST00000173821.8 ENSMUST00000174319.8 ENSMUST00000173733.8 ENSMUST00000172866.8 ENSMUST00000165417.9 ENSMUST00000107825.9 ENSMUST00000102932.10 ENSMUST00000107827.9 ENSMUST00000107826.9 ENSMUST00000014174.14 |
Pax5
|
paired box 5 |
| chr12_+_112645237 | 4.07 |
ENSMUST00000174780.2
ENSMUST00000169593.2 ENSMUST00000173942.2 |
Zbtb42
|
zinc finger and BTB domain containing 42 |
| chr6_-_96143217 | 3.99 |
ENSMUST00000090061.6
|
1700123L14Rik
|
RIKEN cDNA 1700123L14 gene |
| chr17_+_28749808 | 3.94 |
ENSMUST00000233837.2
ENSMUST00000025060.4 |
Armc12
|
armadillo repeat containing 12 |
| chr14_-_75368321 | 3.91 |
ENSMUST00000022574.5
|
Lrrc63
|
leucine rich repeat containing 63 |
| chr7_-_101518484 | 3.90 |
ENSMUST00000140584.2
ENSMUST00000134145.8 |
Folr1
|
folate receptor 1 (adult) |
| chr3_-_106126794 | 3.63 |
ENSMUST00000082219.6
|
Chil4
|
chitinase-like 4 |
| chr2_+_68490186 | 3.56 |
ENSMUST00000055930.6
|
4932414N04Rik
|
RIKEN cDNA 4932414N04 gene |
| chr17_+_28749780 | 3.47 |
ENSMUST00000233923.2
|
Armc12
|
armadillo repeat containing 12 |
| chr3_+_65435825 | 3.46 |
ENSMUST00000047906.10
|
Tiparp
|
TCDD-inducible poly(ADP-ribose) polymerase |
| chr17_-_31363245 | 3.36 |
ENSMUST00000024826.8
|
Tff2
|
trefoil factor 2 (spasmolytic protein 1) |
| chr3_-_101195213 | 3.32 |
ENSMUST00000029456.5
|
Cd2
|
CD2 antigen |
| chr1_+_136395673 | 3.29 |
ENSMUST00000189413.7
ENSMUST00000047817.12 |
Kif14
|
kinesin family member 14 |
| chr8_+_68729219 | 3.17 |
ENSMUST00000066594.4
|
Sh2d4a
|
SH2 domain containing 4A |
| chr4_+_134042423 | 3.13 |
ENSMUST00000105875.8
ENSMUST00000030638.7 |
Trim63
|
tripartite motif-containing 63 |
| chr1_+_130793406 | 3.13 |
ENSMUST00000038829.7
|
Fcmr
|
Fc fragment of IgM receptor |
| chr9_-_100388857 | 3.08 |
ENSMUST00000112874.4
|
Nck1
|
non-catalytic region of tyrosine kinase adaptor protein 1 |
| chr19_+_58748132 | 3.06 |
ENSMUST00000026081.5
|
Pnliprp2
|
pancreatic lipase-related protein 2 |
| chr7_+_45164005 | 3.02 |
ENSMUST00000210532.2
ENSMUST00000085331.14 ENSMUST00000210299.2 |
Tulp2
|
tubby-like protein 2 |
| chr4_-_43499608 | 3.01 |
ENSMUST00000136005.3
ENSMUST00000054538.13 |
Arhgef39
|
Rho guanine nucleotide exchange factor (GEF) 39 |
| chr16_-_64422716 | 2.99 |
ENSMUST00000209382.3
|
Csnka2ip
|
casein kinase 2, alpha prime interacting protein |
| chr14_+_71011744 | 2.97 |
ENSMUST00000022698.8
|
Dok2
|
docking protein 2 |
| chr2_+_15531281 | 2.94 |
ENSMUST00000146205.3
|
Malrd1
|
MAM and LDL receptor class A domain containing 1 |
| chr3_+_65435878 | 2.92 |
ENSMUST00000130705.2
|
Tiparp
|
TCDD-inducible poly(ADP-ribose) polymerase |
| chrX_+_106132840 | 2.84 |
ENSMUST00000118666.8
ENSMUST00000053375.4 |
P2ry10
|
purinergic receptor P2Y, G-protein coupled 10 |
| chr11_-_99877423 | 2.81 |
ENSMUST00000105050.4
|
Krtap16-1
|
keratin associated protein 16-1 |
| chrX_+_106132055 | 2.74 |
ENSMUST00000150494.2
|
P2ry10
|
purinergic receptor P2Y, G-protein coupled 10 |
| chr12_+_71063431 | 2.73 |
ENSMUST00000125125.2
|
Arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr4_-_55532453 | 2.70 |
ENSMUST00000132746.2
ENSMUST00000107619.3 |
Klf4
|
Kruppel-like factor 4 (gut) |
| chr6_+_8520006 | 2.61 |
ENSMUST00000162567.8
ENSMUST00000161217.8 |
Glcci1
|
glucocorticoid induced transcript 1 |
| chr3_+_67281449 | 2.53 |
ENSMUST00000061322.10
|
Mlf1
|
myeloid leukemia factor 1 |
| chr1_+_131566223 | 2.51 |
ENSMUST00000112411.2
|
Ctse
|
cathepsin E |
| chrX_-_9983836 | 2.50 |
ENSMUST00000115543.3
ENSMUST00000044789.10 ENSMUST00000115544.9 |
Srpx
|
sushi-repeat-containing protein |
| chr3_+_106020545 | 2.37 |
ENSMUST00000079132.12
ENSMUST00000139086.2 |
Chia1
|
chitinase, acidic 1 |
| chr4_+_52607204 | 2.37 |
ENSMUST00000107671.2
|
Toporsl
|
topoisomerase I binding, arginine/serine-rich like |
| chr7_+_30681287 | 2.36 |
ENSMUST00000128384.3
|
Fam187b
|
family with sequence similarity 187, member B |
| chr6_+_41036271 | 2.34 |
ENSMUST00000103265.5
|
Trbv4
|
T cell receptor beta, variable 10 |
| chr15_+_6609322 | 2.34 |
ENSMUST00000090461.12
|
Fyb
|
FYN binding protein |
| chr15_-_98816012 | 2.31 |
ENSMUST00000023736.10
|
Lmbr1l
|
limb region 1 like |
| chr7_-_51665430 | 2.28 |
ENSMUST00000185841.2
ENSMUST00000190307.7 ENSMUST00000185758.2 ENSMUST00000180038.8 |
1700015G11Rik
|
RIKEN cDNA 1700015G11 gene |
| chr6_-_69792108 | 2.26 |
ENSMUST00000103367.3
|
Igkv12-44
|
immunoglobulin kappa variable 12-44 |
| chr13_+_4624074 | 2.25 |
ENSMUST00000021628.4
|
Akr1c21
|
aldo-keto reductase family 1, member C21 |
| chr15_-_78739717 | 2.25 |
ENSMUST00000044584.6
|
Lgals2
|
lectin, galactose-binding, soluble 2 |
| chr1_+_45350698 | 2.24 |
ENSMUST00000087883.13
|
Col3a1
|
collagen, type III, alpha 1 |
| chr1_-_58735106 | 2.23 |
ENSMUST00000055313.14
ENSMUST00000188772.7 |
Flacc1
|
flagellum associated containing coiled-coil domains 1 |
| chr6_+_24733239 | 2.23 |
ENSMUST00000031690.6
|
Hyal6
|
hyaluronoglucosaminidase 6 |
| chrX_-_159518743 | 2.22 |
ENSMUST00000135856.2
|
Ppef1
|
protein phosphatase with EF hand calcium-binding domain 1 |
| chr7_+_90075762 | 2.20 |
ENSMUST00000061391.9
|
Ccdc89
|
coiled-coil domain containing 89 |
| chr2_+_124994425 | 2.20 |
ENSMUST00000110494.9
ENSMUST00000110495.3 ENSMUST00000028630.9 |
Slc12a1
|
solute carrier family 12, member 1 |
| chr1_-_138103021 | 2.19 |
ENSMUST00000182755.8
ENSMUST00000193650.2 ENSMUST00000182283.8 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr13_-_97170849 | 2.16 |
ENSMUST00000091377.5
|
1700029F12Rik
|
RIKEN cDNA 1700029F12 gene |
| chr4_-_116024788 | 2.16 |
ENSMUST00000030465.10
ENSMUST00000143426.2 |
Tspan1
|
tetraspanin 1 |
| chr3_+_67281424 | 2.16 |
ENSMUST00000077916.12
|
Mlf1
|
myeloid leukemia factor 1 |
| chr1_-_138102972 | 2.15 |
ENSMUST00000195533.6
ENSMUST00000183301.8 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr2_-_84652890 | 2.12 |
ENSMUST00000028471.6
|
Smtnl1
|
smoothelin-like 1 |
| chr19_+_37364791 | 2.11 |
ENSMUST00000012587.4
|
Kif11
|
kinesin family member 11 |
| chr12_-_15866763 | 2.10 |
ENSMUST00000020922.8
ENSMUST00000221215.2 ENSMUST00000221518.2 |
Trib2
|
tribbles pseudokinase 2 |
| chr12_+_111937978 | 2.08 |
ENSMUST00000079009.11
|
Tdrd9
|
tudor domain containing 9 |
| chr14_-_59380335 | 2.07 |
ENSMUST00000022548.10
ENSMUST00000162674.8 ENSMUST00000159858.2 ENSMUST00000162271.2 |
1700129C05Rik
|
RIKEN cDNA 1700129C05 gene |
| chr11_-_5753693 | 2.07 |
ENSMUST00000020768.4
|
Pgam2
|
phosphoglycerate mutase 2 |
| chr4_-_111759951 | 2.06 |
ENSMUST00000102719.8
ENSMUST00000102721.8 |
Slc5a9
|
solute carrier family 5 (sodium/glucose cotransporter), member 9 |
| chr4_-_108690741 | 2.05 |
ENSMUST00000102740.8
ENSMUST00000102741.8 |
Btf3l4
|
basic transcription factor 3-like 4 |
| chr17_-_23864237 | 2.03 |
ENSMUST00000024696.9
|
Mmp25
|
matrix metallopeptidase 25 |
| chr7_+_119803181 | 2.02 |
ENSMUST00000084640.3
|
Abca14
|
ATP-binding cassette, sub-family A (ABC1), member 14 |
| chr3_+_159201048 | 2.01 |
ENSMUST00000120272.8
|
Depdc1a
|
DEP domain containing 1a |
| chr6_-_136918885 | 2.01 |
ENSMUST00000111891.4
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr8_-_65489834 | 2.00 |
ENSMUST00000142822.4
|
Apela
|
apelin receptor early endogenous ligand |
| chr13_-_51888737 | 1.99 |
ENSMUST00000110039.2
|
Sema4d
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
| chr6_-_115014777 | 1.96 |
ENSMUST00000174848.8
ENSMUST00000032461.12 |
Tamm41
|
TAM41 mitochondrial translocator assembly and maintenance homolog |
| chr1_-_45542442 | 1.94 |
ENSMUST00000086430.5
|
Col5a2
|
collagen, type V, alpha 2 |
| chr6_+_42241916 | 1.93 |
ENSMUST00000031895.13
|
Casp2
|
caspase 2 |
| chr10_+_127541039 | 1.91 |
ENSMUST00000079590.7
|
Myo1a
|
myosin IA |
| chr4_-_144134967 | 1.88 |
ENSMUST00000123854.2
ENSMUST00000030326.10 |
Pramel13
|
PRAME like 13 |
| chr15_-_76906832 | 1.87 |
ENSMUST00000019037.10
ENSMUST00000169226.9 |
Mb
|
myoglobin |
| chr14_+_96118660 | 1.86 |
ENSMUST00000228913.2
ENSMUST00000045892.3 |
Spertl
|
spermatid associated like |
| chr19_+_39049442 | 1.86 |
ENSMUST00000087236.5
|
Cyp2c65
|
cytochrome P450, family 2, subfamily c, polypeptide 65 |
| chr14_+_54429757 | 1.86 |
ENSMUST00000103714.2
|
Traj27
|
T cell receptor alpha joining 27 |
| chr2_-_118558852 | 1.85 |
ENSMUST00000102524.8
|
Plcb2
|
phospholipase C, beta 2 |
| chr5_+_30824121 | 1.81 |
ENSMUST00000144742.6
ENSMUST00000149759.2 ENSMUST00000199320.5 |
Cenpa
|
centromere protein A |
| chr11_-_34724458 | 1.78 |
ENSMUST00000093191.3
|
Spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr4_+_11191726 | 1.78 |
ENSMUST00000029866.16
ENSMUST00000108324.4 |
Ccne2
|
cyclin E2 |
| chr18_+_67933169 | 1.75 |
ENSMUST00000025425.7
|
Cep192
|
centrosomal protein 192 |
| chr2_-_27365633 | 1.75 |
ENSMUST00000138693.8
ENSMUST00000113941.9 ENSMUST00000077737.13 |
Brd3
|
bromodomain containing 3 |
| chr18_-_78222349 | 1.73 |
ENSMUST00000237290.2
|
Slc14a2
|
solute carrier family 14 (urea transporter), member 2 |
| chr4_-_87724533 | 1.72 |
ENSMUST00000126353.8
ENSMUST00000149357.8 |
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr17_+_48037758 | 1.72 |
ENSMUST00000024782.12
ENSMUST00000144955.2 |
Pgc
|
progastricsin (pepsinogen C) |
| chr2_+_118877594 | 1.71 |
ENSMUST00000152380.8
ENSMUST00000099542.9 |
Knl1
|
kinetochore scaffold 1 |
| chr10_+_41179966 | 1.68 |
ENSMUST00000173494.4
|
Ak9
|
adenylate kinase 9 |
| chr14_+_26616514 | 1.68 |
ENSMUST00000238987.2
ENSMUST00000239004.2 ENSMUST00000165929.4 ENSMUST00000090337.12 |
Asb14
|
ankyrin repeat and SOCS box-containing 14 |
| chr6_-_136918844 | 1.67 |
ENSMUST00000204934.2
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr13_-_43632368 | 1.67 |
ENSMUST00000222651.2
|
Ranbp9
|
RAN binding protein 9 |
| chr6_-_125262974 | 1.67 |
ENSMUST00000088246.6
|
Tuba3a
|
tubulin, alpha 3A |
| chr1_-_156766381 | 1.66 |
ENSMUST00000188656.7
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr4_-_14621805 | 1.65 |
ENSMUST00000042221.14
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr1_-_156766351 | 1.65 |
ENSMUST00000189648.2
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr8_+_106785434 | 1.65 |
ENSMUST00000212742.2
ENSMUST00000211991.2 |
Nfatc3
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 3 |
| chr11_-_46280298 | 1.65 |
ENSMUST00000109237.9
|
Itk
|
IL2 inducible T cell kinase |
| chr18_+_21205386 | 1.64 |
ENSMUST00000082235.5
|
Mep1b
|
meprin 1 beta |
| chr3_+_159201077 | 1.64 |
ENSMUST00000029825.14
|
Depdc1a
|
DEP domain containing 1a |
| chr2_+_67004178 | 1.63 |
ENSMUST00000239009.2
ENSMUST00000238912.2 |
Xirp2
|
xin actin-binding repeat containing 2 |
| chr14_-_61495934 | 1.62 |
ENSMUST00000077954.13
|
Sgcg
|
sarcoglycan, gamma (dystrophin-associated glycoprotein) |
| chr3_+_159201092 | 1.62 |
ENSMUST00000106041.3
|
Depdc1a
|
DEP domain containing 1a |
| chr2_-_118558825 | 1.59 |
ENSMUST00000159756.2
|
Plcb2
|
phospholipase C, beta 2 |
| chr4_+_151012375 | 1.59 |
ENSMUST00000139826.8
ENSMUST00000116257.8 |
Tnfrsf9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr12_-_103409912 | 1.57 |
ENSMUST00000055071.9
|
Ifi27l2a
|
interferon, alpha-inducible protein 27 like 2A |
| chr5_+_115034997 | 1.57 |
ENSMUST00000031542.13
ENSMUST00000146072.8 ENSMUST00000150361.2 |
Oasl2
|
2'-5' oligoadenylate synthetase-like 2 |
| chr7_-_115459082 | 1.56 |
ENSMUST00000206123.2
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr11_-_46280336 | 1.55 |
ENSMUST00000020664.13
|
Itk
|
IL2 inducible T cell kinase |
| chr5_-_115236354 | 1.54 |
ENSMUST00000100848.3
|
Gm10401
|
predicted gene 10401 |
| chr7_+_92524495 | 1.53 |
ENSMUST00000207594.2
|
Prcp
|
prolylcarboxypeptidase (angiotensinase C) |
| chr6_-_41291634 | 1.53 |
ENSMUST00000064324.12
|
Try5
|
trypsin 5 |
| chr11_-_55042529 | 1.51 |
ENSMUST00000020502.9
ENSMUST00000069816.6 |
Slc36a3
|
solute carrier family 36 (proton/amino acid symporter), member 3 |
| chr16_+_48692976 | 1.51 |
ENSMUST00000065666.6
|
Retnlg
|
resistin like gamma |
| chr14_-_61495832 | 1.50 |
ENSMUST00000121148.8
|
Sgcg
|
sarcoglycan, gamma (dystrophin-associated glycoprotein) |
| chr8_+_22459785 | 1.50 |
ENSMUST00000070649.2
|
Ccdc70
|
coiled-coil domain containing 70 |
| chr2_-_162929732 | 1.49 |
ENSMUST00000094653.6
|
Gtsf1l
|
gametocyte specific factor 1-like |
| chr6_+_70648743 | 1.49 |
ENSMUST00000103401.3
|
Igkv3-4
|
immunoglobulin kappa variable 3-4 |
| chr8_-_65489791 | 1.46 |
ENSMUST00000124790.8
|
Apela
|
apelin receptor early endogenous ligand |
| chr13_-_19881903 | 1.45 |
ENSMUST00000039340.15
ENSMUST00000091763.3 |
Nme8
|
NME/NM23 family member 8 |
| chr7_+_30411634 | 1.44 |
ENSMUST00000005692.14
ENSMUST00000170371.2 |
Atp4a
|
ATPase, H+/K+ exchanging, gastric, alpha polypeptide |
| chr7_+_45271229 | 1.43 |
ENSMUST00000033100.5
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr1_-_144052997 | 1.42 |
ENSMUST00000111941.2
ENSMUST00000052375.8 |
Rgs13
|
regulator of G-protein signaling 13 |
| chr7_+_99512701 | 1.42 |
ENSMUST00000207855.2
|
Xrra1
|
X-ray radiation resistance associated 1 |
| chr5_-_21990170 | 1.42 |
ENSMUST00000115193.8
ENSMUST00000115192.2 ENSMUST00000115195.8 ENSMUST00000030771.12 |
Dnajc2
|
DnaJ heat shock protein family (Hsp40) member C2 |
| chr6_+_126830102 | 1.41 |
ENSMUST00000202878.4
ENSMUST00000202574.2 |
Akap3
|
A kinase (PRKA) anchor protein 3 |
| chr17_-_35392254 | 1.41 |
ENSMUST00000025257.12
|
Aif1
|
allograft inflammatory factor 1 |
| chr14_-_58063585 | 1.41 |
ENSMUST00000022536.3
|
Ska3
|
spindle and kinetochore associated complex subunit 3 |
| chr4_+_11191354 | 1.40 |
ENSMUST00000170901.8
|
Ccne2
|
cyclin E2 |
| chr3_+_130904000 | 1.38 |
ENSMUST00000029611.14
ENSMUST00000106341.9 ENSMUST00000066849.13 |
Lef1
|
lymphoid enhancer binding factor 1 |
| chr14_+_75368939 | 1.37 |
ENSMUST00000125833.8
ENSMUST00000124499.8 |
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr6_-_14755249 | 1.37 |
ENSMUST00000045096.6
|
Ppp1r3a
|
protein phosphatase 1, regulatory subunit 3A |
| chr13_-_50597906 | 1.36 |
ENSMUST00000225905.2
|
Gm47429
|
predicted gene, 47429 |
| chr18_-_43506277 | 1.35 |
ENSMUST00000118043.8
|
Dpysl3
|
dihydropyrimidinase-like 3 |
| chr14_+_75368532 | 1.35 |
ENSMUST00000143539.8
ENSMUST00000134114.8 |
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr2_+_118877610 | 1.35 |
ENSMUST00000153300.8
ENSMUST00000028799.12 |
Knl1
|
kinetochore scaffold 1 |
| chr11_+_88964667 | 1.35 |
ENSMUST00000100619.11
|
Gm525
|
predicted gene 525 |
| chr7_+_44866635 | 1.35 |
ENSMUST00000097216.5
ENSMUST00000209343.2 ENSMUST00000209678.2 |
Tead2
|
TEA domain family member 2 |
| chr5_+_115061293 | 1.34 |
ENSMUST00000031540.11
ENSMUST00000112143.4 |
Oasl1
|
2'-5' oligoadenylate synthetase-like 1 |
| chr12_+_71183622 | 1.34 |
ENSMUST00000149564.8
|
2700049A03Rik
|
RIKEN cDNA 2700049A03 gene |
| chr12_-_32000534 | 1.33 |
ENSMUST00000172314.9
|
Hbp1
|
high mobility group box transcription factor 1 |
| chr3_+_153549846 | 1.33 |
ENSMUST00000044089.4
|
Asb17
|
ankyrin repeat and SOCS box-containing 17 |
| chr2_+_3705824 | 1.33 |
ENSMUST00000115054.9
|
Fam107b
|
family with sequence similarity 107, member B |
| chr10_-_62628008 | 1.33 |
ENSMUST00000217768.2
ENSMUST00000020268.7 ENSMUST00000218946.2 ENSMUST00000219527.2 |
Ccar1
|
cell division cycle and apoptosis regulator 1 |
| chr10_+_96452860 | 1.33 |
ENSMUST00000038377.9
|
Btg1
|
BTG anti-proliferation factor 1 |
| chr7_-_23998735 | 1.32 |
ENSMUST00000145131.8
|
Zfp61
|
zinc finger protein 61 |
| chr5_+_114141894 | 1.30 |
ENSMUST00000086599.11
|
Dao
|
D-amino acid oxidase |
| chr17_+_41121979 | 1.30 |
ENSMUST00000024721.8
ENSMUST00000233740.2 |
Rhag
|
Rhesus blood group-associated A glycoprotein |
| chr9_-_44473146 | 1.29 |
ENSMUST00000215293.2
|
Cxcr5
|
chemokine (C-X-C motif) receptor 5 |
| chr1_+_107456731 | 1.29 |
ENSMUST00000182198.8
|
Serpinb10
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr10_+_17672004 | 1.28 |
ENSMUST00000037964.7
|
Txlnb
|
taxilin beta |
| chr2_-_163486998 | 1.28 |
ENSMUST00000017851.4
|
Serinc3
|
serine incorporator 3 |
| chr5_+_114141739 | 1.27 |
ENSMUST00000112292.9
|
Dao
|
D-amino acid oxidase |
| chr1_-_171434882 | 1.27 |
ENSMUST00000111277.2
ENSMUST00000004827.14 |
Ly9
|
lymphocyte antigen 9 |
| chr19_+_58717319 | 1.26 |
ENSMUST00000048644.6
ENSMUST00000236445.2 |
Pnliprp1
|
pancreatic lipase related protein 1 |
| chr10_+_111919250 | 1.25 |
ENSMUST00000148897.8
ENSMUST00000020434.4 |
Glipr1l2
|
GLI pathogenesis-related 1 like 2 |
| chr15_-_74920518 | 1.25 |
ENSMUST00000185372.2
ENSMUST00000187347.7 ENSMUST00000188845.7 ENSMUST00000185200.7 ENSMUST00000179762.8 ENSMUST00000191216.7 ENSMUST00000065408.16 |
Ly6c1
|
lymphocyte antigen 6 complex, locus C1 |
| chr7_-_101749433 | 1.25 |
ENSMUST00000106937.8
|
Art5
|
ADP-ribosyltransferase 5 |
| chr11_-_46280281 | 1.24 |
ENSMUST00000101306.4
|
Itk
|
IL2 inducible T cell kinase |
| chr17_+_35413415 | 1.24 |
ENSMUST00000025262.6
ENSMUST00000173600.2 |
Ltb
|
lymphotoxin B |
| chr15_+_102875229 | 1.24 |
ENSMUST00000001699.8
|
Hoxc10
|
homeobox C10 |
| chr7_+_45204317 | 1.24 |
ENSMUST00000107752.12
|
Hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr13_+_94954202 | 1.24 |
ENSMUST00000220825.2
|
Tbca
|
tubulin cofactor A |
| chr1_+_9918599 | 1.24 |
ENSMUST00000097826.6
|
Sgk3
|
serum/glucocorticoid regulated kinase 3 |
| chr12_-_84031622 | 1.24 |
ENSMUST00000164935.3
|
Heatr4
|
HEAT repeat containing 4 |
| chr15_-_36140539 | 1.23 |
ENSMUST00000172831.8
|
Rgs22
|
regulator of G-protein signalling 22 |
| chr15_-_42540363 | 1.23 |
ENSMUST00000022921.7
|
Angpt1
|
angiopoietin 1 |
| chr5_+_150446064 | 1.23 |
ENSMUST00000044620.11
ENSMUST00000202003.4 |
Brca2
|
breast cancer 2, early onset |
| chr19_+_53186430 | 1.22 |
ENSMUST00000237099.2
|
Add3
|
adducin 3 (gamma) |
| chr4_-_87724512 | 1.22 |
ENSMUST00000148059.2
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr7_-_119122681 | 1.22 |
ENSMUST00000033267.4
|
Pdilt
|
protein disulfide isomerase-like, testis expressed |
| chr5_+_92479687 | 1.22 |
ENSMUST00000125462.8
ENSMUST00000113083.9 ENSMUST00000121096.8 |
Art3
|
ADP-ribosyltransferase 3 |
| chr5_+_136067350 | 1.22 |
ENSMUST00000062606.8
|
Upk3b
|
uroplakin 3B |
| chr5_+_122347792 | 1.21 |
ENSMUST00000072602.14
|
Hvcn1
|
hydrogen voltage-gated channel 1 |
| chr1_-_53226756 | 1.21 |
ENSMUST00000190748.2
ENSMUST00000072235.10 |
1700019A02Rik
|
RIKEN cDNA 1700019A02 gene |
| chr13_-_32967937 | 1.21 |
ENSMUST00000238977.3
|
Mylk4
|
myosin light chain kinase family, member 4 |
| chr11_+_101473062 | 1.20 |
ENSMUST00000039581.14
ENSMUST00000100403.9 ENSMUST00000107194.8 ENSMUST00000128614.2 |
Tmem106a
|
transmembrane protein 106A |
| chr11_+_76795292 | 1.20 |
ENSMUST00000142166.8
|
Tmigd1
|
transmembrane and immunoglobulin domain containing 1 |
| chr11_+_87685032 | 1.20 |
ENSMUST00000121303.8
|
Mpo
|
myeloperoxidase |
| chr13_+_55932387 | 1.20 |
ENSMUST00000109898.3
|
Catsper3
|
cation channel, sperm associated 3 |
| chr10_-_23112973 | 1.19 |
ENSMUST00000218049.2
|
Eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr8_-_70975813 | 1.18 |
ENSMUST00000121623.8
ENSMUST00000093456.12 ENSMUST00000118850.8 |
Kxd1
|
KxDL motif containing 1 |
| chr13_+_55932369 | 1.18 |
ENSMUST00000021961.12
|
Catsper3
|
cation channel, sperm associated 3 |
| chr3_-_113325938 | 1.18 |
ENSMUST00000132353.2
|
Amy2a1
|
amylase 2a1 |
| chr7_+_97569156 | 1.18 |
ENSMUST00000041860.13
|
Gdpd4
|
glycerophosphodiester phosphodiesterase domain containing 4 |
| chr5_+_66092265 | 1.17 |
ENSMUST00000201814.4
|
Chrna9
|
cholinergic receptor, nicotinic, alpha polypeptide 9 |
| chr14_+_26414422 | 1.16 |
ENSMUST00000022433.12
|
Dnah12
|
dynein, axonemal, heavy chain 12 |
| chr5_-_44256528 | 1.15 |
ENSMUST00000196178.2
ENSMUST00000197750.5 |
Prom1
|
prominin 1 |
| chr5_+_93354377 | 1.15 |
ENSMUST00000031330.5
|
2010109A12Rik
|
RIKEN cDNA 2010109A12 gene |
| chr6_-_70318437 | 1.14 |
ENSMUST00000196599.2
|
Igkv8-19
|
immunoglobulin kappa variable 8-19 |
| chr7_+_79836581 | 1.14 |
ENSMUST00000032754.9
|
Sema4b
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr6_+_78402956 | 1.14 |
ENSMUST00000079926.6
|
Reg1
|
regenerating islet-derived 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0032487 | cerebellar cortex structural organization(GO:0021698) regulation of Rap protein signal transduction(GO:0032487) negative regulation of integrin activation(GO:0033624) |
| 1.1 | 4.3 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 1.0 | 3.1 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.9 | 5.4 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.9 | 2.6 | GO:0036088 | D-serine catabolic process(GO:0036088) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.8 | 4.1 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.8 | 3.1 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.7 | 2.2 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.7 | 2.1 | GO:1990426 | attachment of spindle microtubules to kinetochore involved in homologous chromosome segregation(GO:0051455) microtubule cytoskeleton organization involved in homologous chromosome segregation(GO:0090172) homologous recombination-dependent replication fork processing(GO:1990426) |
| 0.7 | 2.1 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.7 | 6.2 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.6 | 5.0 | GO:0003147 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.5 | 2.0 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.5 | 2.9 | GO:0070858 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.5 | 1.9 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.5 | 3.4 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.5 | 0.5 | GO:2000387 | positive regulation of ovarian follicle development(GO:2000386) regulation of antral ovarian follicle growth(GO:2000387) positive regulation of antral ovarian follicle growth(GO:2000388) |
| 0.5 | 2.3 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.4 | 2.2 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.4 | 1.7 | GO:0046066 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) purine deoxyribonucleoside triphosphate biosynthetic process(GO:0009216) dGDP metabolic process(GO:0046066) |
| 0.4 | 3.7 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.4 | 1.2 | GO:0002148 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 0.4 | 1.6 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.4 | 1.5 | GO:0002353 | plasma kallikrein-kinin cascade(GO:0002353) |
| 0.4 | 1.5 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.4 | 1.1 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.4 | 1.1 | GO:0030887 | positive regulation of myeloid dendritic cell activation(GO:0030887) |
| 0.4 | 1.1 | GO:0007225 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.3 | 2.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.3 | 1.4 | GO:0061153 | negative regulation of interleukin-13 production(GO:0032696) trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) odontoblast differentiation(GO:0071895) |
| 0.3 | 1.7 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.3 | 1.0 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.3 | 6.6 | GO:0071459 | protein localization to chromosome, centromeric region(GO:0071459) |
| 0.3 | 3.3 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.3 | 1.3 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.3 | 4.7 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.3 | 1.6 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.3 | 1.2 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.3 | 0.9 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.3 | 2.6 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.3 | 1.7 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.3 | 1.1 | GO:0010840 | regulation of circadian sleep/wake cycle, wakefulness(GO:0010840) positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) circadian sleep/wake cycle, wakefulness(GO:0042746) |
| 0.3 | 1.7 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.3 | 2.7 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.3 | 4.1 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.3 | 3.3 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.3 | 1.3 | GO:0009597 | detection of virus(GO:0009597) |
| 0.3 | 2.0 | GO:0060022 | hard palate development(GO:0060022) |
| 0.2 | 2.2 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.2 | 0.7 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) |
| 0.2 | 1.7 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.2 | 0.9 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.2 | 0.4 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.2 | 1.9 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.2 | 0.8 | GO:2000521 | negative regulation of immunological synapse formation(GO:2000521) regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001188) negative regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001189) |
| 0.2 | 1.9 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 4.1 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.2 | 0.8 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.2 | 1.6 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.2 | 9.9 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.2 | 0.8 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.2 | 2.9 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.2 | 4.1 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.2 | 1.0 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.2 | 0.9 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 0.2 | 0.7 | GO:0009608 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.2 | 0.9 | GO:0070543 | response to linoleic acid(GO:0070543) |
| 0.2 | 0.5 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.2 | 3.1 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.2 | 2.9 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.2 | 2.0 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.2 | 1.2 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.2 | 3.5 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.2 | 1.0 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 0.2 | 0.5 | GO:0048822 | enucleate erythrocyte development(GO:0048822) |
| 0.2 | 0.7 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) actin polymerization-dependent cell motility(GO:0070358) positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.2 | 1.8 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.2 | 2.1 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.2 | 1.6 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 | 1.0 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.1 | 3.1 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.1 | 1.3 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 | 2.1 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.1 | 0.6 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.1 | 3.3 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 1.6 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 | 2.2 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.1 | 0.7 | GO:0043056 | forward locomotion(GO:0043056) |
| 0.1 | 2.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.5 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.1 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 | 0.5 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.1 | 0.5 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.1 | 0.8 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 0.1 | GO:0045423 | granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0042253) regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) |
| 0.1 | 1.0 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.1 | 0.6 | GO:0060931 | sinoatrial node cell differentiation(GO:0060921) sinoatrial node cell development(GO:0060931) |
| 0.1 | 0.4 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.1 | 6.0 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 0.7 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.1 | 0.6 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 1.4 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 2.8 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 0.5 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 0.6 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.1 | 0.9 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.1 | 2.4 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.1 | 0.5 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.9 | GO:0007128 | meiotic prophase I(GO:0007128) prophase(GO:0051324) meiotic cell cycle phase(GO:0098762) meiosis I cell cycle phase(GO:0098764) |
| 0.1 | 9.7 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 1.0 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.1 | 0.8 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 0.3 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.1 | 0.5 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.1 | 0.8 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.1 | 0.6 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.1 | 1.5 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 0.9 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.1 | 0.8 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.1 | 0.4 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 1.2 | GO:0045141 | meiotic telomere clustering(GO:0045141) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) |
| 0.1 | 0.9 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.1 | 0.4 | GO:0010957 | negative regulation of vitamin D biosynthetic process(GO:0010957) regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 0.7 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 1.6 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.1 | 1.2 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.1 | 0.3 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.1 | 3.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 0.8 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.1 | 1.5 | GO:0015816 | glycine transport(GO:0015816) |
| 0.1 | 0.6 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.1 | 0.5 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.1 | 0.9 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
| 0.1 | 1.2 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 3.5 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 0.5 | GO:0009180 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.1 | 0.5 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 1.8 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.1 | 1.1 | GO:0035646 | endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.1 | 1.4 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.1 | 1.1 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 0.5 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.6 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.1 | 0.3 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 1.2 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 0.5 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.1 | 0.2 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.1 | 1.0 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.1 | 2.4 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 0.3 | GO:0021815 | modulation of microtubule cytoskeleton involved in cerebral cortex radial glia guided migration(GO:0021815) nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration(GO:0021817) nuclear migration along microtubule(GO:0030473) nuclear migration along microfilament(GO:0031022) |
| 0.1 | 1.0 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 0.3 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 0.1 | 1.3 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.1 | 1.2 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.1 | 0.1 | GO:0061738 | late endosomal microautophagy(GO:0061738) |
| 0.1 | 0.4 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.1 | 1.2 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 0.2 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.1 | 0.8 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 0.7 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.1 | 1.4 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 2.0 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 1.4 | GO:0010155 | regulation of proton transport(GO:0010155) |
| 0.1 | 0.4 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 | 0.4 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.1 | 1.4 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.1 | 0.4 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.0 | 1.0 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 0.8 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 1.5 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 1.4 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 5.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.2 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 3.5 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 0.4 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.0 | 1.1 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.0 | 0.1 | GO:0090673 | endothelial cell-matrix adhesion(GO:0090673) |
| 0.0 | 1.4 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 1.6 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.2 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.0 | 0.2 | GO:0015744 | succinate transport(GO:0015744) |
| 0.0 | 1.7 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 1.2 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) |
| 0.0 | 0.3 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.9 | GO:0001783 | B cell apoptotic process(GO:0001783) |
| 0.0 | 0.5 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.6 | GO:0051608 | histamine transport(GO:0051608) |
| 0.0 | 1.2 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.3 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 1.3 | GO:2000279 | negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.0 | 0.5 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 1.0 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 1.0 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 0.2 | GO:1903847 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.0 | 1.4 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.4 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.6 | GO:0036035 | osteoclast development(GO:0036035) |
| 0.0 | 0.2 | GO:0097461 | ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) positive regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0090282) |
| 0.0 | 2.5 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.1 | GO:0042560 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.0 | 0.3 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 1.0 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.7 | GO:0032402 | melanosome transport(GO:0032402) |
| 0.0 | 1.5 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.5 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.0 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.2 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.1 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.0 | 1.5 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.1 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.3 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.3 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 | 1.6 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 0.1 | GO:1901894 | regulation of calcium-transporting ATPase activity(GO:1901894) |
| 0.0 | 0.5 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 0.8 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 2.6 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 6.5 | GO:0051321 | meiotic cell cycle(GO:0051321) |
| 0.0 | 0.4 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 1.1 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.0 | 0.3 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.4 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.2 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.6 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 1.1 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.5 | GO:0050922 | negative regulation of chemotaxis(GO:0050922) |
| 0.0 | 0.2 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.1 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.2 | GO:2000580 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.2 | GO:0010663 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.0 | 0.3 | GO:0018126 | protein hydroxylation(GO:0018126) |
| 0.0 | 0.8 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.2 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.0 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.8 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.0 | 0.3 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 1.1 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 2.2 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 1.2 | GO:0005977 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 | 0.3 | GO:1903963 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.4 | GO:0097192 | signal transduction in absence of ligand(GO:0038034) extrinsic apoptotic signaling pathway in absence of ligand(GO:0097192) |
| 0.0 | 0.6 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.3 | GO:0031424 | keratinization(GO:0031424) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.9 | 3.4 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.8 | 0.8 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.6 | 1.9 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.5 | 2.1 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.4 | 1.1 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.4 | 1.4 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.3 | 2.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.3 | 3.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.3 | 1.3 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.3 | 2.3 | GO:0071914 | prominosome(GO:0071914) |
| 0.2 | 2.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 0.9 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.2 | 3.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.2 | 3.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 1.0 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.2 | 4.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.2 | 5.0 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.2 | 2.4 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.2 | 3.1 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 1.2 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 1.3 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 3.1 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 10.7 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 4.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.6 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 4.7 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 1.4 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 1.3 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 0.5 | GO:0034687 | integrin alphaL-beta2 complex(GO:0034687) |
| 0.1 | 0.8 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.1 | 1.9 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.1 | 1.2 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 1.2 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 1.0 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 0.5 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.1 | 3.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 10.7 | GO:0000800 | lateral element(GO:0000800) |
| 0.1 | 1.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 0.8 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 3.8 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.6 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 5.9 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 0.4 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.1 | 1.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.4 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 0.2 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.1 | 0.3 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 1.8 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 1.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 1.0 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 2.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.9 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 0.9 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 1.7 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 0.5 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.1 | 5.0 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 2.7 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 1.4 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 0.5 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 7.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 1.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.2 | GO:0008623 | CHRAC(GO:0008623) |
| 0.0 | 2.9 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.5 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 1.5 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.5 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.4 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.3 | GO:0061689 | paranodal junction(GO:0033010) tricellular tight junction(GO:0061689) |
| 0.0 | 0.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.5 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.5 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 1.6 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 1.0 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 1.2 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.5 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.8 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.0 | 5.1 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.3 | GO:0044447 | axoneme part(GO:0044447) |
| 0.0 | 4.0 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 1.8 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.2 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 13.9 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.2 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 0.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 1.0 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 4.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.4 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.9 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 3.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 8.7 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.5 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 1.9 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 1.0 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.8 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.7 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.6 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0070401 | NADP+ binding(GO:0070401) lithocholic acid binding(GO:1902121) |
| 0.7 | 4.5 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.7 | 6.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.7 | 2.1 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.7 | 2.0 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.6 | 2.6 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.5 | 2.2 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.5 | 2.7 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.5 | 1.4 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.5 | 5.0 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.4 | 2.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.4 | 1.7 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.4 | 3.7 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.4 | 3.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.4 | 1.9 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.4 | 1.1 | GO:0032394 | MHC class Ib receptor activity(GO:0032394) |
| 0.3 | 1.2 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.3 | 1.5 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.3 | 4.0 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.3 | 1.4 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.3 | 0.8 | GO:0050785 | advanced glycation end-product receptor activity(GO:0050785) |
| 0.3 | 1.9 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.3 | 1.0 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.2 | 2.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.2 | 5.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 2.9 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.2 | 3.5 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.2 | 6.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.2 | 2.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.2 | 2.3 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.2 | 3.1 | GO:0031432 | titin binding(GO:0031432) |
| 0.2 | 1.0 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.2 | 1.9 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.2 | 0.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 1.2 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.2 | 2.3 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.2 | 5.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 3.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.2 | 0.4 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.2 | 0.9 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.2 | 2.5 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.2 | 1.2 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.2 | 1.7 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.2 | 2.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.2 | 1.2 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.2 | 0.5 | GO:0042936 | dipeptide transporter activity(GO:0042936) |
| 0.2 | 0.8 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.2 | 4.9 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.2 | 0.5 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.2 | 0.8 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.2 | 1.4 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 4.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 1.0 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 0.5 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.1 | 1.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.8 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.1 | 3.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 2.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 1.4 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 0.6 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.1 | 1.0 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 2.5 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.7 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.1 | 2.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.4 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.1 | 6.2 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 1.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 0.9 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.1 | 0.5 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.1 | 2.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 1.0 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 1.2 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 0.3 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.1 | 0.4 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.1 | 2.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 1.5 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 0.5 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 0.4 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 1.8 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.1 | 0.6 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 1.6 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 0.3 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.1 | 2.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 1.9 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 1.1 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.1 | 2.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 1.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 1.1 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.1 | 3.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.2 | GO:0004320 | oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) |
| 0.1 | 1.6 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.9 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.1 | 1.3 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.1 | 2.5 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 2.3 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 0.7 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.6 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 1.6 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.1 | 1.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.2 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.1 | 0.7 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 0.2 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.1 | 0.1 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.1 | 0.8 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 1.2 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.1 | 0.5 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 0.4 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 1.3 | GO:0022842 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.1 | 0.3 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.8 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 1.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.2 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 4.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 1.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.4 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 6.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.5 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.0 | 0.1 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 4.0 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.2 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.7 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 1.1 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.7 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.5 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 1.2 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.4 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 1.7 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.5 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 3.0 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 1.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.9 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.0 | 0.8 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 2.9 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.4 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 1.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 6.4 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.9 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.0 | 0.5 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.2 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.1 | GO:0034597 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.8 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.5 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 6.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.8 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 1.2 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 1.6 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 5.6 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 0.1 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 0.0 | 0.6 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 2.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.2 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 5.1 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.5 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 1.0 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.9 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 1.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.4 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 1.1 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.2 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.4 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 5.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 6.5 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 1.2 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 3.7 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.3 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 1.0 | GO:0008565 | protein transporter activity(GO:0008565) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 3.4 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 12.0 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.1 | 9.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 6.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 3.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 3.8 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 4.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 6.3 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 7.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 2.5 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.6 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 3.4 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 2.7 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 1.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 2.5 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 2.1 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.6 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.9 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.1 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.0 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 7.7 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.8 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 1.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.4 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.5 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.4 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.2 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.3 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.9 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.3 | 9.4 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.3 | 3.4 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.2 | 7.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 10.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.2 | 2.9 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 3.0 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 2.6 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 3.6 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 1.6 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.4 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.1 | 1.8 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 4.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.5 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.1 | 1.0 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 2.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 1.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 2.9 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 1.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 2.0 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 4.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 1.8 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 1.7 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.5 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.8 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.3 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.4 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 1.8 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.8 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 3.9 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 1.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.9 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 2.6 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.9 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.3 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 1.0 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.3 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 1.0 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.4 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.7 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 4.5 | REACTOME CLASS I MHC MEDIATED ANTIGEN PROCESSING PRESENTATION | Genes involved in Class I MHC mediated antigen processing & presentation |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 1.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |