Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Foxl1
Z-value: 0.53
Transcription factors associated with Foxl1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxl1
|
ENSMUSG00000097084.2 | Foxl1 |
|
Foxl1
|
ENSMUSG00000097084.2 | Foxl1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxl1 | mm39_v1_chr8_+_121854566_121854679 | -0.16 | 1.8e-01 | Click! |
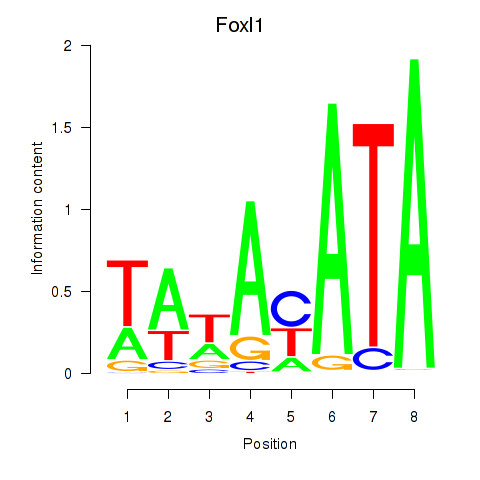
Activity profile of Foxl1 motif
Sorted Z-values of Foxl1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxl1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_15107646 | 4.03 |
ENSMUST00000033842.4
|
Myom2
|
myomesin 2 |
| chr3_-_122828592 | 3.08 |
ENSMUST00000029761.14
|
Myoz2
|
myozenin 2 |
| chr7_-_119078472 | 2.91 |
ENSMUST00000209095.2
ENSMUST00000033263.6 ENSMUST00000207261.2 |
Umod
|
uromodulin |
| chr15_-_11905697 | 2.48 |
ENSMUST00000066529.5
ENSMUST00000228603.2 |
Npr3
|
natriuretic peptide receptor 3 |
| chr5_-_83502966 | 2.29 |
ENSMUST00000053543.11
|
Tecrl
|
trans-2,3-enoyl-CoA reductase-like |
| chr9_+_71123061 | 2.28 |
ENSMUST00000034723.6
|
Aldh1a2
|
aldehyde dehydrogenase family 1, subfamily A2 |
| chr7_-_119078330 | 2.24 |
ENSMUST00000207460.2
|
Umod
|
uromodulin |
| chr18_-_15284476 | 2.21 |
ENSMUST00000025992.7
|
Kctd1
|
potassium channel tetramerisation domain containing 1 |
| chr9_+_121606750 | 2.07 |
ENSMUST00000098272.4
|
Klhl40
|
kelch-like 40 |
| chr10_+_56253418 | 1.79 |
ENSMUST00000068581.9
ENSMUST00000217789.2 |
Gja1
|
gap junction protein, alpha 1 |
| chr6_-_101176147 | 1.77 |
ENSMUST00000239140.2
|
Pdzrn3
|
PDZ domain containing RING finger 3 |
| chr1_+_66426127 | 1.58 |
ENSMUST00000145419.8
|
Map2
|
microtubule-associated protein 2 |
| chr6_-_144978557 | 1.57 |
ENSMUST00000136819.3
|
Bcat1
|
branched chain aminotransferase 1, cytosolic |
| chr18_+_37637317 | 1.56 |
ENSMUST00000052179.8
|
Pcdhb20
|
protocadherin beta 20 |
| chr18_+_37864045 | 1.53 |
ENSMUST00000192535.2
|
Pcdhgb5
|
protocadherin gamma subfamily B, 5 |
| chr5_+_102629240 | 1.52 |
ENSMUST00000073302.12
ENSMUST00000094559.9 |
Arhgap24
|
Rho GTPase activating protein 24 |
| chr9_-_101076198 | 1.50 |
ENSMUST00000066773.9
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr15_-_98705791 | 1.37 |
ENSMUST00000075444.8
|
Ddn
|
dendrin |
| chr5_+_102629365 | 1.33 |
ENSMUST00000112854.8
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr18_-_31580436 | 1.28 |
ENSMUST00000025110.5
|
Syt4
|
synaptotagmin IV |
| chr5_-_83502793 | 1.27 |
ENSMUST00000146669.2
|
Tecrl
|
trans-2,3-enoyl-CoA reductase-like |
| chr1_-_169938060 | 1.24 |
ENSMUST00000027985.8
ENSMUST00000194690.6 |
Ddr2
|
discoidin domain receptor family, member 2 |
| chr16_-_22258469 | 1.04 |
ENSMUST00000079601.13
|
Etv5
|
ets variant 5 |
| chr6_-_14755249 | 1.01 |
ENSMUST00000045096.6
|
Ppp1r3a
|
protein phosphatase 1, regulatory subunit 3A |
| chr1_-_169938298 | 0.99 |
ENSMUST00000192312.6
|
Ddr2
|
discoidin domain receptor family, member 2 |
| chr13_-_23806530 | 0.80 |
ENSMUST00000062045.4
|
H1f4
|
H1.4 linker histone, cluster member |
| chr9_+_43980718 | 0.79 |
ENSMUST00000114830.9
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr3_-_146475974 | 0.73 |
ENSMUST00000106137.8
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr18_-_25886750 | 0.70 |
ENSMUST00000224553.2
ENSMUST00000025117.14 |
Celf4
|
CUGBP, Elav-like family member 4 |
| chr18_+_36431732 | 0.68 |
ENSMUST00000210490.3
|
Igip
|
IgA inducing protein |
| chr16_-_22258320 | 0.67 |
ENSMUST00000170393.2
|
Etv5
|
ets variant 5 |
| chr6_+_42263609 | 0.63 |
ENSMUST00000238845.2
ENSMUST00000031894.13 |
Clcn1
|
chloride channel, voltage-sensitive 1 |
| chrX_+_132751729 | 0.56 |
ENSMUST00000033602.9
|
Tnmd
|
tenomodulin |
| chr8_+_93553901 | 0.51 |
ENSMUST00000034187.9
|
Mmp2
|
matrix metallopeptidase 2 |
| chr7_+_90739904 | 0.49 |
ENSMUST00000107196.10
ENSMUST00000074273.10 |
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr1_+_179788037 | 0.49 |
ENSMUST00000097453.9
ENSMUST00000111117.8 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr17_-_91400142 | 0.41 |
ENSMUST00000160800.9
|
Nrxn1
|
neurexin I |
| chr2_+_91086489 | 0.40 |
ENSMUST00000154959.8
ENSMUST00000059566.11 |
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr6_-_102441628 | 0.35 |
ENSMUST00000032159.7
|
Cntn3
|
contactin 3 |
| chr6_-_57877975 | 0.30 |
ENSMUST00000228322.2
|
Vmn1r22
|
vomeronasal 1 receptor 22 |
| chr2_+_119156265 | 0.29 |
ENSMUST00000102517.4
|
Dll4
|
delta like canonical Notch ligand 4 |
| chr6_-_147165623 | 0.24 |
ENSMUST00000052296.9
ENSMUST00000204197.2 |
Pthlh
|
parathyroid hormone-like peptide |
| chr10_-_102326286 | 0.02 |
ENSMUST00000020040.5
|
Nts
|
neurotensin |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.2 | GO:0072023 | ascending thin limb development(GO:0072021) thick ascending limb development(GO:0072023) metanephric ascending thin limb development(GO:0072218) metanephric thick ascending limb development(GO:0072233) |
| 0.6 | 1.8 | GO:0060156 | vascular transport(GO:0010232) milk ejection(GO:0060156) |
| 0.3 | 2.3 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.3 | 2.8 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.2 | 2.5 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.2 | 4.0 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.2 | 2.1 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.2 | 1.5 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.2 | 1.6 | GO:0009099 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.2 | 1.7 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 3.6 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.1 | 1.3 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 | 2.2 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 | 0.5 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.1 | 0.3 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.1 | 0.2 | GO:0060618 | nipple development(GO:0060618) |
| 0.1 | 0.4 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 0.1 | 0.7 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 | 0.7 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 0.6 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 1.6 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.8 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.0 | 3.1 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 0.5 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.0 | 1.8 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.6 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.5 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 4.0 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 1.8 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 1.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 1.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 2.1 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 5.2 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.4 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 1.8 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 3.1 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.5 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.4 | 5.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.4 | 1.3 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.4 | 2.5 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.3 | 2.3 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.3 | 1.8 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.2 | 2.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.2 | 1.6 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.2 | 4.0 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.2 | 1.6 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 0.7 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 3.6 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.4 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.5 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.8 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 1.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.7 | GO:0036002 | pre-mRNA binding(GO:0036002) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.7 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.0 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.7 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 1.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 1.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.7 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 2.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |