Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
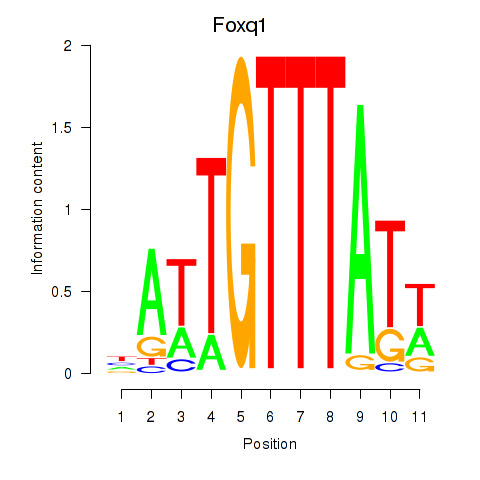
Results for Foxq1
Z-value: 1.00
Transcription factors associated with Foxq1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxq1
|
ENSMUSG00000038415.11 | Foxq1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxq1 | mm39_v1_chr13_+_31740117_31740117 | 0.09 | 4.5e-01 | Click! |
Activity profile of Foxq1 motif
Sorted Z-values of Foxq1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxq1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_138464065 | 8.17 |
ENSMUST00000113027.8
|
Rnf128
|
ring finger protein 128 |
| chr3_+_129326004 | 6.91 |
ENSMUST00000199910.5
ENSMUST00000197070.5 ENSMUST00000071402.7 |
Elovl6
|
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
| chr4_-_129015682 | 6.71 |
ENSMUST00000139450.8
ENSMUST00000125931.9 ENSMUST00000116444.10 |
Hpca
|
hippocalcin |
| chr7_+_45433306 | 5.13 |
ENSMUST00000072580.12
|
Lmtk3
|
lemur tyrosine kinase 3 |
| chr3_+_7431717 | 4.89 |
ENSMUST00000192468.2
ENSMUST00000028999.12 |
Pkia
|
protein kinase inhibitor, alpha |
| chr11_+_16207705 | 4.85 |
ENSMUST00000109645.9
ENSMUST00000109647.3 |
Vstm2a
|
V-set and transmembrane domain containing 2A |
| chr2_+_76236870 | 4.50 |
ENSMUST00000077972.11
ENSMUST00000111929.8 ENSMUST00000111930.9 |
Osbpl6
|
oxysterol binding protein-like 6 |
| chr1_+_17215581 | 4.31 |
ENSMUST00000026879.8
|
Gdap1
|
ganglioside-induced differentiation-associated-protein 1 |
| chr6_-_83513222 | 4.13 |
ENSMUST00000075161.12
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr3_+_68491487 | 3.80 |
ENSMUST00000182997.3
|
Schip1
|
schwannomin interacting protein 1 |
| chr16_-_88360037 | 3.79 |
ENSMUST00000049697.5
|
Cldn8
|
claudin 8 |
| chr6_-_83513184 | 3.74 |
ENSMUST00000205926.2
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr15_-_12549350 | 3.72 |
ENSMUST00000190929.2
|
Pdzd2
|
PDZ domain containing 2 |
| chr7_+_35148461 | 3.70 |
ENSMUST00000118969.8
|
Slc7a9
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 9 |
| chr17_+_31652073 | 3.70 |
ENSMUST00000237363.2
|
Pde9a
|
phosphodiesterase 9A |
| chr3_+_129326285 | 3.59 |
ENSMUST00000197235.5
|
Elovl6
|
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
| chr17_+_31652029 | 3.53 |
ENSMUST00000136384.9
|
Pde9a
|
phosphodiesterase 9A |
| chr18_+_37630044 | 3.52 |
ENSMUST00000059571.7
|
Pcdhb19
|
protocadherin beta 19 |
| chr7_+_35148579 | 3.37 |
ENSMUST00000032703.10
|
Slc7a9
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 9 |
| chr18_+_37651393 | 3.36 |
ENSMUST00000097609.3
|
Pcdhb22
|
protocadherin beta 22 |
| chr10_+_59942274 | 3.33 |
ENSMUST00000165024.3
|
Spock2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 2 |
| chr8_+_47439948 | 3.33 |
ENSMUST00000119686.2
|
Enpp6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6 |
| chr7_+_35148188 | 3.31 |
ENSMUST00000118383.8
|
Slc7a9
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 9 |
| chr2_-_102282844 | 3.29 |
ENSMUST00000099678.5
|
Fjx1
|
four jointed box 1 |
| chr1_+_66426127 | 3.28 |
ENSMUST00000145419.8
|
Map2
|
microtubule-associated protein 2 |
| chr4_-_131565542 | 3.25 |
ENSMUST00000030741.9
ENSMUST00000105987.9 |
Ptpru
|
protein tyrosine phosphatase, receptor type, U |
| chr2_+_164328763 | 3.24 |
ENSMUST00000109349.9
|
Dbndd2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr1_-_43235914 | 3.10 |
ENSMUST00000187357.2
|
Fhl2
|
four and a half LIM domains 2 |
| chr10_+_69761784 | 3.07 |
ENSMUST00000181974.8
ENSMUST00000182795.8 ENSMUST00000182437.8 |
Ank3
|
ankyrin 3, epithelial |
| chr8_+_46111703 | 3.05 |
ENSMUST00000134675.8
ENSMUST00000139869.8 ENSMUST00000126067.8 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr6_+_36364990 | 3.04 |
ENSMUST00000172278.8
|
Chrm2
|
cholinergic receptor, muscarinic 2, cardiac |
| chr8_+_59365291 | 3.03 |
ENSMUST00000160055.2
|
BC030500
|
cDNA sequence BC030500 |
| chr9_+_109865810 | 3.00 |
ENSMUST00000163190.8
|
Map4
|
microtubule-associated protein 4 |
| chr5_-_31265562 | 2.94 |
ENSMUST00000201396.2
ENSMUST00000202740.4 |
Slc30a3
|
solute carrier family 30 (zinc transporter), member 3 |
| chr1_-_169938298 | 2.92 |
ENSMUST00000192312.6
|
Ddr2
|
discoidin domain receptor family, member 2 |
| chr18_+_37827413 | 2.85 |
ENSMUST00000193414.2
|
Pcdhga5
|
protocadherin gamma subfamily A, 5 |
| chr8_+_47439916 | 2.84 |
ENSMUST00000039840.15
|
Enpp6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6 |
| chr19_-_17814984 | 2.84 |
ENSMUST00000025618.16
ENSMUST00000050715.10 |
Pcsk5
|
proprotein convertase subtilisin/kexin type 5 |
| chr9_+_15150341 | 2.81 |
ENSMUST00000034413.8
|
Vstm5
|
V-set and transmembrane domain containing 5 |
| chr10_+_34173426 | 2.65 |
ENSMUST00000047935.8
|
Tspyl4
|
TSPY-like 4 |
| chr18_+_37575553 | 2.65 |
ENSMUST00000056915.3
|
Pcdhb13
|
protocadherin beta 13 |
| chr7_+_18910340 | 2.64 |
ENSMUST00000117338.8
|
Eml2
|
echinoderm microtubule associated protein like 2 |
| chr1_-_169938060 | 2.63 |
ENSMUST00000027985.8
ENSMUST00000194690.6 |
Ddr2
|
discoidin domain receptor family, member 2 |
| chr18_+_13107535 | 2.62 |
ENSMUST00000234035.2
ENSMUST00000235053.2 |
Impact
|
impact, RWD domain protein |
| chr14_-_104081827 | 2.60 |
ENSMUST00000022718.11
|
Ednrb
|
endothelin receptor type B |
| chr14_+_58310143 | 2.59 |
ENSMUST00000022545.14
|
Fgf9
|
fibroblast growth factor 9 |
| chr11_+_77656414 | 2.58 |
ENSMUST00000164315.2
|
Myo18a
|
myosin XVIIIA |
| chr14_-_9015639 | 2.55 |
ENSMUST00000112656.4
|
Synpr
|
synaptoporin |
| chr18_+_37610858 | 2.50 |
ENSMUST00000051442.7
|
Pcdhb16
|
protocadherin beta 16 |
| chr9_-_118917275 | 2.42 |
ENSMUST00000214470.2
|
Plcd1
|
phospholipase C, delta 1 |
| chr5_-_122140403 | 2.40 |
ENSMUST00000134326.2
|
Cux2
|
cut-like homeobox 2 |
| chr7_+_23969822 | 2.37 |
ENSMUST00000108438.10
|
Zfp93
|
zinc finger protein 93 |
| chr3_-_141637245 | 2.34 |
ENSMUST00000106232.8
|
Bmpr1b
|
bone morphogenetic protein receptor, type 1B |
| chr10_-_5144699 | 2.33 |
ENSMUST00000215467.2
|
Syne1
|
spectrin repeat containing, nuclear envelope 1 |
| chr1_-_82746169 | 2.31 |
ENSMUST00000027331.3
|
Tm4sf20
|
transmembrane 4 L six family member 20 |
| chr19_+_28941292 | 2.28 |
ENSMUST00000045674.4
|
Plpp6
|
phospholipid phosphatase 6 |
| chr5_+_24679154 | 2.25 |
ENSMUST00000199856.2
|
Agap3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr7_-_100581314 | 2.21 |
ENSMUST00000107032.3
|
Arhgef17
|
Rho guanine nucleotide exchange factor (GEF) 17 |
| chr4_+_97660971 | 2.21 |
ENSMUST00000152023.8
|
Nfia
|
nuclear factor I/A |
| chr6_+_58573656 | 2.19 |
ENSMUST00000031822.13
|
Abcg2
|
ATP binding cassette subfamily G member 2 (Junior blood group) |
| chr17_-_12894716 | 2.18 |
ENSMUST00000024596.10
|
Slc22a1
|
solute carrier family 22 (organic cation transporter), member 1 |
| chr18_-_89787603 | 2.17 |
ENSMUST00000097495.5
|
Dok6
|
docking protein 6 |
| chr1_+_156666485 | 2.15 |
ENSMUST00000111720.2
|
Angptl1
|
angiopoietin-like 1 |
| chr2_+_109522781 | 2.07 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr15_+_39061612 | 2.05 |
ENSMUST00000082054.12
ENSMUST00000227243.2 ENSMUST00000042917.10 |
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chr2_-_77349909 | 2.05 |
ENSMUST00000111830.9
|
Zfp385b
|
zinc finger protein 385B |
| chr8_+_94537910 | 2.02 |
ENSMUST00000138659.9
|
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chr18_+_37622518 | 1.99 |
ENSMUST00000055949.4
|
Pcdhb18
|
protocadherin beta 18 |
| chr5_-_73787646 | 1.98 |
ENSMUST00000152215.3
|
Lrrc66
|
leucine rich repeat containing 66 |
| chr9_+_50405817 | 1.98 |
ENSMUST00000114474.8
ENSMUST00000188047.2 |
Plet1
|
placenta expressed transcript 1 |
| chr19_-_8700728 | 1.98 |
ENSMUST00000170157.8
|
Slc3a2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 |
| chr3_-_108062172 | 1.95 |
ENSMUST00000062028.8
|
Gpr61
|
G protein-coupled receptor 61 |
| chr7_-_133384449 | 1.94 |
ENSMUST00000063669.8
|
Dhx32
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32 |
| chr12_+_37292029 | 1.91 |
ENSMUST00000160390.2
|
Agmo
|
alkylglycerol monooxygenase |
| chr19_+_43770619 | 1.88 |
ENSMUST00000026208.6
|
Abcc2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr18_+_37801971 | 1.87 |
ENSMUST00000193869.2
|
Pcdhga2
|
protocadherin gamma subfamily A, 2 |
| chr1_+_45350698 | 1.87 |
ENSMUST00000087883.13
|
Col3a1
|
collagen, type III, alpha 1 |
| chr18_+_57601541 | 1.82 |
ENSMUST00000091892.4
ENSMUST00000209782.2 |
Ctxn3
|
cortexin 3 |
| chr19_+_23881821 | 1.77 |
ENSMUST00000237688.2
|
Apba1
|
amyloid beta (A4) precursor protein binding, family A, member 1 |
| chr17_+_12803019 | 1.75 |
ENSMUST00000046959.9
ENSMUST00000233066.2 |
Slc22a2
|
solute carrier family 22 (organic cation transporter), member 2 |
| chr16_+_5703134 | 1.75 |
ENSMUST00000230658.2
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr2_-_25498459 | 1.71 |
ENSMUST00000058137.9
|
Rabl6
|
RAB, member RAS oncogene family-like 6 |
| chr13_+_41071077 | 1.70 |
ENSMUST00000067778.8
ENSMUST00000225759.2 |
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chrX_-_69408627 | 1.68 |
ENSMUST00000101509.9
|
Ids
|
iduronate 2-sulfatase |
| chr11_-_74238498 | 1.63 |
ENSMUST00000080365.6
|
Olfr411
|
olfactory receptor 411 |
| chr12_+_69954506 | 1.62 |
ENSMUST00000223456.2
|
Atl1
|
atlastin GTPase 1 |
| chr2_+_61634797 | 1.62 |
ENSMUST00000048934.15
|
Tbr1
|
T-box brain transcription factor 1 |
| chr6_-_28134544 | 1.58 |
ENSMUST00000115323.8
|
Grm8
|
glutamate receptor, metabotropic 8 |
| chr18_+_69477541 | 1.57 |
ENSMUST00000114985.10
ENSMUST00000128706.8 ENSMUST00000201781.4 ENSMUST00000202674.4 |
Tcf4
|
transcription factor 4 |
| chr8_+_46111778 | 1.56 |
ENSMUST00000143820.8
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr5_+_67473069 | 1.55 |
ENSMUST00000202770.2
|
Slc30a9
|
solute carrier family 30 (zinc transporter), member 9 |
| chr10_+_115653152 | 1.53 |
ENSMUST00000080630.11
ENSMUST00000179196.3 ENSMUST00000035563.15 |
Tspan8
|
tetraspanin 8 |
| chr3_+_31150982 | 1.48 |
ENSMUST00000118204.2
|
Skil
|
SKI-like |
| chr5_+_104350475 | 1.46 |
ENSMUST00000066708.7
|
Dmp1
|
dentin matrix protein 1 |
| chr3_-_26386609 | 1.43 |
ENSMUST00000193603.6
|
Nlgn1
|
neuroligin 1 |
| chr16_-_92196954 | 1.43 |
ENSMUST00000023672.10
|
Rcan1
|
regulator of calcineurin 1 |
| chr16_-_59138611 | 1.39 |
ENSMUST00000216261.2
|
Olfr204
|
olfactory receptor 204 |
| chr7_+_23833572 | 1.37 |
ENSMUST00000205680.2
ENSMUST00000056549.9 |
Zfp235
|
zinc finger protein 235 |
| chr4_+_134042423 | 1.36 |
ENSMUST00000105875.8
ENSMUST00000030638.7 |
Trim63
|
tripartite motif-containing 63 |
| chr4_+_139350152 | 1.33 |
ENSMUST00000039818.10
|
Aldh4a1
|
aldehyde dehydrogenase 4 family, member A1 |
| chrX_+_94942639 | 1.33 |
ENSMUST00000082183.8
|
Zc3h12b
|
zinc finger CCCH-type containing 12B |
| chr18_+_37880027 | 1.31 |
ENSMUST00000193404.2
|
Pcdhga10
|
protocadherin gamma subfamily A, 10 |
| chr11_+_70430870 | 1.31 |
ENSMUST00000157075.8
|
Pld2
|
phospholipase D2 |
| chr7_+_110371811 | 1.30 |
ENSMUST00000005829.13
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr12_-_4957212 | 1.28 |
ENSMUST00000142867.8
|
Ubxn2a
|
UBX domain protein 2A |
| chr18_+_37100672 | 1.27 |
ENSMUST00000193777.6
ENSMUST00000193389.2 |
Pcdha6
|
protocadherin alpha 6 |
| chr1_-_132635042 | 1.26 |
ENSMUST00000043189.14
|
Nfasc
|
neurofascin |
| chr12_+_8027640 | 1.26 |
ENSMUST00000171271.8
ENSMUST00000037811.13 |
Apob
|
apolipoprotein B |
| chr8_+_26401698 | 1.25 |
ENSMUST00000120653.8
ENSMUST00000126226.2 |
Kcnu1
|
potassium channel, subfamily U, member 1 |
| chr14_-_30213408 | 1.24 |
ENSMUST00000112250.6
|
Cacna1d
|
calcium channel, voltage-dependent, L type, alpha 1D subunit |
| chr1_-_132635078 | 1.21 |
ENSMUST00000187861.7
|
Nfasc
|
neurofascin |
| chr18_+_37093321 | 1.21 |
ENSMUST00000192168.2
|
Pcdha5
|
protocadherin alpha 5 |
| chr16_+_43184191 | 1.20 |
ENSMUST00000156367.8
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr4_-_15945359 | 1.20 |
ENSMUST00000029877.9
|
Decr1
|
2,4-dienoyl CoA reductase 1, mitochondrial |
| chr12_-_110807330 | 1.18 |
ENSMUST00000177224.2
ENSMUST00000084974.11 ENSMUST00000070565.15 |
Mok
|
MOK protein kinase |
| chr16_+_43184507 | 1.17 |
ENSMUST00000148775.8
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr12_+_25024913 | 1.15 |
ENSMUST00000066652.7
ENSMUST00000220459.2 ENSMUST00000222941.2 |
Kidins220
|
kinase D-interacting substrate 220 |
| chr2_+_164328375 | 1.15 |
ENSMUST00000069385.15
ENSMUST00000143690.8 |
Dbndd2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr16_-_26345493 | 1.14 |
ENSMUST00000165687.3
|
Tmem207
|
transmembrane protein 207 |
| chr13_+_83723743 | 1.13 |
ENSMUST00000198217.5
ENSMUST00000199210.5 |
Mef2c
|
myocyte enhancer factor 2C |
| chr6_+_103487541 | 1.12 |
ENSMUST00000203912.3
|
Chl1
|
cell adhesion molecule L1-like |
| chr9_-_105398346 | 1.08 |
ENSMUST00000176770.8
ENSMUST00000085133.13 |
Atp2c1
|
ATPase, Ca++-sequestering |
| chr2_+_177783713 | 1.05 |
ENSMUST00000103066.10
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr17_+_19724994 | 1.05 |
ENSMUST00000166081.3
ENSMUST00000231465.2 |
Vmn2r100
|
vomeronasal 2, receptor 100 |
| chr14_-_36657517 | 1.04 |
ENSMUST00000183038.8
|
Ccser2
|
coiled-coil serine rich 2 |
| chr6_-_98319684 | 1.04 |
ENSMUST00000164491.3
|
Mdfic2
|
MyoD family inhibitor domain containing 2 |
| chr3_-_129126362 | 1.04 |
ENSMUST00000029658.14
|
Enpep
|
glutamyl aminopeptidase |
| chr18_+_37580692 | 1.00 |
ENSMUST00000052387.5
|
Pcdhb14
|
protocadherin beta 14 |
| chr2_-_104324035 | 0.98 |
ENSMUST00000111124.8
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chr14_-_9015757 | 0.97 |
ENSMUST00000153954.8
|
Synpr
|
synaptoporin |
| chr3_-_57209357 | 0.96 |
ENSMUST00000196979.5
ENSMUST00000029376.13 |
Tm4sf1
|
transmembrane 4 superfamily member 1 |
| chr6_+_39569502 | 0.95 |
ENSMUST00000135671.8
ENSMUST00000119379.2 |
Ndufb2
|
NADH:ubiquinone oxidoreductase subunit B2 |
| chr16_-_19341016 | 0.93 |
ENSMUST00000214315.2
|
Olfr167
|
olfactory receptor 167 |
| chr11_+_96920751 | 0.92 |
ENSMUST00000021249.11
|
Scrn2
|
secernin 2 |
| chr8_-_34574970 | 0.92 |
ENSMUST00000118811.8
|
Dctn6
|
dynactin 6 |
| chr11_-_107238956 | 0.91 |
ENSMUST00000134763.2
|
Pitpnc1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr16_+_13176238 | 0.90 |
ENSMUST00000149359.2
|
Mrtfb
|
myocardin related transcription factor B |
| chrX_+_71025128 | 0.86 |
ENSMUST00000114569.2
|
Fate1
|
fetal and adult testis expressed 1 |
| chr15_-_12549963 | 0.86 |
ENSMUST00000189324.2
|
Pdzd2
|
PDZ domain containing 2 |
| chr2_-_86061745 | 0.84 |
ENSMUST00000216056.2
|
Olfr1047
|
olfactory receptor 1047 |
| chrM_+_5319 | 0.84 |
ENSMUST00000082402.1
|
mt-Co1
|
mitochondrially encoded cytochrome c oxidase I |
| chr2_+_173579285 | 0.81 |
ENSMUST00000067530.6
|
Vapb
|
vesicle-associated membrane protein, associated protein B and C |
| chr18_-_88945571 | 0.81 |
ENSMUST00000147313.2
|
Socs6
|
suppressor of cytokine signaling 6 |
| chr2_+_24166920 | 0.80 |
ENSMUST00000168941.8
ENSMUST00000028360.8 ENSMUST00000123053.8 |
Il1f5
|
interleukin 1 family, member 5 (delta) |
| chr5_+_102629240 | 0.79 |
ENSMUST00000073302.12
ENSMUST00000094559.9 |
Arhgap24
|
Rho GTPase activating protein 24 |
| chr1_+_24717793 | 0.77 |
ENSMUST00000186190.2
|
Lmbrd1
|
LMBR1 domain containing 1 |
| chr5_-_145816774 | 0.75 |
ENSMUST00000035918.8
|
Cyp3a11
|
cytochrome P450, family 3, subfamily a, polypeptide 11 |
| chr15_+_28203872 | 0.75 |
ENSMUST00000067048.8
|
Dnah5
|
dynein, axonemal, heavy chain 5 |
| chr4_-_88798438 | 0.70 |
ENSMUST00000056014.3
|
Ifne
|
interferon epsilon |
| chr5_+_4073343 | 0.67 |
ENSMUST00000238634.2
|
Akap9
|
A kinase (PRKA) anchor protein (yotiao) 9 |
| chr12_+_71936500 | 0.66 |
ENSMUST00000221317.2
|
Daam1
|
dishevelled associated activator of morphogenesis 1 |
| chr5_+_88465146 | 0.66 |
ENSMUST00000170832.3
|
Prol1
|
proline rich, lacrimal 1 |
| chr1_+_13738967 | 0.65 |
ENSMUST00000088542.4
|
Xkr9
|
X-linked Kx blood group related 9 |
| chr5_-_124717146 | 0.65 |
ENSMUST00000031334.15
|
Eif2b1
|
eukaryotic translation initiation factor 2B, subunit 1 (alpha) |
| chr6_+_41837173 | 0.65 |
ENSMUST00000014248.11
|
Sval2
|
seminal vesicle antigen-like 2 |
| chr9_+_45230370 | 0.62 |
ENSMUST00000034597.8
|
Tmprss13
|
transmembrane protease, serine 13 |
| chr5_+_135754568 | 0.62 |
ENSMUST00000127096.2
|
Por
|
P450 (cytochrome) oxidoreductase |
| chr14_-_54651442 | 0.61 |
ENSMUST00000227334.2
|
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr4_-_36056726 | 0.56 |
ENSMUST00000108124.4
|
Lingo2
|
leucine rich repeat and Ig domain containing 2 |
| chr6_+_132824105 | 0.55 |
ENSMUST00000071696.2
|
Tas2r123
|
taste receptor, type 2, member 123 |
| chr12_-_85317359 | 0.53 |
ENSMUST00000166821.8
ENSMUST00000019378.8 ENSMUST00000220854.2 |
Mlh3
|
mutL homolog 3 |
| chr3_+_79792238 | 0.50 |
ENSMUST00000135021.2
|
Gask1b
|
golgi associated kinase 1B |
| chr1_+_24717722 | 0.48 |
ENSMUST00000186096.7
|
Lmbrd1
|
LMBR1 domain containing 1 |
| chr6_+_128376729 | 0.47 |
ENSMUST00000001561.12
|
Nrip2
|
nuclear receptor interacting protein 2 |
| chr6_+_56933451 | 0.47 |
ENSMUST00000096612.4
|
Vmn1r4
|
vomeronasal 1 receptor 4 |
| chr13_+_22534534 | 0.47 |
ENSMUST00000226909.2
ENSMUST00000227167.2 ENSMUST00000226786.2 |
Vmn1r198
|
vomeronasal 1 receptor 198 |
| chr7_+_102731092 | 0.46 |
ENSMUST00000214215.2
|
Olfr584
|
olfactory receptor 584 |
| chr7_-_103217838 | 0.45 |
ENSMUST00000214806.2
|
Olfr616
|
olfactory receptor 616 |
| chr11_+_46415039 | 0.45 |
ENSMUST00000109228.2
|
Gm12169
|
predicted gene 12169 |
| chr17_+_18269686 | 0.45 |
ENSMUST00000176802.3
|
Vmn2r124
|
vomeronasal 2, receptor 124 |
| chr2_+_120807498 | 0.43 |
ENSMUST00000067582.14
|
Tmem62
|
transmembrane protein 62 |
| chr15_+_102368510 | 0.43 |
ENSMUST00000164688.2
|
Prr13
|
proline rich 13 |
| chr5_+_102629365 | 0.42 |
ENSMUST00000112854.8
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr11_+_96920956 | 0.41 |
ENSMUST00000153482.2
|
Scrn2
|
secernin 2 |
| chr11_+_113540154 | 0.40 |
ENSMUST00000063776.14
|
Cog1
|
component of oligomeric golgi complex 1 |
| chr3_-_130503041 | 0.40 |
ENSMUST00000043937.9
|
Ostc
|
oligosaccharyltransferase complex subunit (non-catalytic) |
| chr16_+_91188609 | 0.39 |
ENSMUST00000160764.2
|
Gm21970
|
predicted gene 21970 |
| chr11_-_62283431 | 0.39 |
ENSMUST00000151498.9
|
Ncor1
|
nuclear receptor co-repressor 1 |
| chr6_+_128376844 | 0.38 |
ENSMUST00000120405.4
|
Nrip2
|
nuclear receptor interacting protein 2 |
| chr7_+_141765529 | 0.36 |
ENSMUST00000097943.2
|
Gm7579
|
predicted gene 7579 |
| chr3_+_79791798 | 0.35 |
ENSMUST00000118853.8
ENSMUST00000145992.2 |
Gask1b
|
golgi associated kinase 1B |
| chr1_-_82748964 | 0.35 |
ENSMUST00000223838.2
|
Gm47791
|
predicted gene, 47791 |
| chr3_+_53396120 | 0.35 |
ENSMUST00000029307.4
|
Stoml3
|
stomatin (Epb7.2)-like 3 |
| chr15_+_54274151 | 0.34 |
ENSMUST00000036737.4
|
Colec10
|
collectin sub-family member 10 |
| chr2_+_87853118 | 0.34 |
ENSMUST00000214438.2
|
Olfr1161
|
olfactory receptor 1161 |
| chr2_+_24167275 | 0.33 |
ENSMUST00000114490.7
ENSMUST00000147885.4 |
Il1f5
|
interleukin 1 family, member 5 (delta) |
| chr10_-_85752932 | 0.33 |
ENSMUST00000218969.2
|
Prdm4
|
PR domain containing 4 |
| chr5_-_130284366 | 0.31 |
ENSMUST00000026387.11
|
Sbds
|
SBDS ribosome maturation factor |
| chr2_-_45000250 | 0.30 |
ENSMUST00000201211.4
ENSMUST00000177302.8 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr14_-_50380137 | 0.28 |
ENSMUST00000213685.2
|
Olfr728
|
olfactory receptor 728 |
| chr2_+_89708781 | 0.27 |
ENSMUST00000111519.3
|
Olfr1257
|
olfactory receptor 1257 |
| chr9_-_56151334 | 0.27 |
ENSMUST00000188142.7
|
Peak1
|
pseudopodium-enriched atypical kinase 1 |
| chr19_-_28941264 | 0.25 |
ENSMUST00000161813.2
|
Spata6l
|
spermatogenesis associated 6 like |
| chr19_-_36896999 | 0.25 |
ENSMUST00000238948.2
ENSMUST00000057337.9 |
Fgfbp3
|
fibroblast growth factor binding protein 3 |
| chr7_+_41909477 | 0.24 |
ENSMUST00000166131.2
|
Vmn2r61
|
vomeronasal 2, receptor 61 |
| chr9_-_60048503 | 0.24 |
ENSMUST00000034829.6
|
Thsd4
|
thrombospondin, type I, domain containing 4 |
| chr9_-_38577138 | 0.24 |
ENSMUST00000076542.2
|
Olfr917
|
olfactory receptor 917 |
| chr13_-_67442285 | 0.23 |
ENSMUST00000224325.2
|
Zfp457
|
zinc finger protein 457 |
| chr11_-_59484115 | 0.22 |
ENSMUST00000215626.2
|
Olfr223
|
olfactory receptor 223 |
| chr12_-_91745903 | 0.22 |
ENSMUST00000170077.2
|
Ston2
|
stonin 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 10.4 | GO:0015811 | L-cystine transport(GO:0015811) |
| 2.2 | 6.7 | GO:0030827 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 1.0 | 4.9 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) |
| 0.9 | 2.6 | GO:1901561 | cellular response to benomyl(GO:0072755) response to benomyl(GO:1901561) |
| 0.9 | 10.5 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.9 | 2.6 | GO:1903027 | regulation of opsonization(GO:1903027) positive regulation of opsonization(GO:1903028) |
| 0.8 | 3.1 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.7 | 2.2 | GO:0019389 | glucuronoside metabolic process(GO:0019389) |
| 0.7 | 2.1 | GO:0061193 | taste bud development(GO:0061193) |
| 0.7 | 6.2 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.6 | 3.0 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.5 | 1.4 | GO:0048789 | cytoskeletal matrix organization at active zone(GO:0048789) neurexin clustering involved in presynaptic membrane assembly(GO:0097115) retrograde trans-synaptic signaling by trans-synaptic protein complex(GO:0098942) |
| 0.5 | 1.9 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.5 | 2.3 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.5 | 1.4 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.4 | 1.3 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.4 | 2.6 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.4 | 3.0 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.4 | 2.0 | GO:0060356 | leucine import(GO:0060356) |
| 0.4 | 2.6 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.4 | 1.9 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.4 | 2.2 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.3 | 5.6 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.3 | 2.4 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.3 | 2.3 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.3 | 7.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.3 | 1.5 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.3 | 1.1 | GO:0038016 | insulin receptor internalization(GO:0038016) |
| 0.3 | 2.8 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 0.3 | 3.3 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.3 | 1.1 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.3 | 1.6 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.3 | 2.9 | GO:0097501 | stress response to metal ion(GO:0097501) |
| 0.2 | 2.5 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.2 | 1.2 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.2 | 1.7 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.2 | 3.1 | GO:1900827 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 | 4.9 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.2 | 2.2 | GO:0030578 | PML body organization(GO:0030578) |
| 0.2 | 0.8 | GO:0044830 | modulation by host of viral RNA genome replication(GO:0044830) negative regulation by virus of viral protein levels in host cell(GO:0046725) |
| 0.2 | 8.2 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.2 | 1.6 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.2 | 3.8 | GO:0007614 | short-term memory(GO:0007614) |
| 0.2 | 3.8 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.2 | 2.1 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.2 | 0.6 | GO:0046210 | nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.2 | 2.6 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.1 | 1.0 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 0.1 | 1.9 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 1.1 | GO:1902714 | negative regulation of interferon-gamma secretion(GO:1902714) |
| 0.1 | 4.3 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 1.3 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 0.6 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.1 | 1.2 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 | 2.2 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 1.3 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 0.4 | GO:0072362 | regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.1 | 1.3 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.1 | 1.7 | GO:0051608 | histamine transport(GO:0051608) |
| 0.1 | 1.1 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 1.5 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.1 | 2.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 3.3 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 0.6 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.1 | 2.0 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) |
| 0.1 | 1.9 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.1 | 4.6 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.5 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 1.1 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.1 | 1.7 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.1 | 0.5 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.1 | 1.3 | GO:0033008 | G-protein coupled receptor internalization(GO:0002031) positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.1 | 11.2 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.1 | 1.0 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 0.8 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 2.4 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 2.4 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.3 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.7 | GO:0031280 | negative regulation of cyclase activity(GO:0031280) |
| 0.0 | 0.3 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 1.6 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) |
| 0.0 | 0.2 | GO:0070893 | transposon integration(GO:0070893) regulation of transposon integration(GO:0070894) negative regulation of transposon integration(GO:0070895) |
| 0.0 | 1.7 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.7 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 2.0 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 3.2 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 3.1 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 1.5 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.6 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 0.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.7 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 1.6 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 1.8 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 1.0 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 2.0 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 5.4 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.0 | 5.4 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 1.2 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.2 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.3 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 1.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 2.2 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.5 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.4 | 6.7 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.4 | 1.3 | GO:0034359 | mature chylomicron(GO:0034359) |
| 0.4 | 3.3 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.2 | 2.3 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.2 | 2.5 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.2 | 0.5 | GO:0005712 | chiasma(GO:0005712) |
| 0.1 | 0.7 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.1 | 4.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 4.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 3.5 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 1.9 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 7.3 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 3.8 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 3.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 2.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 1.6 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 1.0 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.1 | 5.0 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 11.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 1.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.6 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 1.4 | GO:0032433 | NMDA selective glutamate receptor complex(GO:0017146) filopodium tip(GO:0032433) |
| 0.1 | 0.9 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 9.7 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 2.1 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 5.6 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 7.2 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 9.3 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 2.6 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 7.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.2 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 1.1 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 2.6 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 2.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 1.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 11.8 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 3.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 6.3 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 2.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 3.1 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.0 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 5.5 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 2.0 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.8 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 1.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 3.9 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 4.3 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.8 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 2.0 | GO:0001650 | fibrillar center(GO:0001650) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 10.4 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 1.3 | 3.9 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) norepinephrine transmembrane transporter activity(GO:0005333) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.9 | 10.5 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.9 | 2.6 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.8 | 3.0 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.7 | 2.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.6 | 1.9 | GO:0050479 | glyceryl-ether monooxygenase activity(GO:0050479) |
| 0.6 | 6.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.5 | 5.6 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.4 | 1.7 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.4 | 1.2 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.4 | 3.3 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.3 | 1.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.3 | 0.8 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.3 | 1.1 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.3 | 2.0 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.2 | 7.2 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.2 | 2.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.2 | 1.6 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.2 | 2.5 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.2 | 4.9 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.2 | 2.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.2 | 1.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.2 | 1.4 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 1.3 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.2 | 0.6 | GO:0008941 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) iron-cytochrome-c reductase activity(GO:0047726) |
| 0.2 | 2.4 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.1 | 0.5 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.1 | 1.3 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 1.9 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 1.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 1.6 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 1.3 | GO:0035473 | lipase binding(GO:0035473) |
| 0.1 | 1.7 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 1.4 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 2.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 3.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.9 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 4.3 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 1.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 2.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 1.3 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 4.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.4 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.1 | 1.2 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 1.3 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 1.3 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.4 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 1.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 2.6 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 1.0 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 2.9 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 2.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.7 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.6 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.8 | GO:0016675 | oxidoreductase activity, acting on a heme group of donors(GO:0016675) |
| 0.0 | 0.9 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 1.9 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 1.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 1.5 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 2.0 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.9 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 1.2 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.7 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.3 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 2.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 5.8 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 1.1 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 5.8 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 2.2 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.1 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.3 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 8.4 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 3.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.9 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 2.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 2.7 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.2 | 2.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 7.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 2.0 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 1.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 3.1 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 6.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 3.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 4.5 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 3.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 1.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 2.2 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 3.3 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 2.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 2.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.7 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.3 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.0 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.3 | 10.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.3 | 13.0 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.2 | 7.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.2 | 7.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.6 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 1.7 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 3.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 3.8 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 3.0 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 1.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 2.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 3.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 2.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 2.0 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 1.9 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 2.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.8 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 2.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.7 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 1.0 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 2.8 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |