Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for GGCAAGA
Z-value: 0.38
miRNA associated with seed GGCAAGA
| Name | miRBASE accession |
|---|---|
|
mmu-miR-31-5p
|
MIMAT0000538 |
Activity profile of GGCAAGA motif
Sorted Z-values of GGCAAGA motif
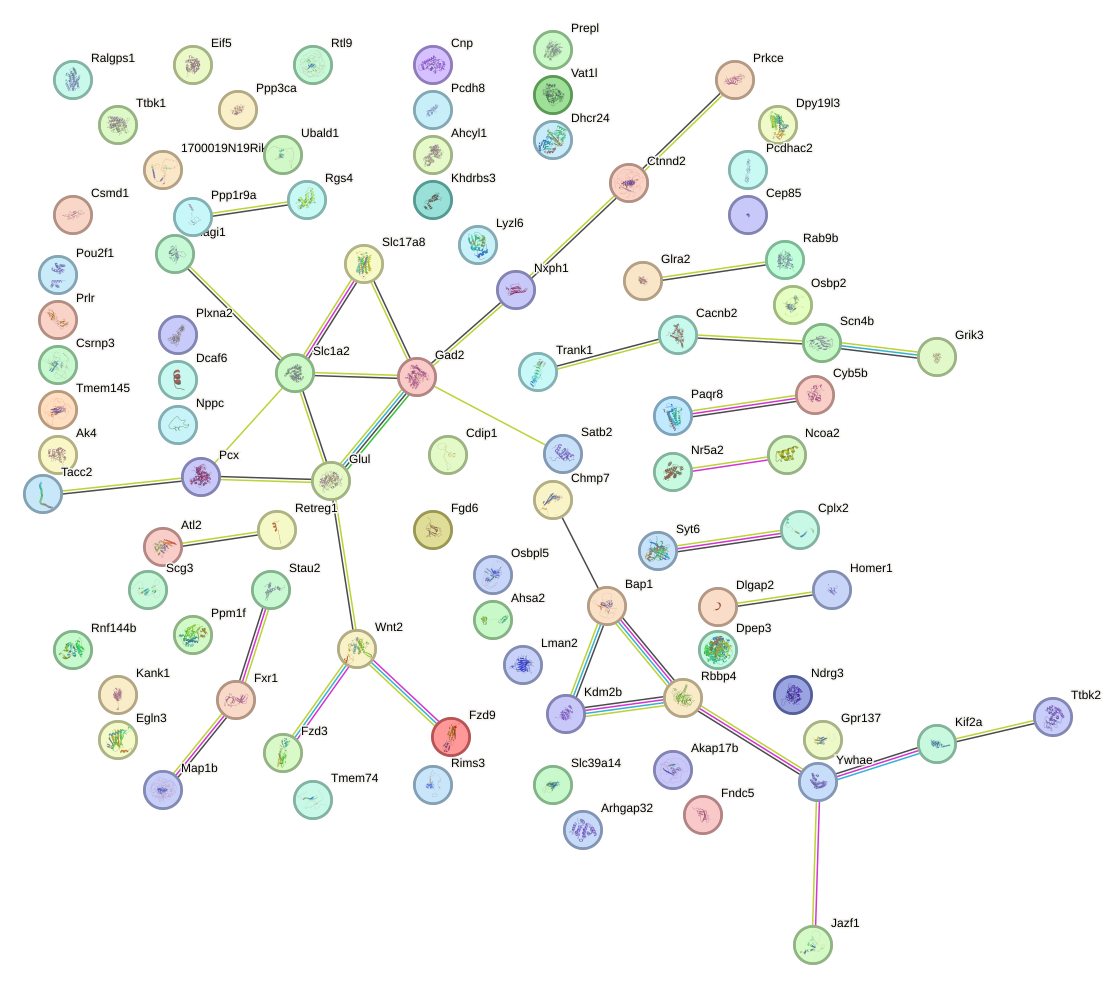
Network of associatons between targets according to the STRING database.
First level regulatory network of GGCAAGA
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_102488985 | 4.01 |
ENSMUST00000080210.10
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr4_+_101276474 | 2.07 |
ENSMUST00000102780.8
ENSMUST00000106946.8 ENSMUST00000106945.8 |
Ak4
|
adenylate kinase 4 |
| chr11_-_3672188 | 1.80 |
ENSMUST00000102950.10
ENSMUST00000101632.4 |
Osbp2
|
oxysterol binding protein 2 |
| chr9_-_75591274 | 1.60 |
ENSMUST00000214244.2
ENSMUST00000213324.2 ENSMUST00000034699.8 |
Scg3
|
secretogranin III |
| chr19_-_58782874 | 1.22 |
ENSMUST00000028299.11
|
1700019N19Rik
|
RIKEN cDNA 1700019N19 gene |
| chr9_+_111140741 | 1.18 |
ENSMUST00000078626.8
|
Trank1
|
tetratricopeptide repeat and ankyrin repeat containing 1 |
| chr19_+_4560500 | 1.09 |
ENSMUST00000068004.13
ENSMUST00000224726.3 |
Pcx
|
pyruvate carboxylase |
| chr8_-_106706035 | 1.08 |
ENSMUST00000034371.9
|
Dpep3
|
dipeptidase 3 |
| chr17_-_46798566 | 0.92 |
ENSMUST00000047034.9
|
Ttbk1
|
tau tubulin kinase 1 |
| chr4_-_91260265 | 0.81 |
ENSMUST00000107110.8
ENSMUST00000008633.15 ENSMUST00000107118.8 |
Elavl2
|
ELAV like RNA binding protein 1 |
| chr16_+_41353360 | 0.80 |
ENSMUST00000099761.10
|
Lsamp
|
limbic system-associated membrane protein |
| chr15_-_43733389 | 0.79 |
ENSMUST00000067469.6
|
Tmem74
|
transmembrane protein 74 |
| chr14_-_80008745 | 0.79 |
ENSMUST00000039568.11
ENSMUST00000195355.2 |
Pcdh8
|
protocadherin 8 |
| chr7_+_25005510 | 0.69 |
ENSMUST00000119703.8
ENSMUST00000205639.3 ENSMUST00000108409.2 |
Tmem145
|
transmembrane protein 145 |
| chr9_-_52590686 | 0.69 |
ENSMUST00000098768.3
ENSMUST00000213843.2 |
AI593442
|
expressed sequence AI593442 |
| chr7_+_24776630 | 0.67 |
ENSMUST00000179556.2
ENSMUST00000053410.11 |
Zfp574
|
zinc finger protein 574 |
| chrX_-_164110372 | 0.65 |
ENSMUST00000058787.9
|
Glra2
|
glycine receptor, alpha 2 subunit |
| chr6_-_18030584 | 0.63 |
ENSMUST00000010941.6
|
Wnt2
|
wingless-type MMTV integration site family, member 2 |
| chr15_+_82159398 | 0.61 |
ENSMUST00000023095.14
ENSMUST00000230365.2 |
Septin3
|
septin 3 |
| chr8_+_14145848 | 0.61 |
ENSMUST00000152652.8
ENSMUST00000133298.8 |
Dlgap2
|
DLG associated protein 2 |
| chr14_-_70588803 | 0.61 |
ENSMUST00000143153.2
ENSMUST00000127000.2 ENSMUST00000068044.14 ENSMUST00000022688.10 |
Slc39a14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr6_+_4903299 | 0.60 |
ENSMUST00000035813.9
|
Ppp1r9a
|
protein phosphatase 1, regulatory subunit 9A |
| chr11_-_103529678 | 0.60 |
ENSMUST00000107014.8
ENSMUST00000021328.8 |
Lyzl6
|
lysozyme-like 6 |
| chr2_+_22512195 | 0.55 |
ENSMUST00000028123.4
|
Gad2
|
glutamic acid decarboxylase 2 |
| chr8_+_114932312 | 0.54 |
ENSMUST00000049509.7
ENSMUST00000150963.2 |
Vat1l
|
vesicle amine transport protein 1 like |
| chr6_-_53045546 | 0.52 |
ENSMUST00000074541.6
|
Jazf1
|
JAZF zinc finger 1 |
| chr9_+_32027335 | 0.51 |
ENSMUST00000174641.8
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr6_+_8948608 | 0.50 |
ENSMUST00000160300.2
|
Nxph1
|
neurexophilin 1 |
| chr1_-_57011595 | 0.50 |
ENSMUST00000042857.14
|
Satb2
|
special AT-rich sequence binding protein 2 |
| chr4_+_106418224 | 0.49 |
ENSMUST00000047973.4
|
Dhcr24
|
24-dehydrocholesterol reductase |
| chr12_+_111504450 | 0.49 |
ENSMUST00000166123.9
ENSMUST00000222441.2 |
Eif5
|
eukaryotic translation initiation factor 5 |
| chr2_+_65676111 | 0.48 |
ENSMUST00000122912.8
|
Csrnp3
|
cysteine-serine-rich nuclear protein 3 |
| chr1_-_165287999 | 0.48 |
ENSMUST00000027856.13
|
Dcaf6
|
DDB1 and CUL4 associated factor 6 |
| chr4_+_129030710 | 0.48 |
ENSMUST00000102600.4
|
Fndc5
|
fibronectin type III domain containing 5 |
| chr15_+_30172716 | 0.48 |
ENSMUST00000081728.7
|
Ctnnd2
|
catenin (cadherin associated protein), delta 2 |
| chr9_-_75518585 | 0.47 |
ENSMUST00000098552.10
ENSMUST00000064433.11 |
Tmod2
|
tropomodulin 2 |
| chr4_+_120711974 | 0.46 |
ENSMUST00000071093.9
|
Rims3
|
regulating synaptic membrane exocytosis 3 |
| chr15_+_10177709 | 0.46 |
ENSMUST00000124470.8
|
Prlr
|
prolactin receptor |
| chr1_-_16589511 | 0.45 |
ENSMUST00000162751.8
ENSMUST00000027052.13 ENSMUST00000149320.9 |
Stau2
|
staufen double-stranded RNA binding protein 2 |
| chr5_+_30868908 | 0.45 |
ENSMUST00000114729.8
|
Dpysl5
|
dihydropyrimidinase-like 5 |
| chr4_+_125384481 | 0.44 |
ENSMUST00000030676.8
|
Grik3
|
glutamate receptor, ionotropic, kainate 3 |
| chr3_+_34074048 | 0.43 |
ENSMUST00000001620.13
|
Fxr1
|
fragile X mental retardation gene 1, autosomal homolog |
| chr16_-_4698148 | 0.42 |
ENSMUST00000037843.7
|
Ubald1
|
UBA-like domain containing 1 |
| chr6_-_124391994 | 0.41 |
ENSMUST00000035861.6
ENSMUST00000112532.8 ENSMUST00000080557.12 |
Pex5
|
peroxisomal biogenesis factor 5 |
| chr1_-_136888118 | 0.40 |
ENSMUST00000192357.6
ENSMUST00000027649.14 |
Nr5a2
|
nuclear receptor subfamily 5, group A, member 2 |
| chrX_-_135769285 | 0.39 |
ENSMUST00000058814.7
|
Rab9b
|
RAB9B, member RAS oncogene family |
| chr14_+_30973407 | 0.38 |
ENSMUST00000022458.11
|
Bap1
|
Brca1 associated protein 1 |
| chr3_+_103482591 | 0.38 |
ENSMUST00000090697.11
ENSMUST00000239027.2 |
Syt6
|
synaptotagmin VI |
| chr1_-_165762469 | 0.37 |
ENSMUST00000069609.12
ENSMUST00000111427.9 ENSMUST00000111426.11 |
Pou2f1
|
POU domain, class 2, transcription factor 1 |
| chr13_+_47276132 | 0.36 |
ENSMUST00000068891.12
|
Rnf144b
|
ring finger protein 144B |
| chr10_-_89457115 | 0.36 |
ENSMUST00000020102.14
|
Slc17a8
|
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 8 |
| chr13_+_93444514 | 0.35 |
ENSMUST00000079086.8
|
Homer1
|
homer scaffolding protein 1 |
| chr9_+_45049687 | 0.35 |
ENSMUST00000060125.7
|
Scn4b
|
sodium channel, type IV, beta |
| chr4_-_20778530 | 0.34 |
ENSMUST00000119374.8
|
Nkain3
|
Na+/K+ transporting ATPase interacting 3 |
| chr4_-_133914329 | 0.33 |
ENSMUST00000040271.12
|
Cep85
|
centrosomal protein 85 |
| chr14_-_65499835 | 0.32 |
ENSMUST00000131309.3
|
Fzd3
|
frizzled class receptor 3 |
| chr1_+_153775682 | 0.32 |
ENSMUST00000086199.12
|
Glul
|
glutamate-ammonia ligase (glutamine synthetase) |
| chr7_-_134539993 | 0.31 |
ENSMUST00000171394.3
|
Insyn2a
|
inhibitory synaptic factor 2A |
| chr12_-_98867431 | 0.31 |
ENSMUST00000065716.8
|
Eml5
|
echinoderm microtubule associated protein like 5 |
| chr13_+_54519161 | 0.31 |
ENSMUST00000026985.9
|
Cplx2
|
complexin 2 |
| chr14_-_69969959 | 0.31 |
ENSMUST00000036381.10
|
Chmp7
|
charged multivesicular body protein 7 |
| chr17_+_86475205 | 0.31 |
ENSMUST00000097275.9
|
Prkce
|
protein kinase C, epsilon |
| chr13_-_99653045 | 0.30 |
ENSMUST00000064762.6
|
Map1b
|
microtubule-associated protein 1B |
| chr18_+_37130860 | 0.29 |
ENSMUST00000115659.6
|
Pcdha9
|
protocadherin alpha 9 |
| chr1_-_169575203 | 0.29 |
ENSMUST00000027991.12
ENSMUST00000111357.2 |
Rgs4
|
regulator of G-protein signaling 4 |
| chr18_+_38383297 | 0.28 |
ENSMUST00000025314.7
ENSMUST00000236078.2 |
Dele1
|
DAP3 binding cell death enhancer 1 |
| chr3_-_107603778 | 0.28 |
ENSMUST00000029490.15
|
Ahcyl1
|
S-adenosylhomocysteine hydrolase-like 1 |
| chr10_+_93871863 | 0.27 |
ENSMUST00000020208.5
|
Fgd6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr5_-_76452365 | 0.27 |
ENSMUST00000075159.5
|
Clock
|
circadian locomotor output cycles kaput |
| chr5_-_123038329 | 0.26 |
ENSMUST00000031435.14
|
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr2_-_33261411 | 0.26 |
ENSMUST00000131298.7
ENSMUST00000091039.5 ENSMUST00000042615.13 |
Ralgps1
|
Ral GEF with PH domain and SH3 binding motif 1 |
| chr17_-_80203457 | 0.26 |
ENSMUST00000068282.7
ENSMUST00000112437.8 |
Atl2
|
atlastin GTPase 2 |
| chr2_+_14609063 | 0.26 |
ENSMUST00000114723.9
|
Cacnb2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr16_+_16714333 | 0.26 |
ENSMUST00000027373.12
ENSMUST00000232247.2 |
Ppm1f
|
protein phosphatase 1F (PP2C domain containing) |
| chr1_+_194302123 | 0.25 |
ENSMUST00000027952.12
|
Plxna2
|
plexin A2 |
| chr19_-_6918796 | 0.25 |
ENSMUST00000025909.11
ENSMUST00000099774.10 |
Gpr137
|
G protein-coupled receptor 137 |
| chr1_+_20960819 | 0.25 |
ENSMUST00000189400.7
|
Paqr8
|
progestin and adipoQ receptor family member VIII |
| chr7_+_130179063 | 0.25 |
ENSMUST00000207918.2
ENSMUST00000215492.2 ENSMUST00000084513.12 ENSMUST00000059145.14 |
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr8_+_107877252 | 0.25 |
ENSMUST00000034400.5
|
Cyb5b
|
cytochrome b5 type B |
| chr13_-_107158535 | 0.24 |
ENSMUST00000117539.8
ENSMUST00000122233.8 ENSMUST00000022204.16 ENSMUST00000159772.8 |
Kif2a
|
kinesin family member 2A |
| chr17_-_85397627 | 0.23 |
ENSMUST00000072406.5
ENSMUST00000171795.9 ENSMUST00000234676.2 |
Prepl
|
prolyl endopeptidase-like |
| chr15_+_68800261 | 0.22 |
ENSMUST00000022954.7
|
Khdrbs3
|
KH domain containing, RNA binding, signal transduction associated 3 |
| chr8_-_17585263 | 0.22 |
ENSMUST00000082104.7
|
Csmd1
|
CUB and Sushi multiple domains 1 |
| chr7_-_35453818 | 0.22 |
ENSMUST00000051377.15
|
Dpy19l3
|
dpy-19-like 3 (C. elegans) |
| chr4_-_129229159 | 0.20 |
ENSMUST00000102598.4
|
Rbbp4
|
retinoblastoma binding protein 4, chromatin remodeling factor |
| chr3_+_136375839 | 0.20 |
ENSMUST00000070198.14
|
Ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isoform |
| chr12_-_54250646 | 0.20 |
ENSMUST00000039516.4
|
Egln3
|
egl-9 family hypoxia-inducible factor 3 |
| chr15_+_25843225 | 0.20 |
ENSMUST00000022881.15
|
Retreg1
|
reticulophagy regulator 1 |
| chr5_+_76988444 | 0.19 |
ENSMUST00000120639.9
ENSMUST00000163347.8 ENSMUST00000121851.2 |
Cracd
|
capping protein inhibiting regulator of actin |
| chr16_-_4607848 | 0.18 |
ENSMUST00000004173.12
|
Cdip1
|
cell death inducing Trp53 target 1 |
| chr11_-_23447866 | 0.18 |
ENSMUST00000128559.2
ENSMUST00000147157.8 ENSMUST00000109539.8 |
Ahsa2
|
AHA1, activator of heat shock protein ATPase 2 |
| chr11_+_100465730 | 0.18 |
ENSMUST00000103120.5
|
Cnp
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr11_+_75623695 | 0.17 |
ENSMUST00000067664.10
|
Ywhae
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide |
| chrX_+_141882590 | 0.17 |
ENSMUST00000165829.3
|
Rtl9
|
retrotransposon Gag like 9 |
| chrX_-_35909011 | 0.17 |
ENSMUST00000051906.13
|
Akap17b
|
A kinase (PRKA) anchor protein 17B |
| chr13_-_55510595 | 0.17 |
ENSMUST00000021940.8
|
Lman2
|
lectin, mannose-binding 2 |
| chr19_+_25214322 | 0.16 |
ENSMUST00000049400.15
|
Kank1
|
KN motif and ankyrin repeat domains 1 |
| chr1_-_13442658 | 0.15 |
ENSMUST00000081713.11
|
Ncoa2
|
nuclear receptor coactivator 2 |
| chr2_-_156833932 | 0.15 |
ENSMUST00000109558.2
ENSMUST00000069600.13 ENSMUST00000072298.13 |
Ndrg3
|
N-myc downstream regulated gene 3 |
| chr3_+_102377234 | 0.15 |
ENSMUST00000035952.5
ENSMUST00000198168.5 ENSMUST00000106925.9 |
Ngf
|
nerve growth factor |
| chr13_+_38010203 | 0.15 |
ENSMUST00000128570.9
|
Rreb1
|
ras responsive element binding protein 1 |
| chr7_-_136916123 | 0.15 |
ENSMUST00000106118.10
ENSMUST00000168203.2 ENSMUST00000169486.9 ENSMUST00000033378.13 |
Ebf3
|
early B cell factor 3 |
| chr16_+_42727926 | 0.15 |
ENSMUST00000151244.8
ENSMUST00000114694.9 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr7_+_96981517 | 0.14 |
ENSMUST00000054107.6
|
Kctd21
|
potassium channel tetramerisation domain containing 21 |
| chr10_-_25076008 | 0.14 |
ENSMUST00000100012.3
|
Akap7
|
A kinase (PRKA) anchor protein 7 |
| chr6_+_29917004 | 0.14 |
ENSMUST00000151738.4
ENSMUST00000115224.8 ENSMUST00000046028.12 |
Strip2
|
striatin interacting protein 2 |
| chr1_+_162398256 | 0.14 |
ENSMUST00000194810.6
ENSMUST00000050010.11 ENSMUST00000150040.8 |
Vamp4
|
vesicle-associated membrane protein 4 |
| chr18_+_65022035 | 0.14 |
ENSMUST00000224385.3
ENSMUST00000163516.9 |
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like |
| chr5_+_65694552 | 0.13 |
ENSMUST00000142407.8
|
Ube2k
|
ubiquitin-conjugating enzyme E2K |
| chr3_+_62245765 | 0.13 |
ENSMUST00000079300.13
|
Arhgef26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr6_-_3763618 | 0.13 |
ENSMUST00000171613.8
|
Calcr
|
calcitonin receptor |
| chr2_-_63928339 | 0.12 |
ENSMUST00000131615.9
|
Fign
|
fidgetin |
| chr1_-_16727067 | 0.12 |
ENSMUST00000188641.7
|
Eloc
|
elongin C |
| chr17_-_47998953 | 0.12 |
ENSMUST00000113301.2
ENSMUST00000113302.10 |
Tomm6
|
translocase of outer mitochondrial membrane 6 |
| chr9_+_51958453 | 0.11 |
ENSMUST00000163153.9
|
Rdx
|
radixin |
| chr17_+_36134122 | 0.11 |
ENSMUST00000001569.15
ENSMUST00000174080.8 |
Flot1
|
flotillin 1 |
| chr9_-_21421458 | 0.11 |
ENSMUST00000213167.2
ENSMUST00000034698.9 |
Tmed1
|
transmembrane p24 trafficking protein 1 |
| chr12_+_110452222 | 0.10 |
ENSMUST00000084985.11
ENSMUST00000221074.2 |
Ppp2r5c
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr9_-_89505178 | 0.10 |
ENSMUST00000044491.13
|
Minar1
|
membrane integral NOTCH2 associated receptor 1 |
| chr8_-_25691380 | 0.10 |
ENSMUST00000084512.11
|
Tacc1
|
transforming, acidic coiled-coil containing protein 1 |
| chr5_-_67585137 | 0.10 |
ENSMUST00000169190.5
|
Bend4
|
BEN domain containing 4 |
| chrX_+_81992467 | 0.10 |
ENSMUST00000114000.8
|
Dmd
|
dystrophin, muscular dystrophy |
| chr3_+_58484057 | 0.09 |
ENSMUST00000107924.3
|
Selenot
|
selenoprotein T |
| chr3_-_116456292 | 0.09 |
ENSMUST00000029570.9
|
Mfsd14a
|
major facilitator superfamily domain containing 14A |
| chrX_+_104807868 | 0.09 |
ENSMUST00000033581.4
|
Fgf16
|
fibroblast growth factor 16 |
| chr19_+_27299939 | 0.08 |
ENSMUST00000056708.4
|
Kcnv2
|
potassium channel, subfamily V, member 2 |
| chr9_-_26941306 | 0.08 |
ENSMUST00000034470.11
|
Vps26b
|
VPS26 retromer complex component B |
| chr4_+_116578117 | 0.07 |
ENSMUST00000045542.13
ENSMUST00000106459.8 |
Tesk2
|
testis-specific kinase 2 |
| chrX_-_72868544 | 0.07 |
ENSMUST00000002080.12
ENSMUST00000114438.3 |
Pdzd4
|
PDZ domain containing 4 |
| chr11_-_86884507 | 0.07 |
ENSMUST00000018571.5
|
Ypel2
|
yippee like 2 |
| chr10_+_128212953 | 0.07 |
ENSMUST00000014642.10
|
Ankrd52
|
ankyrin repeat domain 52 |
| chr8_+_83589979 | 0.06 |
ENSMUST00000078525.7
|
Rnf150
|
ring finger protein 150 |
| chr13_+_81031512 | 0.06 |
ENSMUST00000099356.10
|
Arrdc3
|
arrestin domain containing 3 |
| chr9_+_113641615 | 0.06 |
ENSMUST00000111838.10
ENSMUST00000166734.10 ENSMUST00000214522.2 ENSMUST00000163895.3 |
Clasp2
|
CLIP associating protein 2 |
| chr6_-_32565127 | 0.06 |
ENSMUST00000115096.4
|
Plxna4
|
plexin A4 |
| chr10_-_107321938 | 0.06 |
ENSMUST00000000445.2
|
Myf5
|
myogenic factor 5 |
| chr2_-_33321306 | 0.06 |
ENSMUST00000113158.8
|
Zbtb34
|
zinc finger and BTB domain containing 34 |
| chr19_-_24454720 | 0.05 |
ENSMUST00000099556.2
|
Fam122a
|
family with sequence similarity 122, member A |
| chr4_-_141450710 | 0.05 |
ENSMUST00000102484.5
ENSMUST00000177592.2 |
Ddi2
|
DNA-damage inducible protein 2 |
| chr7_-_63588610 | 0.05 |
ENSMUST00000063694.10
|
Klf13
|
Kruppel-like factor 13 |
| chr2_-_120183575 | 0.05 |
ENSMUST00000028752.8
ENSMUST00000102501.10 |
Vps39
|
VPS39 HOPS complex subunit |
| chr4_-_141412910 | 0.05 |
ENSMUST00000105782.2
|
Rsc1a1
|
regulatory solute carrier protein, family 1, member 1 |
| chr15_+_44291470 | 0.05 |
ENSMUST00000226827.2
ENSMUST00000060652.5 |
Eny2
|
ENY2 transcription and export complex 2 subunit |
| chr4_-_14826587 | 0.05 |
ENSMUST00000117268.9
ENSMUST00000236953.2 |
Otud6b
|
OTU domain containing 6B |
| chr9_+_102988940 | 0.04 |
ENSMUST00000189134.2
ENSMUST00000035155.8 |
Rab6b
|
RAB6B, member RAS oncogene family |
| chr8_-_61436249 | 0.04 |
ENSMUST00000004430.14
ENSMUST00000110301.2 ENSMUST00000093490.9 |
Clcn3
|
chloride channel, voltage-sensitive 3 |
| chr8_-_111532495 | 0.04 |
ENSMUST00000150680.2
ENSMUST00000076846.11 |
Il34
|
interleukin 34 |
| chr2_-_53081199 | 0.04 |
ENSMUST00000239398.2
ENSMUST00000076313.14 ENSMUST00000210789.3 |
Prpf40a
|
pre-mRNA processing factor 40A |
| chr5_-_92231314 | 0.04 |
ENSMUST00000169094.8
ENSMUST00000167918.8 |
G3bp2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr2_+_124452493 | 0.04 |
ENSMUST00000103239.10
ENSMUST00000103240.9 |
Sema6d
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| chr11_-_106084334 | 0.04 |
ENSMUST00000007444.14
ENSMUST00000152008.2 ENSMUST00000103072.10 ENSMUST00000106867.2 |
Strada
|
STE20-related kinase adaptor alpha |
| chr12_+_101942222 | 0.03 |
ENSMUST00000047357.10
|
Cpsf2
|
cleavage and polyadenylation specific factor 2 |
| chrX_+_150127171 | 0.02 |
ENSMUST00000073364.6
|
Fam120c
|
family with sequence similarity 120, member C |
| chr16_-_46317318 | 0.02 |
ENSMUST00000023335.13
ENSMUST00000023334.15 |
Nectin3
|
nectin cell adhesion molecule 3 |
| chr12_+_80690985 | 0.02 |
ENSMUST00000219405.2
ENSMUST00000085245.7 |
Slc39a9
|
solute carrier family 39 (zinc transporter), member 9 |
| chr16_-_4497496 | 0.02 |
ENSMUST00000038552.13
ENSMUST00000090480.6 |
Coro7
|
coronin 7 |
| chr13_-_85437118 | 0.02 |
ENSMUST00000109552.3
|
Rasa1
|
RAS p21 protein activator 1 |
| chr19_+_57441332 | 0.01 |
ENSMUST00000026073.14
ENSMUST00000026072.5 ENSMUST00000238107.2 |
Trub1
|
TruB pseudouridine (psi) synthase family member 1 |
| chr4_-_66322695 | 0.01 |
ENSMUST00000084496.3
|
Astn2
|
astrotactin 2 |
| chr4_+_17853452 | 0.01 |
ENSMUST00000029881.10
|
Mmp16
|
matrix metallopeptidase 16 |
| chr6_-_59403264 | 0.00 |
ENSMUST00000051065.6
|
Gprin3
|
GPRIN family member 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.0 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.2 | 1.1 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.2 | 0.6 | GO:1904954 | canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904954) |
| 0.2 | 0.6 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.1 | 0.4 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.1 | 0.5 | GO:0098964 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.1 | 0.6 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.1 | 0.3 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.4 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 2.1 | GO:0046033 | AMP metabolic process(GO:0046033) |
| 0.1 | 0.3 | GO:0021934 | hindbrain tangential cell migration(GO:0021934) |
| 0.1 | 0.3 | GO:0045976 | negative regulation of mitotic cell cycle, embryonic(GO:0045976) |
| 0.1 | 0.5 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 0.2 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.1 | 0.3 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.1 | 0.3 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.1 | 0.6 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.1 | 0.4 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.1 | 0.3 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.1 | 0.2 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.1 | 0.2 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.0 | 0.2 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.5 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.6 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.5 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.3 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.4 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0021627 | olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 0.0 | 0.6 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.8 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.5 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.5 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.8 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.3 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.3 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.1 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) |
| 0.0 | 0.2 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.0 | 0.2 | GO:1904016 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.3 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.2 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.3 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.3 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.0 | 0.1 | GO:0021644 | vagus nerve morphogenesis(GO:0021644) |
| 0.0 | 0.2 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.3 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.5 | GO:0030539 | male genitalia development(GO:0030539) |
| 0.0 | 1.1 | GO:0007140 | male meiosis(GO:0007140) |
| 0.0 | 0.1 | GO:1902966 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.0 | 0.1 | GO:0038042 | dimeric G-protein coupled receptor signaling pathway(GO:0038042) calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.1 | GO:0090327 | negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.0 | 0.3 | GO:0061162 | establishment of monopolar cell polarity(GO:0061162) |
| 0.0 | 0.2 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.3 | GO:0086016 | AV node cell action potential(GO:0086016) AV node cell to bundle of His cell signaling(GO:0086027) |
| 0.0 | 0.1 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.0 | 0.4 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.0 | 0.0 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.0 | 0.3 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.4 | GO:2000637 | positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.0 | 0.1 | GO:0090003 | regulation of Golgi to plasma membrane protein transport(GO:0042996) regulation of establishment of protein localization to plasma membrane(GO:0090003) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.0 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 0.3 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.1 | 0.6 | GO:0044326 | dendritic spine neck(GO:0044326) growth cone lamellipodium(GO:1990761) |
| 0.1 | 0.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.4 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.6 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 1.8 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.2 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.0 | 0.3 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.0 | 0.1 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 0.1 | GO:1903754 | cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 0.0 | 0.5 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.2 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.9 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.4 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.6 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.3 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.3 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.0 | 0.4 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.7 | 4.0 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.3 | 1.1 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.1 | 0.9 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.1 | 0.6 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.5 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.1 | 0.5 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.3 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) ethanol binding(GO:0035276) |
| 0.1 | 0.4 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.4 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 0.5 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 1.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 0.4 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.3 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.3 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.6 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.3 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.3 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.3 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.6 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.6 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.4 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.2 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 1.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.4 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.0 | 0.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0031699 | beta-3 adrenergic receptor binding(GO:0031699) |
| 0.0 | 0.5 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.0 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.2 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.2 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.0 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 3.1 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.7 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.6 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.6 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 1.6 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |