Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Foxm1
Z-value: 1.34
Transcription factors associated with Foxm1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxm1
|
ENSMUSG00000001517.15 | Foxm1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxm1 | mm39_v1_chr6_+_128339882_128340022 | -0.24 | 4.2e-02 | Click! |
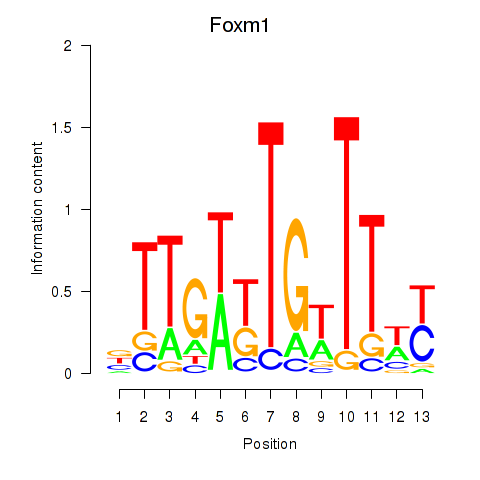
Activity profile of Foxm1 motif
Sorted Z-values of Foxm1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxm1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_15149755 | 15.38 |
ENSMUST00000108273.2
|
Necab1
|
N-terminal EF-hand calcium binding protein 1 |
| chr1_-_79417732 | 14.92 |
ENSMUST00000185234.2
ENSMUST00000049972.6 |
Scg2
|
secretogranin II |
| chr2_+_65676111 | 12.89 |
ENSMUST00000122912.8
|
Csrnp3
|
cysteine-serine-rich nuclear protein 3 |
| chr2_+_65676176 | 11.41 |
ENSMUST00000053910.10
|
Csrnp3
|
cysteine-serine-rich nuclear protein 3 |
| chr16_+_17093941 | 11.40 |
ENSMUST00000164950.11
|
Tmem191c
|
transmembrane protein 191C |
| chr10_-_81066607 | 10.93 |
ENSMUST00000047408.6
|
Atcay
|
ataxia, cerebellar, Cayman type |
| chr5_+_137551774 | 10.68 |
ENSMUST00000136088.8
ENSMUST00000139395.8 |
Actl6b
|
actin-like 6B |
| chr3_-_80710097 | 10.55 |
ENSMUST00000075316.10
ENSMUST00000107745.8 |
Gria2
|
glutamate receptor, ionotropic, AMPA2 (alpha 2) |
| chr11_-_100244866 | 10.36 |
ENSMUST00000173630.8
|
Hap1
|
huntingtin-associated protein 1 |
| chr9_-_75504926 | 10.20 |
ENSMUST00000164100.2
|
Tmod2
|
tropomodulin 2 |
| chrX_-_74621828 | 9.82 |
ENSMUST00000033545.6
|
Rab39b
|
RAB39B, member RAS oncogene family |
| chr2_+_96148418 | 9.77 |
ENSMUST00000135431.8
ENSMUST00000162807.9 |
Lrrc4c
|
leucine rich repeat containing 4C |
| chr15_+_41615084 | 9.70 |
ENSMUST00000229511.2
ENSMUST00000229836.2 |
Oxr1
|
oxidation resistance 1 |
| chr2_+_135894099 | 9.63 |
ENSMUST00000144403.8
|
Lamp5
|
lysosomal-associated membrane protein family, member 5 |
| chr10_-_49659355 | 9.17 |
ENSMUST00000105484.10
ENSMUST00000218598.2 ENSMUST00000079751.9 ENSMUST00000218441.2 |
Grik2
|
glutamate receptor, ionotropic, kainate 2 (beta 2) |
| chr8_-_58249608 | 9.02 |
ENSMUST00000204067.3
|
Galntl6
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 6 |
| chr3_-_152373997 | 8.99 |
ENSMUST00000045262.11
|
Ak5
|
adenylate kinase 5 |
| chr5_-_103359117 | 8.93 |
ENSMUST00000112846.8
ENSMUST00000170792.9 ENSMUST00000112847.9 ENSMUST00000238446.3 ENSMUST00000133069.8 |
Mapk10
|
mitogen-activated protein kinase 10 |
| chr10_+_90665639 | 8.33 |
ENSMUST00000179337.9
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr18_+_37085673 | 8.18 |
ENSMUST00000192512.6
ENSMUST00000192295.2 ENSMUST00000115661.5 |
Pcdha4
Gm42416
|
protocadherin alpha 4 predicted gene, 42416 |
| chr5_+_137551790 | 8.14 |
ENSMUST00000136565.8
ENSMUST00000149292.8 ENSMUST00000125489.2 |
Actl6b
|
actin-like 6B |
| chr6_-_138399896 | 7.93 |
ENSMUST00000161450.8
ENSMUST00000163024.8 ENSMUST00000162185.8 |
Lmo3
|
LIM domain only 3 |
| chr2_+_67948057 | 7.75 |
ENSMUST00000112346.3
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr11_-_41891359 | 7.66 |
ENSMUST00000070735.10
|
Gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 2 |
| chr14_+_76192449 | 7.64 |
ENSMUST00000050120.4
|
Kctd4
|
potassium channel tetramerisation domain containing 4 |
| chr11_-_41891111 | 7.62 |
ENSMUST00000109290.2
|
Gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 2 |
| chr16_-_23709564 | 7.46 |
ENSMUST00000004480.5
|
Sst
|
somatostatin |
| chr1_+_158189831 | 7.45 |
ENSMUST00000193042.6
ENSMUST00000046110.16 |
Astn1
|
astrotactin 1 |
| chr18_+_37100672 | 7.35 |
ENSMUST00000193777.6
ENSMUST00000193389.2 |
Pcdha6
|
protocadherin alpha 6 |
| chr12_-_41536430 | 7.14 |
ENSMUST00000043884.6
|
Lrrn3
|
leucine rich repeat protein 3, neuronal |
| chr8_+_83891972 | 7.09 |
ENSMUST00000034145.11
|
Tbc1d9
|
TBC1 domain family, member 9 |
| chr10_+_123099945 | 6.83 |
ENSMUST00000238972.2
ENSMUST00000050756.8 |
Tafa2
|
TAFA chemokine like family member 2 |
| chr2_+_107120934 | 6.81 |
ENSMUST00000037012.3
|
Kcna4
|
potassium voltage-gated channel, shaker-related subfamily, member 4 |
| chr14_-_24537067 | 6.41 |
ENSMUST00000026322.9
|
Polr3a
|
polymerase (RNA) III (DNA directed) polypeptide A |
| chr2_+_91376650 | 6.32 |
ENSMUST00000099716.11
ENSMUST00000046769.16 ENSMUST00000111337.3 |
Ckap5
|
cytoskeleton associated protein 5 |
| chr11_-_99313078 | 6.27 |
ENSMUST00000017741.4
|
Krt12
|
keratin 12 |
| chr9_+_22365586 | 6.03 |
ENSMUST00000168332.2
|
Gm17545
|
predicted gene, 17545 |
| chr8_+_59365291 | 5.98 |
ENSMUST00000160055.2
|
BC030500
|
cDNA sequence BC030500 |
| chr1_+_158190090 | 5.98 |
ENSMUST00000194369.6
ENSMUST00000195311.6 |
Astn1
|
astrotactin 1 |
| chr12_+_29890205 | 5.76 |
ENSMUST00000218198.2
|
Myt1l
|
myelin transcription factor 1-like |
| chr6_+_142359099 | 5.47 |
ENSMUST00000126521.9
ENSMUST00000211094.2 |
Spx
|
spexin hormone |
| chr14_+_14986011 | 5.35 |
ENSMUST00000164366.8
|
Gm3752
|
predicted gene 3752 |
| chr18_+_37453427 | 5.10 |
ENSMUST00000078271.4
|
Pcdhb5
|
protocadherin beta 5 |
| chr6_-_128558560 | 4.97 |
ENSMUST00000060574.9
|
A2ml1
|
alpha-2-macroglobulin like 1 |
| chr4_-_42874178 | 4.77 |
ENSMUST00000107978.2
|
Fam205c
|
family with sequence similarity 205, member C |
| chr18_+_37884653 | 4.67 |
ENSMUST00000194928.2
|
Pcdhgb7
|
protocadherin gamma subfamily B, 7 |
| chr9_+_72832904 | 4.65 |
ENSMUST00000038489.6
|
Pygo1
|
pygopus 1 |
| chr14_-_9015757 | 4.51 |
ENSMUST00000153954.8
|
Synpr
|
synaptoporin |
| chr19_-_58444336 | 4.48 |
ENSMUST00000131877.2
|
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr4_-_42874197 | 4.44 |
ENSMUST00000055944.11
|
Fam205c
|
family with sequence similarity 205, member C |
| chr18_-_12429048 | 4.38 |
ENSMUST00000234212.2
|
Ankrd29
|
ankyrin repeat domain 29 |
| chr10_-_5019044 | 4.35 |
ENSMUST00000095899.5
|
Syne1
|
spectrin repeat containing, nuclear envelope 1 |
| chr1_+_158189900 | 4.33 |
ENSMUST00000170718.7
|
Astn1
|
astrotactin 1 |
| chr8_-_25086976 | 4.33 |
ENSMUST00000033956.7
|
Ido1
|
indoleamine 2,3-dioxygenase 1 |
| chr3_+_75982890 | 4.08 |
ENSMUST00000160261.8
|
Fstl5
|
follistatin-like 5 |
| chr2_+_138120401 | 4.08 |
ENSMUST00000075410.5
|
Btbd3
|
BTB (POZ) domain containing 3 |
| chr3_+_33853941 | 4.03 |
ENSMUST00000099153.10
|
Ttc14
|
tetratricopeptide repeat domain 14 |
| chr13_+_83720457 | 3.87 |
ENSMUST00000196730.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chrX_+_94942639 | 3.72 |
ENSMUST00000082183.8
|
Zc3h12b
|
zinc finger CCCH-type containing 12B |
| chr10_+_115979787 | 3.72 |
ENSMUST00000105271.9
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr1_-_153719876 | 3.63 |
ENSMUST00000238926.2
ENSMUST00000124558.10 |
Rgsl1
|
regulator of G-protein signaling like 1 |
| chr11_-_40586029 | 3.60 |
ENSMUST00000101347.10
|
Mat2b
|
methionine adenosyltransferase II, beta |
| chr13_+_83720484 | 3.56 |
ENSMUST00000196207.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr9_+_88462944 | 3.29 |
ENSMUST00000164661.4
ENSMUST00000215498.2 ENSMUST00000216686.2 |
Trim43a
|
tripartite motif-containing 43A |
| chr4_-_62389098 | 3.23 |
ENSMUST00000135811.2
ENSMUST00000120095.8 ENSMUST00000030087.14 ENSMUST00000107452.8 ENSMUST00000155522.8 |
Wdr31
|
WD repeat domain 31 |
| chr18_+_67266784 | 3.16 |
ENSMUST00000236918.2
|
Gnal
|
guanine nucleotide binding protein, alpha stimulating, olfactory type |
| chr3_+_133942244 | 3.11 |
ENSMUST00000181904.3
|
Cxxc4
|
CXXC finger 4 |
| chr3_-_123483772 | 3.00 |
ENSMUST00000172537.3
|
Ndst3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr4_-_36136463 | 2.68 |
ENSMUST00000098151.9
|
Lingo2
|
leucine rich repeat and Ig domain containing 2 |
| chr2_-_102231208 | 2.68 |
ENSMUST00000102573.8
|
Trim44
|
tripartite motif-containing 44 |
| chr1_-_127782735 | 2.66 |
ENSMUST00000208183.3
|
Map3k19
|
mitogen-activated protein kinase kinase kinase 19 |
| chr5_+_31684331 | 2.62 |
ENSMUST00000114533.9
ENSMUST00000202214.4 ENSMUST00000201858.4 ENSMUST00000202950.4 |
Slc4a1ap
|
solute carrier family 4 (anion exchanger), member 1, adaptor protein |
| chr12_-_85143852 | 2.61 |
ENSMUST00000177289.9
|
Prox2
|
prospero homeobox 2 |
| chr3_+_106020545 | 2.55 |
ENSMUST00000079132.12
ENSMUST00000139086.2 |
Chia1
|
chitinase, acidic 1 |
| chr10_-_20600797 | 2.49 |
ENSMUST00000020165.14
|
Pde7b
|
phosphodiesterase 7B |
| chr19_-_58443593 | 2.44 |
ENSMUST00000135730.2
ENSMUST00000152507.8 |
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr7_+_75351713 | 2.35 |
ENSMUST00000207239.2
|
Akap13
|
A kinase (PRKA) anchor protein 13 |
| chr2_-_29677634 | 2.31 |
ENSMUST00000177467.8
ENSMUST00000113807.10 |
Trub2
|
TruB pseudouridine (psi) synthase family member 2 |
| chr13_-_21722197 | 2.30 |
ENSMUST00000168629.2
ENSMUST00000218154.2 |
Olfr1366
|
olfactory receptor 1366 |
| chr17_-_10501816 | 2.30 |
ENSMUST00000233684.2
|
Qk
|
quaking, KH domain containing RNA binding |
| chr12_+_98594388 | 2.25 |
ENSMUST00000048402.12
ENSMUST00000101144.10 ENSMUST00000101146.4 |
Spata7
|
spermatogenesis associated 7 |
| chr5_-_31684036 | 2.13 |
ENSMUST00000202421.2
ENSMUST00000201769.4 ENSMUST00000065388.11 |
Supt7l
|
SPT7-like, STAGA complex gamma subunit |
| chr13_+_96524802 | 2.10 |
ENSMUST00000099295.6
|
Poc5
|
POC5 centriolar protein |
| chr7_-_86044743 | 2.05 |
ENSMUST00000053958.6
|
Olfr303
|
olfactory receptor 303 |
| chr2_-_32314017 | 2.05 |
ENSMUST00000113307.9
|
Slc25a25
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 |
| chr16_+_17149235 | 1.90 |
ENSMUST00000023450.15
ENSMUST00000231884.2 |
Serpind1
|
serine (or cysteine) peptidase inhibitor, clade D, member 1 |
| chr9_+_38738911 | 1.86 |
ENSMUST00000051238.7
ENSMUST00000219798.2 |
Olfr923
|
olfactory receptor 923 |
| chr17_-_65847731 | 1.77 |
ENSMUST00000233117.2
ENSMUST00000062161.7 |
Tmem232
|
transmembrane protein 232 |
| chr8_-_76133212 | 1.77 |
ENSMUST00000212864.2
|
Gm10358
|
predicted gene 10358 |
| chr7_-_106423873 | 1.74 |
ENSMUST00000208895.3
|
Olfr702
|
olfactory receptor 702 |
| chr14_-_40730180 | 1.73 |
ENSMUST00000136661.8
|
Prxl2a
|
peroxiredoxin like 2A |
| chr19_-_32717138 | 1.72 |
ENSMUST00000236985.2
|
Atad1
|
ATPase family, AAA domain containing 1 |
| chr6_+_126916956 | 1.63 |
ENSMUST00000201617.2
|
D6Wsu163e
|
DNA segment, Chr 6, Wayne State University 163, expressed |
| chr11_+_120489358 | 1.61 |
ENSMUST00000093140.5
|
Anapc11
|
anaphase promoting complex subunit 11 |
| chr8_-_46577183 | 1.55 |
ENSMUST00000170416.8
|
Snx25
|
sorting nexin 25 |
| chr1_-_36982747 | 1.52 |
ENSMUST00000185964.3
|
Tmem131
|
transmembrane protein 131 |
| chr4_+_143139571 | 1.52 |
ENSMUST00000155157.2
|
Pramel12
|
PRAME like 12 |
| chr6_+_32027216 | 1.50 |
ENSMUST00000031778.6
|
1700012A03Rik
|
RIKEN cDNA 1700012A03 gene |
| chr7_-_103778992 | 1.49 |
ENSMUST00000053743.6
|
Ubqln5
|
ubiquilin 5 |
| chr12_+_3944798 | 1.48 |
ENSMUST00000172879.2
ENSMUST00000174774.8 |
Dnmt3a
|
DNA methyltransferase 3A |
| chr5_-_125467240 | 1.48 |
ENSMUST00000156249.2
|
Ubc
|
ubiquitin C |
| chr2_-_29938841 | 1.47 |
ENSMUST00000113711.3
|
Dync2i2
|
dynein 2 intermediate chain 2 |
| chr9_+_78020554 | 1.47 |
ENSMUST00000009972.6
ENSMUST00000117330.8 ENSMUST00000044551.8 |
Cilk1
|
ciliogenesis associated kinase 1 |
| chr1_+_179788037 | 1.46 |
ENSMUST00000097453.9
ENSMUST00000111117.8 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr15_+_94527117 | 1.46 |
ENSMUST00000080141.6
|
Tmem117
|
transmembrane protein 117 |
| chr17_+_26042600 | 1.45 |
ENSMUST00000026833.6
|
Wdr24
|
WD repeat domain 24 |
| chr4_+_98434930 | 1.42 |
ENSMUST00000102792.10
|
Patj
|
PATJ, crumbs cell polarity complex component |
| chr19_-_32717166 | 1.37 |
ENSMUST00000235142.2
ENSMUST00000070210.6 ENSMUST00000236011.2 |
Atad1
|
ATPase family, AAA domain containing 1 |
| chr11_-_40583493 | 1.36 |
ENSMUST00000040167.11
|
Mat2b
|
methionine adenosyltransferase II, beta |
| chr4_+_143139490 | 1.31 |
ENSMUST00000132915.2
ENSMUST00000037356.8 |
Pramel12
|
PRAME like 12 |
| chr1_+_75119419 | 1.28 |
ENSMUST00000097694.11
ENSMUST00000190240.7 |
Retreg2
|
reticulophagy regulator family member 2 |
| chr13_-_77279395 | 1.27 |
ENSMUST00000159300.8
|
Slf1
|
SMC5-SMC6 complex localization factor 1 |
| chr10_-_88440869 | 1.25 |
ENSMUST00000119185.8
ENSMUST00000238199.2 |
Mybpc1
|
myosin binding protein C, slow-type |
| chr12_-_85317359 | 1.25 |
ENSMUST00000166821.8
ENSMUST00000019378.8 ENSMUST00000220854.2 |
Mlh3
|
mutL homolog 3 |
| chr11_-_69706124 | 1.19 |
ENSMUST00000210714.2
|
Gm39566
|
predicted gene, 39566 |
| chr2_+_111158766 | 1.19 |
ENSMUST00000090326.2
|
Olfr1281
|
olfactory receptor 1281 |
| chr2_+_131052421 | 1.10 |
ENSMUST00000110206.2
|
Ap5s1
|
adaptor-related protein 5 complex, sigma 1 subunit |
| chr5_-_33011530 | 1.10 |
ENSMUST00000130134.3
ENSMUST00000120129.9 |
Prr14l
|
proline rich 14-like |
| chr10_+_94412116 | 1.09 |
ENSMUST00000117929.2
|
Tmcc3
|
transmembrane and coiled coil domains 3 |
| chr6_+_106746138 | 1.08 |
ENSMUST00000205163.2
|
Trnt1
|
tRNA nucleotidyl transferase, CCA-adding, 1 |
| chr7_-_103718362 | 1.07 |
ENSMUST00000077417.3
|
Olfr644
|
olfactory receptor 644 |
| chr19_-_58443830 | 1.03 |
ENSMUST00000026076.14
|
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr2_+_131052283 | 1.02 |
ENSMUST00000110210.8
ENSMUST00000089506.12 ENSMUST00000110208.8 |
Ap5s1
|
adaptor-related protein 5 complex, sigma 1 subunit |
| chr19_-_4171536 | 1.02 |
ENSMUST00000025767.14
|
Aip
|
aryl-hydrocarbon receptor-interacting protein |
| chr19_+_56450085 | 0.99 |
ENSMUST00000225909.2
|
Plekhs1
|
pleckstrin homology domain containing, family S member 1 |
| chr11_+_93934940 | 0.98 |
ENSMUST00000132079.8
|
Spag9
|
sperm associated antigen 9 |
| chr17_+_29899420 | 0.96 |
ENSMUST00000130052.9
|
Cmtr1
|
cap methyltransferase 1 |
| chr11_+_120489247 | 0.89 |
ENSMUST00000026128.10
|
Anapc11
|
anaphase promoting complex subunit 11 |
| chr16_-_78887971 | 0.89 |
ENSMUST00000023566.11
ENSMUST00000060402.6 |
Tmprss15
|
transmembrane protease, serine 15 |
| chr19_+_56450062 | 0.89 |
ENSMUST00000178590.9
ENSMUST00000039666.8 |
Plekhs1
|
pleckstrin homology domain containing, family S member 1 |
| chr19_-_8196196 | 0.87 |
ENSMUST00000113298.9
|
Slc22a29
|
solute carrier family 22. member 29 |
| chr11_+_74255497 | 0.86 |
ENSMUST00000077794.4
|
Olfr412
|
olfactory receptor 412 |
| chr18_-_39062201 | 0.84 |
ENSMUST00000134864.2
|
Fgf1
|
fibroblast growth factor 1 |
| chr4_-_12171971 | 0.84 |
ENSMUST00000087052.11
ENSMUST00000108285.9 ENSMUST00000177837.8 |
Cibar1
|
CBY1 interacting BAR domain containing 1 |
| chr11_+_54517164 | 0.84 |
ENSMUST00000239168.2
|
Rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr5_+_135661472 | 0.83 |
ENSMUST00000043707.7
|
Rhbdd2
|
rhomboid domain containing 2 |
| chr8_-_21946576 | 0.82 |
ENSMUST00000110752.4
|
Defa42
|
defensin, alpha, 42 |
| chr17_-_65847777 | 0.81 |
ENSMUST00000086722.10
|
Tmem232
|
transmembrane protein 232 |
| chr3_-_75389047 | 0.75 |
ENSMUST00000193989.4
ENSMUST00000203169.3 |
Wdr49
|
WD repeat domain 49 |
| chr8_-_37200051 | 0.75 |
ENSMUST00000098826.10
|
Dlc1
|
deleted in liver cancer 1 |
| chr5_-_125467063 | 0.74 |
ENSMUST00000136312.2
|
Ubc
|
ubiquitin C |
| chr8_-_21817031 | 0.72 |
ENSMUST00000098890.4
|
Defa29
|
defensin, alpha, 29 |
| chr9_+_19624125 | 0.68 |
ENSMUST00000077023.4
|
Olfr857
|
olfactory receptor 857 |
| chr16_+_92089268 | 0.67 |
ENSMUST00000047383.10
|
Kcne2
|
potassium voltage-gated channel, Isk-related subfamily, gene 2 |
| chr7_-_106491314 | 0.63 |
ENSMUST00000088687.3
|
Olfr707
|
olfactory receptor 707 |
| chr2_+_112114906 | 0.62 |
ENSMUST00000053666.8
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr5_+_67125759 | 0.61 |
ENSMUST00000238993.2
ENSMUST00000038188.14 |
Limch1
|
LIM and calponin homology domains 1 |
| chr5_+_138083345 | 0.60 |
ENSMUST00000019660.11
ENSMUST00000066617.12 ENSMUST00000110963.8 |
Zkscan1
|
zinc finger with KRAB and SCAN domains 1 |
| chr10_+_127256993 | 0.59 |
ENSMUST00000170336.8
|
R3hdm2
|
R3H domain containing 2 |
| chr2_+_3514071 | 0.58 |
ENSMUST00000036350.3
|
Cdnf
|
cerebral dopamine neurotrophic factor |
| chr13_+_119599287 | 0.50 |
ENSMUST00000026519.10
ENSMUST00000225186.2 ENSMUST00000224081.2 ENSMUST00000224312.2 |
4833420G17Rik
|
RIKEN cDNA 4833420G17 gene |
| chr2_+_158466802 | 0.49 |
ENSMUST00000045644.9
|
Actr5
|
ARP5 actin-related protein 5 |
| chr5_+_123366253 | 0.48 |
ENSMUST00000100729.9
|
Psmd9
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 9 |
| chr17_+_8591228 | 0.47 |
ENSMUST00000163578.8
|
T2
|
brachyury 2 |
| chr6_-_71800788 | 0.45 |
ENSMUST00000065103.4
|
Mrpl35
|
mitochondrial ribosomal protein L35 |
| chr2_-_86061745 | 0.44 |
ENSMUST00000216056.2
|
Olfr1047
|
olfactory receptor 1047 |
| chr10_+_111919250 | 0.43 |
ENSMUST00000148897.8
ENSMUST00000020434.4 |
Glipr1l2
|
GLI pathogenesis-related 1 like 2 |
| chr13_+_24118417 | 0.43 |
ENSMUST00000072391.2
|
H2ac1
|
H2A clustered histone 1 |
| chr5_-_86666408 | 0.42 |
ENSMUST00000140095.2
ENSMUST00000134179.8 |
Tmprss11g
|
transmembrane protease, serine 11g |
| chr13_-_23420117 | 0.42 |
ENSMUST00000235574.2
ENSMUST00000226672.3 |
Vmn1r222
|
vomeronasal 1 receptor 222 |
| chr7_+_5037117 | 0.41 |
ENSMUST00000076791.4
|
4632433K11Rik
|
RIKEN cDNA 4632433K11 gene |
| chr11_-_58434332 | 0.38 |
ENSMUST00000108823.3
|
Olfr329
|
olfactory receptor 329 |
| chr10_-_130116088 | 0.35 |
ENSMUST00000061571.5
|
Neurod4
|
neurogenic differentiation 4 |
| chrX_+_101847829 | 0.34 |
ENSMUST00000131451.2
|
Dmrtc1c1
|
DMRT-like family C1c1 |
| chr12_+_84147428 | 0.31 |
ENSMUST00000056822.4
|
Acot6
|
acyl-CoA thioesterase 6 |
| chr10_-_78588884 | 0.30 |
ENSMUST00000105383.4
|
Ccdc105
|
coiled-coil domain containing 105 |
| chrX_+_37689503 | 0.30 |
ENSMUST00000000365.3
|
Mcts1
|
malignant T cell amplified sequence 1 |
| chrX_-_6907858 | 0.29 |
ENSMUST00000115752.8
|
Ccnb3
|
cyclin B3 |
| chr4_+_98435155 | 0.29 |
ENSMUST00000030290.8
|
Patj
|
PATJ, crumbs cell polarity complex component |
| chr6_-_142332700 | 0.29 |
ENSMUST00000129694.8
ENSMUST00000203772.3 |
Recql
|
RecQ protein-like |
| chr17_+_48037758 | 0.29 |
ENSMUST00000024782.12
ENSMUST00000144955.2 |
Pgc
|
progastricsin (pepsinogen C) |
| chr17_+_6130205 | 0.28 |
ENSMUST00000100955.3
|
Gtf2h5
|
general transcription factor IIH, polypeptide 5 |
| chr10_-_129951252 | 0.27 |
ENSMUST00000213145.2
|
Olfr823
|
olfactory receptor 823 |
| chr12_-_40087393 | 0.27 |
ENSMUST00000146905.2
|
Arl4a
|
ADP-ribosylation factor-like 4A |
| chr4_-_117749191 | 0.23 |
ENSMUST00000030265.4
|
Dph2
|
DPH2 homolog |
| chr11_-_86698484 | 0.22 |
ENSMUST00000018569.14
|
Dhx40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chrX_+_141011695 | 0.22 |
ENSMUST00000112913.2
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr5_+_135398796 | 0.22 |
ENSMUST00000000940.15
|
Nsun5
|
NOL1/NOP2/Sun domain family, member 5 |
| chr11_+_93935021 | 0.19 |
ENSMUST00000075695.13
ENSMUST00000092777.11 |
Spag9
|
sperm associated antigen 9 |
| chr11_+_49195568 | 0.18 |
ENSMUST00000217290.2
|
Olfr10
|
olfactory receptor 10 |
| chr2_-_111062182 | 0.17 |
ENSMUST00000099620.5
|
Olfr1275
|
olfactory receptor 1275 |
| chr10_-_129010923 | 0.14 |
ENSMUST00000169800.4
|
Olfr772
|
olfactory receptor 772 |
| chr16_-_56533179 | 0.14 |
ENSMUST00000136394.8
|
Tfg
|
Trk-fused gene |
| chr17_+_37670473 | 0.11 |
ENSMUST00000178766.3
ENSMUST00000215398.2 |
Olfr104-ps
|
olfactory receptor 104, pseudogene |
| chr11_+_58668915 | 0.11 |
ENSMUST00000081533.5
|
Olfr315
|
olfactory receptor 315 |
| chr4_-_143026033 | 0.09 |
ENSMUST00000119654.2
|
Pdpn
|
podoplanin |
| chr12_-_98540944 | 0.09 |
ENSMUST00000221305.2
|
Kcnk10
|
potassium channel, subfamily K, member 10 |
| chr5_+_69714279 | 0.07 |
ENSMUST00000087228.11
ENSMUST00000031113.13 ENSMUST00000173205.3 |
Guf1
|
GUF1 homolog, GTPase |
| chr7_-_99512558 | 0.07 |
ENSMUST00000207137.2
ENSMUST00000207063.2 ENSMUST00000207580.2 |
Spcs2
|
signal peptidase complex subunit 2 homolog (S. cerevisiae) |
| chr3_-_95264467 | 0.06 |
ENSMUST00000107171.10
ENSMUST00000015841.12 ENSMUST00000107170.3 |
Setdb1
|
SET domain, bifurcated 1 |
| chr11_-_99433984 | 0.05 |
ENSMUST00000107443.8
ENSMUST00000074253.4 |
Krt40
|
keratin 40 |
| chr2_+_135501605 | 0.04 |
ENSMUST00000134310.8
|
Plcb4
|
phospholipase C, beta 4 |
| chr2_-_88058678 | 0.04 |
ENSMUST00000214386.2
|
Olfr1170
|
olfactory receptor 1170 |
| chrX_+_111404963 | 0.04 |
ENSMUST00000026602.9
ENSMUST00000113412.3 |
2010106E10Rik
|
RIKEN cDNA 2010106E10 gene |
| chr4_+_42969934 | 0.03 |
ENSMUST00000190902.2
ENSMUST00000030163.12 |
1700022I11Rik
|
RIKEN cDNA 1700022I11 gene |
| chrY_-_79161056 | 0.03 |
ENSMUST00000179922.2
|
Gm20917
|
predicted gene, 20917 |
| chr6_-_142332757 | 0.03 |
ENSMUST00000032370.13
ENSMUST00000100832.10 ENSMUST00000128082.2 ENSMUST00000111803.9 |
Recql
|
RecQ protein-like |
| chr2_+_57887896 | 0.02 |
ENSMUST00000112616.8
ENSMUST00000166729.2 |
Galnt5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr6_+_142332941 | 0.02 |
ENSMUST00000032372.7
|
Golt1b
|
golgi transport 1B |
| chr11_+_74150634 | 0.01 |
ENSMUST00000217016.2
ENSMUST00000214303.2 |
Olfr406
|
olfactory receptor 406 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.9 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 2.1 | 10.4 | GO:0032901 | positive regulation of neurotrophin production(GO:0032901) |
| 1.4 | 4.3 | GO:0036269 | swimming behavior(GO:0036269) |
| 1.3 | 8.9 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 1.1 | 6.3 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 1.0 | 14.9 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 1.0 | 17.8 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.9 | 7.7 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.8 | 5.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.8 | 15.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.8 | 10.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.8 | 8.3 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.7 | 5.5 | GO:1904306 | positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 0.7 | 12.3 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.6 | 4.3 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.6 | 3.7 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.6 | 7.4 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.6 | 9.7 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.4 | 6.3 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.4 | 3.2 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.4 | 2.6 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.4 | 1.1 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.3 | 3.0 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.3 | 1.3 | GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.3 | 2.3 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.3 | 10.5 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.3 | 1.0 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.2 | 2.5 | GO:1902101 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.2 | 24.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.2 | 1.9 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.2 | 1.0 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.2 | 0.7 | GO:1902159 | regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 0.2 | 0.5 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.2 | 0.6 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.1 | 4.7 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.1 | 2.2 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 | 1.2 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.1 | 6.4 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 2.1 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.1 | 2.3 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 | 18.8 | GO:0006333 | chromatin assembly or disassembly(GO:0006333) |
| 0.1 | 1.5 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.1 | 0.8 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.1 | 5.8 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.1 | 2.3 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.1 | 0.5 | GO:0014028 | notochord formation(GO:0014028) |
| 0.1 | 9.8 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 7.5 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 1.6 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 7.1 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.1 | 1.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 8.6 | GO:0009142 | nucleoside triphosphate biosynthetic process(GO:0009142) |
| 0.1 | 0.5 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 0.3 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.1 | 0.3 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 | 2.1 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 5.1 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.3 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.2 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.7 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.1 | GO:1904328 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.0 | 0.5 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 11.3 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 1.6 | GO:0043304 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.0 | 8.0 | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation(GO:0050731) |
| 0.0 | 2.8 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.3 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.8 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 6.9 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.4 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 4.1 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 0.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.9 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 3.1 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 0.1 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.9 | GO:0000060 | protein import into nucleus, translocation(GO:0000060) |
| 0.0 | 1.5 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 2.7 | GO:0032147 | activation of protein kinase activity(GO:0032147) |
| 0.0 | 15.7 | GO:0007267 | cell-cell signaling(GO:0007267) |
| 0.0 | 1.3 | GO:0000725 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.0 | 2.9 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.0 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 1.1 | 9.2 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 1.0 | 20.4 | GO:0031045 | dense core granule(GO:0031045) |
| 1.0 | 9.6 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.8 | 17.4 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.7 | 18.8 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.5 | 15.3 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.5 | 4.3 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.4 | 1.2 | GO:0005712 | chiasma(GO:0005712) |
| 0.3 | 6.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.2 | 1.0 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.2 | 6.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.2 | 2.4 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.2 | 10.2 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.2 | 4.5 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 7.1 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 1.5 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 2.1 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 2.1 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.1 | 10.4 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 2.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 1.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 1.2 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 19.7 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 8.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 2.2 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 4.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 3.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.5 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.5 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 19.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.5 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 8.6 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 3.0 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 2.2 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 11.5 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 1.7 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 1.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 2.2 | GO:0043209 | myelin sheath(GO:0043209) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 19.7 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 1.9 | 7.7 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 1.8 | 8.9 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 1.4 | 4.3 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.8 | 10.4 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.6 | 9.0 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.5 | 15.3 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.4 | 3.0 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.4 | 10.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.4 | 1.8 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.3 | 1.0 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.3 | 9.8 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.3 | 5.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) cobalt ion binding(GO:0050897) |
| 0.3 | 13.5 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.3 | 1.2 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.3 | 6.2 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.3 | 6.8 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.3 | 7.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.3 | 6.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 2.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 1.9 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.2 | 2.1 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.2 | 10.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.2 | 1.0 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.2 | 5.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.2 | 0.6 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.1 | 4.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 2.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 2.5 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 1.5 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 8.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 1.6 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.1 | 0.7 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.1 | 3.2 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 2.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 1.2 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 7.5 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.8 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.3 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.5 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 7.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.8 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 1.1 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 2.1 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.7 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.3 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.9 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 3.7 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 17.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 2.2 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 3.1 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 2.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 4.9 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 2.2 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 5.3 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 3.6 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 2.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 9.0 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.2 | 10.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 7.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 6.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 8.0 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 3.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 3.1 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 2.1 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 4.7 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 2.3 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 1.5 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 1.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.8 | PID FGF PATHWAY | FGF signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 15.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.5 | 9.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.5 | 3.2 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.4 | 8.9 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.4 | 10.5 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.3 | 4.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.3 | 6.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.3 | 9.0 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 7.4 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 5.0 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 9.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 2.5 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.1 | 8.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 6.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 6.3 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 3.0 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 2.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.8 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 1.2 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 5.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 2.3 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 2.5 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |