Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
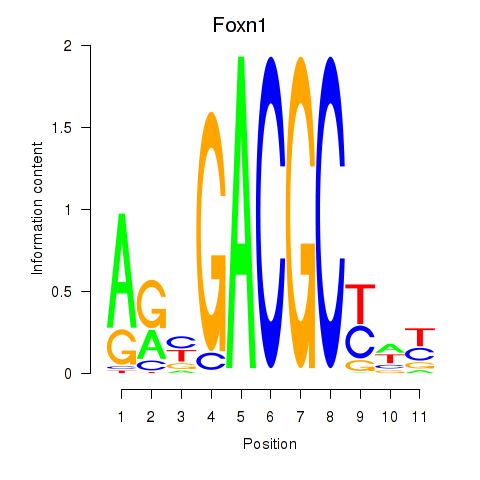
Results for Foxn1
Z-value: 0.48
Transcription factors associated with Foxn1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxn1
|
ENSMUSG00000002057.5 | Foxn1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxn1 | mm39_v1_chr11_-_78277384_78277433 | -0.02 | 8.8e-01 | Click! |
Activity profile of Foxn1 motif
Sorted Z-values of Foxn1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxn1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_32522548 | 2.69 |
ENSMUST00000041859.9
|
Gmds
|
GDP-mannose 4, 6-dehydratase |
| chr13_-_55979191 | 2.28 |
ENSMUST00000021968.7
|
Pitx1
|
paired-like homeodomain transcription factor 1 |
| chr7_-_24997393 | 2.12 |
ENSMUST00000005583.12
|
Pafah1b3
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 3 |
| chr7_-_24997291 | 1.95 |
ENSMUST00000148150.8
ENSMUST00000155118.2 |
Pafah1b3
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 3 |
| chr9_-_35469818 | 1.59 |
ENSMUST00000034612.7
|
Ddx25
|
DEAD box helicase 25 |
| chr17_-_25652750 | 1.47 |
ENSMUST00000159610.8
ENSMUST00000159048.8 ENSMUST00000078496.12 |
Cacna1h
|
calcium channel, voltage-dependent, T type, alpha 1H subunit |
| chr11_-_120239301 | 1.41 |
ENSMUST00000062147.14
ENSMUST00000128055.2 |
Actg1
|
actin, gamma, cytoplasmic 1 |
| chr2_+_109848151 | 1.41 |
ENSMUST00000028580.12
|
Ccdc34
|
coiled-coil domain containing 34 |
| chr7_+_133378586 | 1.37 |
ENSMUST00000065359.12
ENSMUST00000209511.2 ENSMUST00000151031.2 ENSMUST00000121560.2 |
Fank1
|
fibronectin type 3 and ankyrin repeat domains 1 |
| chr19_+_43600738 | 1.36 |
ENSMUST00000057178.11
|
Nkx2-3
|
NK2 homeobox 3 |
| chr19_+_38384428 | 1.16 |
ENSMUST00000054098.4
|
Slc35g1
|
solute carrier family 35, member G1 |
| chr8_-_85705338 | 1.12 |
ENSMUST00000064922.7
|
Junb
|
jun B proto-oncogene |
| chr12_+_86129329 | 1.10 |
ENSMUST00000054565.8
ENSMUST00000222821.2 ENSMUST00000222905.2 |
Ift43
|
intraflagellar transport 43 |
| chrX_-_110372843 | 1.06 |
ENSMUST00000096348.10
ENSMUST00000113428.9 |
Rps6ka6
|
ribosomal protein S6 kinase polypeptide 6 |
| chr8_+_27513819 | 1.03 |
ENSMUST00000033873.9
ENSMUST00000211043.2 |
Erlin2
|
ER lipid raft associated 2 |
| chr5_+_98477157 | 1.01 |
ENSMUST00000080333.8
|
Cfap299
|
cilia and flagella associated protein 299 |
| chr2_+_109848224 | 1.00 |
ENSMUST00000150183.9
|
Ccdc34
|
coiled-coil domain containing 34 |
| chr9_-_75591274 | 1.00 |
ENSMUST00000214244.2
ENSMUST00000213324.2 ENSMUST00000034699.8 |
Scg3
|
secretogranin III |
| chrX_+_72716756 | 0.99 |
ENSMUST00000033752.14
ENSMUST00000114467.9 |
Slc6a8
|
solute carrier family 6 (neurotransmitter transporter, creatine), member 8 |
| chr7_+_28580847 | 0.96 |
ENSMUST00000066880.6
|
Capn12
|
calpain 12 |
| chr14_-_30973164 | 0.94 |
ENSMUST00000226565.2
ENSMUST00000022459.5 |
Phf7
|
PHD finger protein 7 |
| chr1_-_154975376 | 0.88 |
ENSMUST00000055322.6
|
Ier5
|
immediate early response 5 |
| chr8_-_84738761 | 0.86 |
ENSMUST00000191523.2
ENSMUST00000190457.2 ENSMUST00000185457.2 |
Misp3
|
MISP family member 3 |
| chr19_+_46140942 | 0.80 |
ENSMUST00000026254.14
|
Gbf1
|
golgi-specific brefeldin A-resistance factor 1 |
| chr8_-_112603292 | 0.77 |
ENSMUST00000034431.3
|
Tmem170
|
transmembrane protein 170 |
| chr8_+_27513839 | 0.75 |
ENSMUST00000209563.2
ENSMUST00000209520.2 |
Erlin2
|
ER lipid raft associated 2 |
| chr7_-_109380745 | 0.74 |
ENSMUST00000207400.2
ENSMUST00000033331.7 |
Nrip3
|
nuclear receptor interacting protein 3 |
| chrX_-_110372800 | 0.73 |
ENSMUST00000137712.9
|
Rps6ka6
|
ribosomal protein S6 kinase polypeptide 6 |
| chr2_+_18069375 | 0.72 |
ENSMUST00000114671.8
ENSMUST00000114680.9 |
Mllt10
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 10 |
| chr8_+_27513289 | 0.70 |
ENSMUST00000209795.2
ENSMUST00000209976.2 |
Erlin2
|
ER lipid raft associated 2 |
| chr6_-_85490568 | 0.68 |
ENSMUST00000095759.5
|
Egr4
|
early growth response 4 |
| chr7_+_123582021 | 0.66 |
ENSMUST00000106437.2
|
Hs3st4
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 4 |
| chr1_-_161704224 | 0.66 |
ENSMUST00000048377.11
|
Suco
|
SUN domain containing ossification factor |
| chr19_-_14575395 | 0.64 |
ENSMUST00000052011.15
ENSMUST00000167776.3 |
Tle4
|
transducin-like enhancer of split 4 |
| chr11_-_103254257 | 0.60 |
ENSMUST00000092557.6
|
Arhgap27
|
Rho GTPase activating protein 27 |
| chr2_-_57014015 | 0.59 |
ENSMUST00000112629.8
|
Nr4a2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr7_-_126944754 | 0.58 |
ENSMUST00000205266.2
|
Zfp768
|
zinc finger protein 768 |
| chrX_+_98179725 | 0.57 |
ENSMUST00000052839.7
|
Efnb1
|
ephrin B1 |
| chrX_-_110372733 | 0.57 |
ENSMUST00000065976.12
|
Rps6ka6
|
ribosomal protein S6 kinase polypeptide 6 |
| chr10_-_127098932 | 0.56 |
ENSMUST00000217895.2
|
Kif5a
|
kinesin family member 5A |
| chr16_-_90524214 | 0.56 |
ENSMUST00000099554.5
|
Mis18a
|
MIS18 kinetochore protein A |
| chr11_-_100628979 | 0.52 |
ENSMUST00000155500.2
ENSMUST00000107364.8 ENSMUST00000019317.12 |
Rab5c
|
RAB5C, member RAS oncogene family |
| chr5_-_115439016 | 0.51 |
ENSMUST00000009157.4
|
Dynll1
|
dynein light chain LC8-type 1 |
| chr5_-_46013838 | 0.51 |
ENSMUST00000087164.10
ENSMUST00000121573.8 |
Lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr7_-_126944578 | 0.50 |
ENSMUST00000060783.7
|
Zfp768
|
zinc finger protein 768 |
| chr11_+_50101717 | 0.49 |
ENSMUST00000147468.8
|
Mgat4b
|
mannoside acetylglucosaminyltransferase 4, isoenzyme B |
| chr5_-_114828967 | 0.48 |
ENSMUST00000112212.2
ENSMUST00000112214.8 |
Gltp
|
glycolipid transfer protein |
| chr17_-_30795403 | 0.47 |
ENSMUST00000237037.2
ENSMUST00000168787.8 |
Btbd9
|
BTB (POZ) domain containing 9 |
| chr17_-_37262495 | 0.46 |
ENSMUST00000040402.14
ENSMUST00000174711.8 |
Ppp1r11
|
protein phosphatase 1, regulatory inhibitor subunit 11 |
| chr9_+_70114623 | 0.46 |
ENSMUST00000034745.9
|
Myo1e
|
myosin IE |
| chr5_-_115438971 | 0.45 |
ENSMUST00000112090.2
|
Dynll1
|
dynein light chain LC8-type 1 |
| chr18_+_60659257 | 0.45 |
ENSMUST00000223984.2
ENSMUST00000025505.7 ENSMUST00000223590.2 |
Dctn4
|
dynactin 4 |
| chr18_+_33072194 | 0.43 |
ENSMUST00000042868.6
|
Camk4
|
calcium/calmodulin-dependent protein kinase IV |
| chr9_+_45818250 | 0.42 |
ENSMUST00000216672.2
|
Pcsk7
|
proprotein convertase subtilisin/kexin type 7 |
| chr17_+_80681087 | 0.42 |
ENSMUST00000225658.3
ENSMUST00000224966.3 ENSMUST00000225223.3 ENSMUST00000225548.3 |
Arhgef33
|
Rho guanine nucleotide exchange factor (GEF) 33 |
| chr13_-_77283534 | 0.41 |
ENSMUST00000159462.3
ENSMUST00000151524.9 |
Slf1
|
SMC5-SMC6 complex localization factor 1 |
| chr6_+_149310471 | 0.40 |
ENSMUST00000086829.11
ENSMUST00000111513.9 |
Bicd1
|
BICD cargo adaptor 1 |
| chr19_-_58932026 | 0.39 |
ENSMUST00000237297.2
|
Hspa12a
|
heat shock protein 12A |
| chr3_+_5815863 | 0.39 |
ENSMUST00000192045.2
|
Gm8797
|
predicted pseudogene 8797 |
| chr3_-_115800989 | 0.38 |
ENSMUST00000067485.4
|
Slc30a7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr19_+_45352173 | 0.38 |
ENSMUST00000223764.2
ENSMUST00000065601.13 ENSMUST00000224102.2 ENSMUST00000111936.4 |
Btrc
|
beta-transducin repeat containing protein |
| chr17_-_30795136 | 0.38 |
ENSMUST00000079924.8
ENSMUST00000236584.2 ENSMUST00000236825.2 ENSMUST00000235587.2 |
Btbd9
|
BTB (POZ) domain containing 9 |
| chr8_-_11058458 | 0.38 |
ENSMUST00000040514.8
|
Irs2
|
insulin receptor substrate 2 |
| chr9_-_45896075 | 0.37 |
ENSMUST00000217636.2
|
Pafah1b2
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 |
| chr6_-_30873669 | 0.36 |
ENSMUST00000048774.13
ENSMUST00000166192.7 |
Copg2
|
coatomer protein complex, subunit gamma 2 |
| chr2_+_125701054 | 0.35 |
ENSMUST00000028636.13
ENSMUST00000125084.8 |
Galk2
|
galactokinase 2 |
| chr19_+_36325683 | 0.35 |
ENSMUST00000225920.2
|
Pcgf5
|
polycomb group ring finger 5 |
| chr13_-_98453475 | 0.35 |
ENSMUST00000022163.15
ENSMUST00000152704.8 |
Btf3
|
basic transcription factor 3 |
| chr19_+_58931847 | 0.34 |
ENSMUST00000054280.10
ENSMUST00000200910.4 |
Eno4
|
enolase 4 |
| chr17_-_29456750 | 0.30 |
ENSMUST00000137727.3
|
Cpne5
|
copine V |
| chr9_+_45817795 | 0.30 |
ENSMUST00000039059.8
|
Pcsk7
|
proprotein convertase subtilisin/kexin type 7 |
| chr3_+_14643669 | 0.30 |
ENSMUST00000029069.13
ENSMUST00000165922.3 |
E2f5
|
E2F transcription factor 5 |
| chr9_-_72892617 | 0.30 |
ENSMUST00000124565.3
|
Ccpg1os
|
cell cycle progression 1, opposite strand |
| chr17_-_29457064 | 0.30 |
ENSMUST00000024805.15
|
Cpne5
|
copine V |
| chr19_+_53933271 | 0.29 |
ENSMUST00000025932.9
|
Shoc2
|
Shoc2, leucine rich repeat scaffold protein |
| chr14_+_63843943 | 0.29 |
ENSMUST00000119973.3
|
Xkr6
|
X-linked Kx blood group related 6 |
| chr9_-_45896663 | 0.29 |
ENSMUST00000214179.2
|
Pafah1b2
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 |
| chr2_-_74489763 | 0.29 |
ENSMUST00000173623.2
ENSMUST00000001867.13 |
Evx2
|
even-skipped homeobox 2 |
| chr11_-_69649452 | 0.28 |
ENSMUST00000058470.16
|
Polr2a
|
polymerase (RNA) II (DNA directed) polypeptide A |
| chrX_-_110373151 | 0.27 |
ENSMUST00000123213.8
|
Rps6ka6
|
ribosomal protein S6 kinase polypeptide 6 |
| chr19_-_53932867 | 0.27 |
ENSMUST00000235688.2
ENSMUST00000235348.2 |
Bbip1
|
BBSome interacting protein 1 |
| chr9_-_45896110 | 0.27 |
ENSMUST00000215060.2
ENSMUST00000213853.2 ENSMUST00000216334.2 |
Pafah1b2
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 |
| chr11_-_69649004 | 0.26 |
ENSMUST00000071213.4
|
Polr2a
|
polymerase (RNA) II (DNA directed) polypeptide A |
| chr4_+_116414855 | 0.25 |
ENSMUST00000030460.15
|
Gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr13_+_77283632 | 0.24 |
ENSMUST00000168779.3
ENSMUST00000225605.2 |
2210408I21Rik
|
RIKEN cDNA 2210408I21 gene |
| chr5_-_34093678 | 0.24 |
ENSMUST00000030993.8
|
Nelfa
|
negative elongation factor complex member A, Whsc2 |
| chr7_+_126446588 | 0.22 |
ENSMUST00000141805.8
ENSMUST00000064110.14 ENSMUST00000205938.2 ENSMUST00000152051.8 |
Doc2a
|
double C2, alpha |
| chr17_-_65920481 | 0.22 |
ENSMUST00000024897.10
|
Vapa
|
vesicle-associated membrane protein, associated protein A |
| chr9_-_107167046 | 0.22 |
ENSMUST00000035194.8
|
Mapkapk3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr3_-_158267771 | 0.22 |
ENSMUST00000199890.5
ENSMUST00000238317.3 ENSMUST00000200137.5 ENSMUST00000106044.6 |
Lrrc7
|
leucine rich repeat containing 7 |
| chr7_-_133378468 | 0.21 |
ENSMUST00000033290.12
|
Dhx32
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32 |
| chr4_+_116415251 | 0.21 |
ENSMUST00000106475.2
|
Gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr2_-_163486998 | 0.20 |
ENSMUST00000017851.4
|
Serinc3
|
serine incorporator 3 |
| chr19_-_53933052 | 0.20 |
ENSMUST00000135402.4
|
Bbip1
|
BBSome interacting protein 1 |
| chr13_+_81931196 | 0.20 |
ENSMUST00000022009.10
ENSMUST00000223793.2 |
Cetn3
|
centrin 3 |
| chr14_+_111912529 | 0.19 |
ENSMUST00000042767.9
|
Slitrk5
|
SLIT and NTRK-like family, member 5 |
| chr10_+_88215079 | 0.19 |
ENSMUST00000130301.8
ENSMUST00000020251.10 |
Gnptab
|
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chr13_-_64422775 | 0.18 |
ENSMUST00000221634.2
ENSMUST00000039318.16 |
Cdc14b
|
CDC14 cell division cycle 14B |
| chr13_-_64422693 | 0.17 |
ENSMUST00000109770.2
|
Cdc14b
|
CDC14 cell division cycle 14B |
| chr5_+_110692162 | 0.16 |
ENSMUST00000040001.14
|
Galnt9
|
polypeptide N-acetylgalactosaminyltransferase 9 |
| chr8_-_26087552 | 0.16 |
ENSMUST00000210234.2
ENSMUST00000211422.2 |
Letm2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr11_-_100288566 | 0.16 |
ENSMUST00000001592.15
ENSMUST00000107403.2 |
Jup
|
junction plakoglobin |
| chr13_+_81931642 | 0.16 |
ENSMUST00000224574.2
|
Cetn3
|
centrin 3 |
| chr12_+_105750952 | 0.15 |
ENSMUST00000109901.9
ENSMUST00000168186.8 ENSMUST00000163473.8 ENSMUST00000170540.8 ENSMUST00000166735.8 ENSMUST00000170002.8 |
Papola
|
poly (A) polymerase alpha |
| chr15_-_27681584 | 0.15 |
ENSMUST00000226145.2
ENSMUST00000226170.2 |
Otulinl
|
OTU deubiquitinase with linear linkage specificity like |
| chr8_-_33374825 | 0.15 |
ENSMUST00000238791.2
|
Nrg1
|
neuregulin 1 |
| chr7_-_133378410 | 0.13 |
ENSMUST00000130182.2
ENSMUST00000106139.8 |
Dhx32
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32 |
| chr11_+_104441489 | 0.13 |
ENSMUST00000018800.9
|
Myl4
|
myosin, light polypeptide 4 |
| chr11_-_104441218 | 0.13 |
ENSMUST00000106962.9
ENSMUST00000106961.2 ENSMUST00000093923.9 |
Cdc27
|
cell division cycle 27 |
| chr2_+_131052283 | 0.13 |
ENSMUST00000110210.8
ENSMUST00000089506.12 ENSMUST00000110208.8 |
Ap5s1
|
adaptor-related protein 5 complex, sigma 1 subunit |
| chr11_+_67345895 | 0.12 |
ENSMUST00000108681.9
|
Gas7
|
growth arrest specific 7 |
| chr2_-_27365633 | 0.12 |
ENSMUST00000138693.8
ENSMUST00000113941.9 ENSMUST00000077737.13 |
Brd3
|
bromodomain containing 3 |
| chr9_-_110483210 | 0.10 |
ENSMUST00000196488.5
ENSMUST00000133191.8 ENSMUST00000167320.8 |
Nbeal2
|
neurobeachin-like 2 |
| chr10_+_88214989 | 0.09 |
ENSMUST00000127615.8
|
Gnptab
|
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chr9_-_59393893 | 0.09 |
ENSMUST00000171975.8
|
Arih1
|
ariadne RBR E3 ubiquitin protein ligase 1 |
| chr17_-_80680970 | 0.08 |
ENSMUST00000086549.2
|
Gm10190
|
predicted gene 10190 |
| chr8_-_26087475 | 0.08 |
ENSMUST00000210810.2
ENSMUST00000210616.2 ENSMUST00000079160.8 |
Letm2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr15_-_27681643 | 0.08 |
ENSMUST00000100739.5
|
Otulinl
|
OTU deubiquitinase with linear linkage specificity like |
| chr14_-_55919256 | 0.05 |
ENSMUST00000227873.2
ENSMUST00000007733.8 |
Tinf2
|
Terf1 (TRF1)-interacting nuclear factor 2 |
| chr13_+_55612050 | 0.04 |
ENSMUST00000046533.9
|
Prr7
|
proline rich 7 (synaptic) |
| chr1_-_65090278 | 0.03 |
ENSMUST00000161960.2
ENSMUST00000087359.6 |
Cryge
|
crystallin, gamma E |
| chr2_+_131052421 | 0.02 |
ENSMUST00000110206.2
|
Ap5s1
|
adaptor-related protein 5 complex, sigma 1 subunit |
| chr14_+_75521783 | 0.01 |
ENSMUST00000022577.6
ENSMUST00000227049.2 |
Zc3h13
|
zinc finger CCCH type containing 13 |
| chr11_-_70130620 | 0.00 |
ENSMUST00000040428.4
|
Rnasek
|
ribonuclease, RNase K |
| chr8_-_84425724 | 0.00 |
ENSMUST00000005616.16
|
Pkn1
|
protein kinase N1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.3 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.3 | 1.0 | GO:0015881 | creatine transport(GO:0015881) |
| 0.3 | 2.7 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.3 | 0.8 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.2 | 2.5 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.2 | 1.4 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.2 | 1.2 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.1 | 0.6 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.1 | 1.5 | GO:0034651 | cortisol biosynthetic process(GO:0034651) |
| 0.1 | 2.6 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.1 | 2.1 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 0.4 | GO:1900275 | negative regulation of phospholipase C activity(GO:1900275) |
| 0.1 | 0.6 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.1 | 0.6 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 0.3 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.1 | 0.9 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.1 | 1.1 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.1 | 0.4 | GO:1990166 | positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) protein localization to site of double-strand break(GO:1990166) |
| 0.1 | 0.5 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.1 | 0.9 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.1 | 0.5 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.0 | 0.8 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.2 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.4 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.4 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 0.4 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 | 1.6 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.5 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.2 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.4 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.2 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.2 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.7 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 | 0.6 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.0 | 0.5 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.4 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 1.4 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.1 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.1 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.6 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.4 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 5.0 | GO:0016042 | lipid catabolic process(GO:0016042) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 1.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 1.6 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 0.5 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 1.6 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 0.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 0.8 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.4 | GO:0033647 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 0.0 | 1.0 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.5 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 1.4 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 1.4 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 1.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.6 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.4 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 4.0 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.2 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 1.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.4 | GO:0035869 | ciliary transition zone(GO:0035869) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 5.0 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.4 | 1.5 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.3 | 1.0 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.1 | 0.4 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.1 | 0.4 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.1 | 0.2 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 0.3 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 0.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 1.0 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.5 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.5 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.7 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 2.5 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 1.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 2.3 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 1.9 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.7 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.8 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 2.3 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.5 | GO:0051861 | glycolipid binding(GO:0051861) |
| 0.0 | 0.2 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.6 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.0 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.8 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 2.4 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.6 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 2.6 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 1.4 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.5 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.6 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.7 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 1.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.4 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.4 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |