Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for GUAACAG
Z-value: 0.29
miRNA associated with seed GUAACAG
| Name | miRBASE accession |
|---|---|
|
mmu-miR-194-5p
|
MIMAT0000224 |
Activity profile of GUAACAG motif
Sorted Z-values of GUAACAG motif
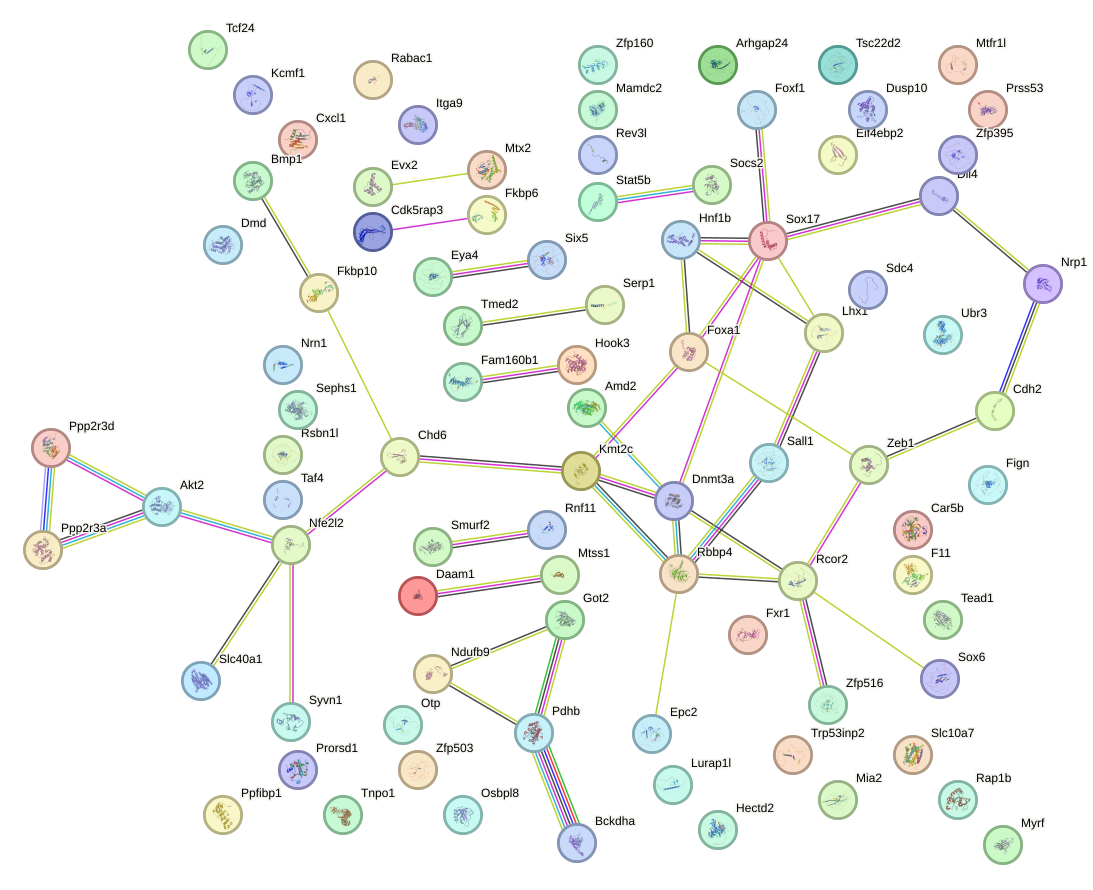
Network of associatons between targets according to the STRING database.
First level regulatory network of GUAACAG
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_64473091 | 1.80 |
ENSMUST00000175965.10
|
Onecut2
|
one cut domain, family member 2 |
| chr12_-_57592907 | 1.16 |
ENSMUST00000044380.8
|
Foxa1
|
forkhead box A1 |
| chr7_+_112278520 | 1.16 |
ENSMUST00000084705.13
ENSMUST00000239442.2 ENSMUST00000239404.2 ENSMUST00000059768.18 |
Tead1
|
TEA domain family member 1 |
| chr16_+_42727926 | 1.14 |
ENSMUST00000151244.8
ENSMUST00000114694.9 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr2_+_155223728 | 0.67 |
ENSMUST00000043237.14
ENSMUST00000174685.8 |
Trp53inp2
|
transformation related protein 53 inducible nuclear protein 2 |
| chr8_-_96615138 | 0.66 |
ENSMUST00000034097.8
|
Got2
|
glutamatic-oxaloacetic transaminase 2, mitochondrial |
| chr13_-_36918424 | 0.65 |
ENSMUST00000037623.15
|
Nrn1
|
neuritin 1 |
| chr10_-_61288437 | 0.62 |
ENSMUST00000167087.2
ENSMUST00000020288.15 |
Eif4ebp2
|
eukaryotic translation initiation factor 4E binding protein 2 |
| chr9_+_118435755 | 0.58 |
ENSMUST00000044165.14
|
Itga9
|
integrin alpha 9 |
| chr7_+_27290969 | 0.54 |
ENSMUST00000108344.9
|
Akt2
|
thymoma viral proto-oncogene 2 |
| chr6_+_145156860 | 0.53 |
ENSMUST00000111725.8
ENSMUST00000111726.10 ENSMUST00000039729.5 ENSMUST00000111723.8 ENSMUST00000111724.8 ENSMUST00000111721.2 ENSMUST00000111719.2 |
Etfrf1
|
electron transfer flavoprotein regulatory factor 1 |
| chr8_-_89770790 | 0.53 |
ENSMUST00000034090.8
|
Sall1
|
spalt like transcription factor 1 |
| chr16_+_43960183 | 0.51 |
ENSMUST00000159514.8
ENSMUST00000161326.8 ENSMUST00000063520.15 ENSMUST00000063542.8 |
Naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr7_-_25358406 | 0.50 |
ENSMUST00000071329.8
|
Bckdha
|
branched chain ketoacid dehydrogenase E1, alpha polypeptide |
| chr18_-_16942289 | 0.50 |
ENSMUST00000025166.14
|
Cdh2
|
cadherin 2 |
| chr8_-_26609153 | 0.49 |
ENSMUST00000037182.14
|
Hook3
|
hook microtubule tethering protein 3 |
| chr8_-_45715049 | 0.49 |
ENSMUST00000034064.5
|
F11
|
coagulation factor XI |
| chr10_-_95252712 | 0.49 |
ENSMUST00000020215.16
|
Socs2
|
suppressor of cytokine signaling 2 |
| chr19_-_23425757 | 0.47 |
ENSMUST00000036069.8
|
Mamdc2
|
MAM domain containing 2 |
| chr14_-_70757601 | 0.47 |
ENSMUST00000022693.9
|
Bmp1
|
bone morphogenetic protein 1 |
| chr12_+_71877838 | 0.46 |
ENSMUST00000223272.2
ENSMUST00000085299.4 |
Daam1
|
dishevelled associated activator of morphogenesis 1 |
| chrX_+_100419965 | 0.46 |
ENSMUST00000119080.8
|
Gjb1
|
gap junction protein, beta 1 |
| chr8_+_129085719 | 0.45 |
ENSMUST00000026917.10
|
Nrp1
|
neuropilin 1 |
| chr2_-_164285097 | 0.45 |
ENSMUST00000017153.4
|
Sdc4
|
syndecan 4 |
| chr1_-_45964730 | 0.45 |
ENSMUST00000027137.11
|
Slc40a1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr2_+_119156265 | 0.40 |
ENSMUST00000102517.4
|
Dll4
|
delta like canonical Notch ligand 4 |
| chr15_+_38869415 | 0.38 |
ENSMUST00000179165.9
|
Fzd6
|
frizzled class receptor 6 |
| chr11_+_83741657 | 0.37 |
ENSMUST00000021016.10
|
Hnf1b
|
HNF1 homeobox B |
| chr14_+_14296748 | 0.36 |
ENSMUST00000022268.10
|
Pdhb
|
pyruvate dehydrogenase (lipoamide) beta |
| chr9_-_101128976 | 0.36 |
ENSMUST00000075941.12
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr8_+_121811091 | 0.35 |
ENSMUST00000181504.2
|
Foxf1
|
forkhead box F1 |
| chr11_-_88609048 | 0.34 |
ENSMUST00000107909.8
|
Msi2
|
musashi RNA-binding protein 2 |
| chr1_-_10038030 | 0.34 |
ENSMUST00000185184.2
|
Tcf24
|
transcription factor 24 |
| chr1_-_4566619 | 0.34 |
ENSMUST00000192913.2
ENSMUST00000027035.10 |
Sox17
|
SRY (sex determining region Y)-box 17 |
| chr2_+_4886298 | 0.33 |
ENSMUST00000027973.14
|
Sephs1
|
selenophosphate synthetase 1 |
| chr11_-_106811507 | 0.33 |
ENSMUST00000103067.10
|
Smurf2
|
SMAD specific E3 ubiquitin protein ligase 2 |
| chr12_+_3857001 | 0.32 |
ENSMUST00000020991.15
ENSMUST00000172509.8 |
Dnmt3a
|
DNA methyltransferase 3A |
| chr7_-_115637970 | 0.32 |
ENSMUST00000166877.8
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr2_-_63928339 | 0.32 |
ENSMUST00000131615.9
|
Fign
|
fidgetin |
| chr7_-_127490139 | 0.31 |
ENSMUST00000205300.2
ENSMUST00000121394.3 |
Prss53
|
protease, serine 53 |
| chr15_-_58953838 | 0.30 |
ENSMUST00000080371.8
|
Mtss1
|
MTSS I-BAR domain containing 1 |
| chr5_-_25703700 | 0.30 |
ENSMUST00000173073.8
ENSMUST00000045291.14 ENSMUST00000173174.2 |
Kmt2c
|
lysine (K)-specific methyltransferase 2C |
| chr15_+_58805605 | 0.29 |
ENSMUST00000022980.5
|
Ndufb9
|
NADH:ubiquinone oxidoreductase subunit B9 |
| chr6_+_146789978 | 0.28 |
ENSMUST00000016631.14
ENSMUST00000203730.3 ENSMUST00000111623.9 |
Ppfibp1
|
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
| chr5_+_91039092 | 0.27 |
ENSMUST00000031327.9
|
Cxcl1
|
chemokine (C-X-C motif) ligand 1 |
| chr19_-_10217968 | 0.27 |
ENSMUST00000189897.2
ENSMUST00000186056.7 ENSMUST00000088013.12 |
Myrf
|
myelin regulatory factor |
| chr10_+_39608094 | 0.26 |
ENSMUST00000019986.13
|
Rev3l
|
REV3 like, DNA directed polymerase zeta catalytic subunit |
| chr14_-_22039543 | 0.25 |
ENSMUST00000043409.9
|
Zfp503
|
zinc finger protein 503 |
| chrX_-_162810959 | 0.25 |
ENSMUST00000033739.5
|
Car5b
|
carbonic anhydrase 5b, mitochondrial |
| chr7_-_45750153 | 0.25 |
ENSMUST00000180081.3
|
Kcnj11
|
potassium inwardly rectifying channel, subfamily J, member 11 |
| chr19_+_36532061 | 0.25 |
ENSMUST00000169036.9
ENSMUST00000047247.12 |
Hectd2
|
HECT domain E3 ubiquitin protein ligase 2 |
| chr11_-_96807273 | 0.25 |
ENSMUST00000103152.11
|
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr18_+_5591864 | 0.24 |
ENSMUST00000025081.13
ENSMUST00000159390.8 |
Zeb1
|
zinc finger E-box binding homeobox 1 |
| chr2_-_160950936 | 0.24 |
ENSMUST00000039782.14
ENSMUST00000134178.8 |
Chd6
|
chromodomain helicase DNA binding protein 6 |
| chr7_+_18828519 | 0.24 |
ENSMUST00000049454.6
|
Six5
|
sine oculis-related homeobox 5 |
| chr2_-_75534985 | 0.24 |
ENSMUST00000102672.5
|
Nfe2l2
|
nuclear factor, erythroid derived 2, like 2 |
| chr10_-_40178182 | 0.23 |
ENSMUST00000099945.6
ENSMUST00000238953.2 ENSMUST00000238969.2 |
Amd1
|
S-adenosylmethionine decarboxylase 1 |
| chr4_-_134262509 | 0.23 |
ENSMUST00000102550.10
|
Mtfr1l
|
mitochondrial fission regulator 1-like |
| chr14_+_65596070 | 0.23 |
ENSMUST00000066994.7
|
Zfp395
|
zinc finger protein 395 |
| chr6_-_72876882 | 0.22 |
ENSMUST00000068697.11
|
Kcmf1
|
potassium channel modulatory factor 1 |
| chr10_+_111000613 | 0.21 |
ENSMUST00000105275.9
|
Osbpl8
|
oxysterol binding protein-like 8 |
| chr11_-_100713348 | 0.21 |
ENSMUST00000107358.9
|
Stat5b
|
signal transducer and activator of transcription 5B |
| chr7_-_24672083 | 0.21 |
ENSMUST00000076961.9
|
Rabac1
|
Rab acceptor 1 (prenylated) |
| chr2_-_179618439 | 0.21 |
ENSMUST00000041618.13
ENSMUST00000227325.2 |
Taf4
|
TATA-box binding protein associated factor 4 |
| chr5_-_21156766 | 0.20 |
ENSMUST00000036489.10
|
Rsbn1l
|
round spermatid basic protein 1-like |
| chr19_+_57349112 | 0.20 |
ENSMUST00000036407.6
|
Fam160b1
|
family with sequence similarity 160, member B1 |
| chr11_-_84416340 | 0.20 |
ENSMUST00000018842.14
|
Lhx1
|
LIM homeobox protein 1 |
| chr2_+_49341498 | 0.20 |
ENSMUST00000092123.11
|
Epc2
|
enhancer of polycomb homolog 2 |
| chr3_-_58433313 | 0.19 |
ENSMUST00000029385.9
|
Serp1
|
stress-associated endoplasmic reticulum protein 1 |
| chr2_-_74489763 | 0.19 |
ENSMUST00000173623.2
ENSMUST00000001867.13 |
Evx2
|
even-skipped homeobox 2 |
| chr10_-_117681864 | 0.19 |
ENSMUST00000064667.9
|
Rap1b
|
RAS related protein 1b |
| chr10_-_121933276 | 0.19 |
ENSMUST00000140299.3
|
Rxylt1
|
ribitol xylosyltransferase 1 |
| chr12_+_59178072 | 0.19 |
ENSMUST00000176464.8
ENSMUST00000170992.9 ENSMUST00000176322.8 |
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr15_+_81284333 | 0.19 |
ENSMUST00000163754.9
ENSMUST00000041609.11 |
Xpnpep3
|
X-prolyl aminopeptidase 3, mitochondrial |
| chr19_+_6096606 | 0.18 |
ENSMUST00000138532.8
ENSMUST00000129081.8 ENSMUST00000156550.8 |
Syvn1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr1_+_183766572 | 0.18 |
ENSMUST00000048655.8
|
Dusp10
|
dual specificity phosphatase 10 |
| chr2_+_74656145 | 0.18 |
ENSMUST00000028511.8
|
Mtx2
|
metaxin 2 |
| chr11_-_95966477 | 0.17 |
ENSMUST00000090541.12
|
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 (subunit 9) |
| chr11_-_29464998 | 0.17 |
ENSMUST00000133103.2
ENSMUST00000039900.4 |
Prorsd1
|
prolyl-tRNA synthetase domain containing 1 |
| chr2_+_69727563 | 0.17 |
ENSMUST00000055758.16
ENSMUST00000112251.9 |
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr4_+_80828883 | 0.17 |
ENSMUST00000055922.4
|
Lurap1l
|
leucine rich adaptor protein 1-like |
| chr8_+_79235946 | 0.17 |
ENSMUST00000209490.2
ENSMUST00000034111.10 |
Slc10a7
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 7 |
| chr13_+_95012107 | 0.17 |
ENSMUST00000022195.13
|
Otp
|
orthopedia homeobox |
| chr3_+_34074048 | 0.17 |
ENSMUST00000001620.13
|
Fxr1
|
fragile X mental retardation gene 1, autosomal homolog |
| chr5_+_124678758 | 0.16 |
ENSMUST00000060226.11
|
Tmed2
|
transmembrane p24 trafficking protein 2 |
| chr18_+_82928959 | 0.16 |
ENSMUST00000171238.8
|
Zfp516
|
zinc finger protein 516 |
| chr10_-_23225781 | 0.16 |
ENSMUST00000092665.12
|
Eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr4_-_129229159 | 0.16 |
ENSMUST00000102598.4
|
Rbbp4
|
retinoblastoma binding protein 4, chromatin remodeling factor |
| chr4_-_109333866 | 0.16 |
ENSMUST00000030284.10
|
Rnf11
|
ring finger protein 11 |
| chr7_+_120276801 | 0.16 |
ENSMUST00000208454.2
ENSMUST00000060175.8 |
Mosmo
|
modulator of smoothened |
| chr5_+_102629240 | 0.15 |
ENSMUST00000073302.12
ENSMUST00000094559.9 |
Arhgap24
|
Rho GTPase activating protein 24 |
| chrX_+_81992467 | 0.15 |
ENSMUST00000114000.8
|
Dmd
|
dystrophin, muscular dystrophy |
| chr13_-_99027544 | 0.15 |
ENSMUST00000109399.9
|
Tnpo1
|
transportin 1 |
| chr12_+_111005768 | 0.15 |
ENSMUST00000084968.14
|
Rcor1
|
REST corepressor 1 |
| chr5_-_135378896 | 0.15 |
ENSMUST00000201534.2
ENSMUST00000044972.11 |
Fkbp6
|
FK506 binding protein 6 |
| chr3_+_58322119 | 0.14 |
ENSMUST00000099090.7
ENSMUST00000199164.2 |
Tsc22d2
|
TSC22 domain family, member 2 |
| chr1_+_131838294 | 0.14 |
ENSMUST00000062264.8
|
Nucks1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr17_+_74835290 | 0.14 |
ENSMUST00000180037.8
|
Birc6
|
baculoviral IAP repeat-containing 6 |
| chr3_-_51184895 | 0.14 |
ENSMUST00000108051.8
ENSMUST00000108053.9 |
Elf2
|
E74-like factor 2 |
| chr16_+_34842764 | 0.13 |
ENSMUST00000061156.10
|
Hacd2
|
3-hydroxyacyl-CoA dehydratase 2 |
| chr3_+_135144202 | 0.13 |
ENSMUST00000166033.6
|
Ube2d3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr2_+_30842966 | 0.12 |
ENSMUST00000028199.12
|
Tor1b
|
torsin family 1, member B |
| chr13_-_47083194 | 0.12 |
ENSMUST00000056978.8
|
Kif13a
|
kinesin family member 13A |
| chr19_+_7471398 | 0.12 |
ENSMUST00000170373.9
ENSMUST00000236308.2 ENSMUST00000235557.2 |
Atl3
|
atlastin GTPase 3 |
| chr17_+_75312520 | 0.12 |
ENSMUST00000234490.2
ENSMUST00000001927.12 |
Ltbp1
|
latent transforming growth factor beta binding protein 1 |
| chr5_-_21260878 | 0.12 |
ENSMUST00000030556.8
|
Ptpn12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr14_+_34395845 | 0.11 |
ENSMUST00000048263.14
|
Wapl
|
WAPL cohesin release factor |
| chr14_+_123897383 | 0.11 |
ENSMUST00000049681.14
|
Itgbl1
|
integrin, beta-like 1 |
| chr9_+_44516140 | 0.11 |
ENSMUST00000170489.2
ENSMUST00000217034.2 |
Ddx6
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 6 |
| chr13_-_38712387 | 0.11 |
ENSMUST00000035988.16
|
Txndc5
|
thioredoxin domain containing 5 |
| chr1_+_93163553 | 0.11 |
ENSMUST00000062202.14
|
Sned1
|
sushi, nidogen and EGF-like domains 1 |
| chr18_+_35731658 | 0.11 |
ENSMUST00000041314.17
ENSMUST00000236666.2 ENSMUST00000236020.2 ENSMUST00000235400.2 |
Paip2
|
polyadenylate-binding protein-interacting protein 2 |
| chr10_+_51553852 | 0.11 |
ENSMUST00000122922.10
ENSMUST00000219364.2 |
Rfx6
|
regulatory factor X, 6 |
| chr9_+_47441471 | 0.11 |
ENSMUST00000114548.8
ENSMUST00000152459.8 ENSMUST00000143026.9 ENSMUST00000085909.9 ENSMUST00000114547.8 ENSMUST00000239368.2 ENSMUST00000214542.2 ENSMUST00000034581.4 |
Cadm1
|
cell adhesion molecule 1 |
| chr3_-_32791296 | 0.10 |
ENSMUST00000043966.8
|
Mrpl47
|
mitochondrial ribosomal protein L47 |
| chr13_+_96679233 | 0.10 |
ENSMUST00000077672.12
ENSMUST00000109444.3 |
Cert1
|
ceramide transporter 1 |
| chr7_-_110682204 | 0.10 |
ENSMUST00000161051.8
ENSMUST00000160132.8 ENSMUST00000162415.9 |
Eif4g2
|
eukaryotic translation initiation factor 4, gamma 2 |
| chr5_-_124387812 | 0.10 |
ENSMUST00000162812.8
|
Pitpnm2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chr8_-_69848167 | 0.10 |
ENSMUST00000072427.7
ENSMUST00000213012.2 ENSMUST00000239456.2 |
Gm10033
|
predicted gene 10033 |
| chr12_-_85421467 | 0.10 |
ENSMUST00000040766.9
|
Tmed10
|
transmembrane p24 trafficking protein 10 |
| chr7_+_58878490 | 0.10 |
ENSMUST00000202945.4
ENSMUST00000107537.5 |
Ube3a
|
ubiquitin protein ligase E3A |
| chr16_-_62607105 | 0.10 |
ENSMUST00000152553.2
ENSMUST00000063089.12 |
Nsun3
|
NOL1/NOP2/Sun domain family member 3 |
| chr1_+_191638854 | 0.10 |
ENSMUST00000044954.7
|
Slc30a1
|
solute carrier family 30 (zinc transporter), member 1 |
| chrX_+_10583629 | 0.09 |
ENSMUST00000115524.8
ENSMUST00000008179.7 ENSMUST00000156321.2 |
Mid1ip1
|
Mid1 interacting protein 1 (gastrulation specific G12-like (zebrafish)) |
| chr14_-_20844074 | 0.09 |
ENSMUST00000080440.14
ENSMUST00000100837.11 ENSMUST00000071816.7 |
Camk2g
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr2_+_32041083 | 0.08 |
ENSMUST00000036691.14
ENSMUST00000069817.15 |
Prrc2b
|
proline-rich coiled-coil 2B |
| chr7_-_99508117 | 0.08 |
ENSMUST00000209032.2
ENSMUST00000036274.8 |
Spcs2
|
signal peptidase complex subunit 2 homolog (S. cerevisiae) |
| chr13_+_49495025 | 0.08 |
ENSMUST00000048544.14
ENSMUST00000110084.4 ENSMUST00000110085.11 |
Bicd2
|
BICD cargo adaptor 2 |
| chr6_-_148113872 | 0.08 |
ENSMUST00000136008.8
|
Ergic2
|
ERGIC and golgi 2 |
| chrX_-_37653396 | 0.08 |
ENSMUST00000016681.15
|
Cul4b
|
cullin 4B |
| chr15_-_96358612 | 0.08 |
ENSMUST00000047835.8
|
Scaf11
|
SR-related CTD-associated factor 11 |
| chr4_-_122854966 | 0.08 |
ENSMUST00000030408.12
ENSMUST00000127047.2 |
Mfsd2a
|
major facilitator superfamily domain containing 2A |
| chr10_-_35587888 | 0.07 |
ENSMUST00000080898.4
|
Amd2
|
S-adenosylmethionine decarboxylase 2 |
| chr5_+_124577952 | 0.07 |
ENSMUST00000059580.11
|
Kmt5a
|
lysine methyltransferase 5A |
| chr2_-_35994072 | 0.07 |
ENSMUST00000112961.10
ENSMUST00000112966.10 |
Lhx6
|
LIM homeobox protein 6 |
| chr1_-_39844467 | 0.07 |
ENSMUST00000171319.4
|
Gm3646
|
predicted gene 3646 |
| chr13_+_55740948 | 0.07 |
ENSMUST00000109905.5
|
Tmed9
|
transmembrane p24 trafficking protein 9 |
| chr15_-_38079089 | 0.07 |
ENSMUST00000110336.4
|
Ubr5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chr5_+_103573367 | 0.07 |
ENSMUST00000048957.11
|
Ptpn13
|
protein tyrosine phosphatase, non-receptor type 13 |
| chr3_-_104960437 | 0.07 |
ENSMUST00000077548.12
|
Cttnbp2nl
|
CTTNBP2 N-terminal like |
| chr2_-_18397547 | 0.07 |
ENSMUST00000091418.12
ENSMUST00000166495.8 |
Dnajc1
|
DnaJ heat shock protein family (Hsp40) member C1 |
| chr2_-_156728988 | 0.07 |
ENSMUST00000029164.9
|
Sla2
|
Src-like-adaptor 2 |
| chr6_+_8259379 | 0.07 |
ENSMUST00000162034.8
|
Umad1
|
UMAP1-MVP12 associated (UMA) domain containing 1 |
| chr3_-_89325594 | 0.07 |
ENSMUST00000029679.4
|
Cks1b
|
CDC28 protein kinase 1b |
| chr11_-_82719850 | 0.06 |
ENSMUST00000021036.13
ENSMUST00000074515.11 ENSMUST00000103218.3 |
Rffl
|
ring finger and FYVE like domain containing protein |
| chr5_-_144902598 | 0.06 |
ENSMUST00000110677.8
ENSMUST00000085684.11 ENSMUST00000100461.7 |
Smurf1
|
SMAD specific E3 ubiquitin protein ligase 1 |
| chr3_-_141687987 | 0.06 |
ENSMUST00000029948.15
|
Bmpr1b
|
bone morphogenetic protein receptor, type 1B |
| chr17_-_65920481 | 0.06 |
ENSMUST00000024897.10
|
Vapa
|
vesicle-associated membrane protein, associated protein A |
| chr1_+_90981144 | 0.06 |
ENSMUST00000068167.13
ENSMUST00000097649.10 ENSMUST00000186762.7 ENSMUST00000097650.10 |
Lrrfip1
|
leucine rich repeat (in FLII) interacting protein 1 |
| chr9_+_72714156 | 0.05 |
ENSMUST00000055535.9
|
Prtg
|
protogenin |
| chr12_-_21423551 | 0.05 |
ENSMUST00000101551.10
|
Adam17
|
a disintegrin and metallopeptidase domain 17 |
| chr5_-_52347826 | 0.05 |
ENSMUST00000199321.5
ENSMUST00000195922.2 ENSMUST00000031061.12 |
Dhx15
|
DEAH (Asp-Glu-Ala-His) box polypeptide 15 |
| chr1_+_132345293 | 0.05 |
ENSMUST00000045110.14
ENSMUST00000188389.2 |
Dstyk
|
dual serine/threonine and tyrosine protein kinase |
| chr18_-_77801624 | 0.04 |
ENSMUST00000074653.6
|
8030462N17Rik
|
RIKEN cDNA 8030462N17 gene |
| chr12_-_84265609 | 0.04 |
ENSMUST00000046266.13
ENSMUST00000220974.2 |
Mideas
|
mitotic deacetylase associated SANT domain protein |
| chr8_+_108020092 | 0.04 |
ENSMUST00000169453.8
|
Nfat5
|
nuclear factor of activated T cells 5 |
| chr4_-_108637979 | 0.04 |
ENSMUST00000106657.8
|
Zfyve9
|
zinc finger, FYVE domain containing 9 |
| chr8_+_85897951 | 0.04 |
ENSMUST00000076896.6
|
Cks1brt
|
CDC28 protein kinase 1b, retrogene |
| chr5_-_123038329 | 0.04 |
ENSMUST00000031435.14
|
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr12_-_102844537 | 0.04 |
ENSMUST00000045652.8
ENSMUST00000223554.2 |
Btbd7
|
BTB (POZ) domain containing 7 |
| chr9_-_50571080 | 0.03 |
ENSMUST00000034567.4
|
Dlat
|
dihydrolipoamide S-acetyltransferase (E2 component of pyruvate dehydrogenase complex) |
| chr16_+_11140740 | 0.03 |
ENSMUST00000180792.8
|
Snx29
|
sorting nexin 29 |
| chr1_-_86287080 | 0.03 |
ENSMUST00000027438.8
|
Ncl
|
nucleolin |
| chr12_-_100486950 | 0.03 |
ENSMUST00000223020.2
ENSMUST00000062957.8 |
Ttc7b
|
tetratricopeptide repeat domain 7B |
| chr9_+_7184514 | 0.03 |
ENSMUST00000215683.2
ENSMUST00000034499.10 |
Dcun1d5
|
DCN1, defective in cullin neddylation 1, domain containing 5 (S. cerevisiae) |
| chr11_-_116197523 | 0.03 |
ENSMUST00000133468.2
ENSMUST00000106411.10 ENSMUST00000106413.10 ENSMUST00000021147.14 |
Exoc7
|
exocyst complex component 7 |
| chr3_+_99048379 | 0.03 |
ENSMUST00000004343.7
|
Wars2
|
tryptophanyl tRNA synthetase 2 (mitochondrial) |
| chr18_+_10725532 | 0.03 |
ENSMUST00000052838.11
|
Mib1
|
mindbomb E3 ubiquitin protein ligase 1 |
| chr14_+_121272950 | 0.02 |
ENSMUST00000026635.8
|
Farp1
|
FERM, RhoGEF (Arhgef) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr16_+_32219324 | 0.02 |
ENSMUST00000115149.3
|
Tm4sf19
|
transmembrane 4 L six family member 19 |
| chr9_+_70450130 | 0.02 |
ENSMUST00000049263.9
|
Sltm
|
SAFB-like, transcription modulator |
| chr13_+_84370405 | 0.02 |
ENSMUST00000057495.10
ENSMUST00000225069.2 |
Tmem161b
|
transmembrane protein 161B |
| chr6_+_51500881 | 0.01 |
ENSMUST00000049152.15
|
Snx10
|
sorting nexin 10 |
| chr7_-_109585649 | 0.01 |
ENSMUST00000094097.12
|
Tmem41b
|
transmembrane protein 41B |
| chr1_-_64776890 | 0.01 |
ENSMUST00000116133.4
ENSMUST00000063982.7 |
Fzd5
|
frizzled class receptor 5 |
| chr7_-_133826817 | 0.01 |
ENSMUST00000067680.11
|
Adam12
|
a disintegrin and metallopeptidase domain 12 (meltrin alpha) |
| chr5_-_64126194 | 0.01 |
ENSMUST00000154169.4
|
Rell1
|
RELT-like 1 |
| chr10_-_128727542 | 0.01 |
ENSMUST00000026408.7
|
Gdf11
|
growth differentiation factor 11 |
| chr1_+_166828982 | 0.01 |
ENSMUST00000165874.8
ENSMUST00000190081.7 |
Fam78b
|
family with sequence similarity 78, member B |
| chr18_+_65564009 | 0.00 |
ENSMUST00000224056.3
ENSMUST00000049248.7 |
Malt1
|
MALT1 paracaspase |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.2 | 0.7 | GO:0006533 | fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) |
| 0.1 | 0.4 | GO:0097374 | cell migration involved in vasculogenesis(GO:0035441) neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) sensory neuron axon guidance(GO:0097374) |
| 0.1 | 0.4 | GO:0070839 | divalent metal ion export(GO:0070839) |
| 0.1 | 0.4 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.1 | 0.5 | GO:2000384 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.1 | 0.5 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 0.4 | GO:0007439 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.1 | 0.3 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 0.5 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.1 | 0.4 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.1 | 0.4 | GO:0061235 | regulation of pronephros size(GO:0035565) mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 0.1 | 0.3 | GO:0070949 | neutrophil mediated killing of fungus(GO:0070947) regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.1 | 0.5 | GO:0071486 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.1 | 1.8 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 0.3 | GO:0060807 | cardiogenic plate morphogenesis(GO:0003142) regulation of transcription from RNA polymerase II promoter involved in definitive endodermal cell fate specification(GO:0060807) |
| 0.1 | 0.4 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.1 | 0.5 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.1 | 0.3 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.1 | 0.2 | GO:0097065 | cervix development(GO:0060067) anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.1 | 1.2 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.1 | 0.3 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 0.3 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.1 | 0.4 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.1 | 0.3 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.2 | GO:0021629 | olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 0.0 | 0.5 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.4 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.2 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.2 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.0 | 0.1 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.0 | 0.5 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.5 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 0.5 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.2 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.1 | GO:0019046 | release from viral latency(GO:0019046) regulation of DNA strand elongation(GO:0060382) |
| 0.0 | 0.4 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.0 | 0.1 | GO:0035037 | sperm entry(GO:0035037) |
| 0.0 | 0.1 | GO:0002946 | tRNA C5-cytosine methylation(GO:0002946) |
| 0.0 | 0.2 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.2 | GO:0046544 | development of secondary male sexual characteristics(GO:0046544) |
| 0.0 | 0.3 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.3 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.7 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.3 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.1 | GO:0051977 | lysophospholipid transport(GO:0051977) |
| 0.0 | 0.5 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.1 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.1 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 1.1 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0071211 | protein targeting to vacuole involved in autophagy(GO:0071211) |
| 0.0 | 0.1 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.2 | GO:0014842 | regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.0 | 0.1 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.5 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.1 | GO:1905077 | negative regulation of interleukin-17 secretion(GO:1905077) |
| 0.0 | 0.2 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.1 | GO:0061687 | detoxification of inorganic compound(GO:0061687) |
| 0.0 | 0.2 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.2 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 0.5 | GO:0061036 | positive regulation of cartilage development(GO:0061036) |
| 0.0 | 0.1 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.2 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.3 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.1 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.1 | 0.6 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) |
| 0.1 | 0.5 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.1 | 0.5 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 0.3 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 0.4 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.4 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.5 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.1 | GO:0038045 | large latent transforming growth factor-beta complex(GO:0038045) |
| 0.0 | 0.2 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.5 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.3 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.5 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.5 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.8 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.3 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.2 | 0.5 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.1 | 0.4 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.1 | 0.3 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.1 | 0.5 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.1 | 0.4 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 1.2 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 0.1 | GO:0080023 | 3R-hydroxyacyl-CoA dehydratase activity(GO:0080023) |
| 0.0 | 0.3 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.4 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.6 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.3 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.5 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.4 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.5 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.5 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.5 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.2 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.0 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.2 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 2.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.6 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.5 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.9 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.5 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.5 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.4 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 1.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.5 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.5 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.7 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.5 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |