Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for GUGCAAA
Z-value: 0.73
miRNA associated with seed GUGCAAA
| Name | miRBASE accession |
|---|---|
|
mmu-miR-19a-3p
|
MIMAT0000651 |
|
mmu-miR-19b-3p
|
MIMAT0000513 |
Activity profile of GUGCAAA motif
Sorted Z-values of GUGCAAA motif
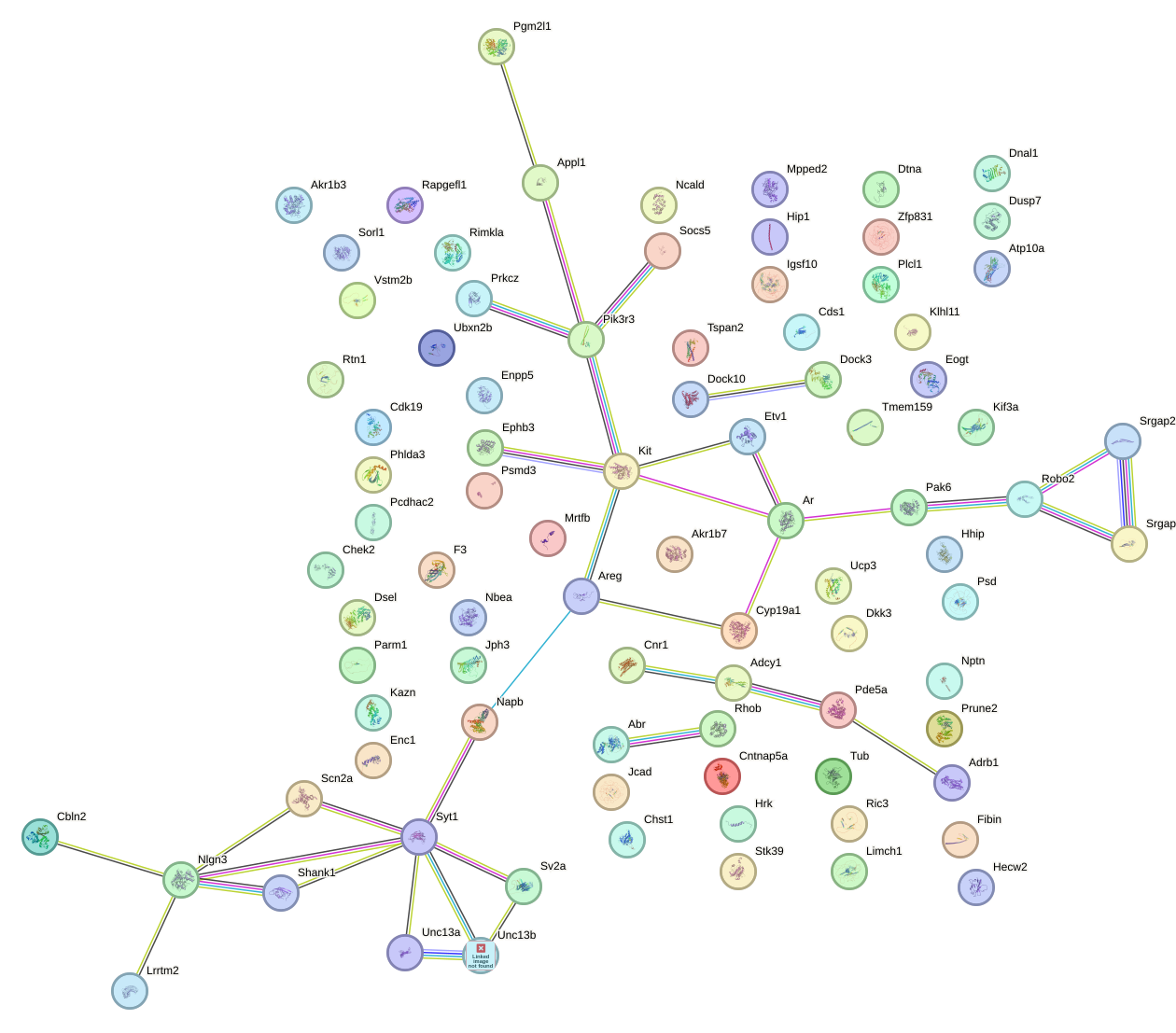
Network of associatons between targets according to the STRING database.
First level regulatory network of GUGCAAA
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_117988399 | 4.64 |
ENSMUST00000164960.4
|
Rasgef1a
|
RasGEF domain family, member 1A |
| chr3_+_102641822 | 4.54 |
ENSMUST00000029451.12
|
Tspan2
|
tetraspanin 2 |
| chr12_-_72455708 | 4.32 |
ENSMUST00000078505.14
|
Rtn1
|
reticulon 1 |
| chr7_-_111758241 | 4.02 |
ENSMUST00000033036.7
|
Dkk3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr2_-_148574353 | 4.01 |
ENSMUST00000028926.13
|
Napb
|
N-ethylmaleimide sensitive fusion protein attachment protein beta |
| chr10_+_56253418 | 3.80 |
ENSMUST00000068581.9
ENSMUST00000217789.2 |
Gja1
|
gap junction protein, alpha 1 |
| chr4_-_141966662 | 3.66 |
ENSMUST00000036476.10
|
Kazn
|
kazrin, periplakin interacting protein |
| chr7_+_108610032 | 3.58 |
ENSMUST00000033341.12
|
Tub
|
tubby bipartite transcription factor |
| chr6_-_92683136 | 3.41 |
ENSMUST00000032093.12
|
Prickle2
|
prickle planar cell polarity protein 2 |
| chr5_+_91665474 | 3.34 |
ENSMUST00000040576.10
|
Parm1
|
prostate androgen-regulated mucin-like protein 1 |
| chr15_-_37792635 | 3.09 |
ENSMUST00000090150.11
ENSMUST00000150453.2 |
Ncald
|
neurocalcin delta |
| chr8_-_72124359 | 2.99 |
ENSMUST00000177517.8
ENSMUST00000030170.15 |
Unc13a
|
unc-13 homolog A |
| chr19_-_46315543 | 2.92 |
ENSMUST00000223917.2
ENSMUST00000224447.2 ENSMUST00000041391.5 ENSMUST00000096029.12 |
Psd
|
pleckstrin and Sec7 domain containing |
| chr1_-_111792599 | 2.92 |
ENSMUST00000035462.7
|
Dsel
|
dermatan sulfate epimerase-like |
| chr7_-_108682491 | 2.88 |
ENSMUST00000120876.2
ENSMUST00000055993.13 |
Ric3
|
RIC3 acetylcholine receptor chaperone |
| chr2_-_110193502 | 2.83 |
ENSMUST00000099626.5
|
Fibin
|
fin bud initiation factor homolog (zebrafish) |
| chr3_+_122522592 | 2.82 |
ENSMUST00000066728.10
|
Pde5a
|
phosphodiesterase 5A, cGMP-specific |
| chr4_+_33924632 | 2.76 |
ENSMUST00000057188.7
|
Cnr1
|
cannabinoid receptor 1 (brain) |
| chr13_+_97377604 | 2.66 |
ENSMUST00000041623.9
|
Enc1
|
ectodermal-neural cortex 1 |
| chr12_+_38830081 | 2.64 |
ENSMUST00000095767.11
|
Etv1
|
ets variant 1 |
| chr18_+_23548455 | 2.62 |
ENSMUST00000115832.4
|
Dtna
|
dystrobrevin alpha |
| chr2_+_92430043 | 2.62 |
ENSMUST00000065797.7
|
Chst1
|
carbohydrate sulfotransferase 1 |
| chr2_+_118493713 | 2.60 |
ENSMUST00000099557.10
|
Pak6
|
p21 (RAC1) activated kinase 6 |
| chr8_+_122457302 | 2.59 |
ENSMUST00000026357.12
|
Jph3
|
junctophilin 3 |
| chr18_+_4634878 | 2.52 |
ENSMUST00000037029.7
|
Jcad
|
junctional cadherin 5 associated |
| chr3_-_56091096 | 2.50 |
ENSMUST00000029374.8
|
Nbea
|
neurobeachin |
| chr11_+_53458188 | 2.48 |
ENSMUST00000057330.15
ENSMUST00000120613.9 ENSMUST00000173744.8 ENSMUST00000118353.9 |
Kif3a
|
kinesin family member 3A |
| chr11_-_61470462 | 2.43 |
ENSMUST00000147501.8
ENSMUST00000146455.8 ENSMUST00000108711.8 ENSMUST00000108712.8 ENSMUST00000001063.15 ENSMUST00000108713.8 ENSMUST00000179936.8 ENSMUST00000178202.8 |
Epn2
|
epsin 2 |
| chr8_-_68427217 | 2.41 |
ENSMUST00000098696.10
ENSMUST00000038959.16 ENSMUST00000093469.11 |
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr16_-_74208180 | 2.37 |
ENSMUST00000117200.8
|
Robo2
|
roundabout guidance receptor 2 |
| chr5_+_75735576 | 2.36 |
ENSMUST00000144270.8
ENSMUST00000005815.7 |
Kit
|
KIT proto-oncogene receptor tyrosine kinase |
| chr7_+_58307930 | 2.36 |
ENSMUST00000168747.3
|
Atp10a
|
ATPase, class V, type 10A |
| chr16_+_13176238 | 2.34 |
ENSMUST00000149359.2
|
Mrtfb
|
myocardin related transcription factor B |
| chr11_+_7013422 | 2.30 |
ENSMUST00000020706.5
|
Adcy1
|
adenylate cyclase 1 |
| chr17_+_44389704 | 2.28 |
ENSMUST00000154166.8
ENSMUST00000024756.5 |
Enpp5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 |
| chr9_-_117081518 | 2.27 |
ENSMUST00000111773.10
ENSMUST00000068962.14 ENSMUST00000044901.14 |
Rbms3
|
RNA binding motif, single stranded interacting protein |
| chr12_-_8550003 | 2.24 |
ENSMUST00000067384.6
|
Rhob
|
ras homolog family member B |
| chr7_+_43959637 | 2.23 |
ENSMUST00000107938.8
|
Shank1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr4_+_116078830 | 2.21 |
ENSMUST00000030464.14
|
Pik3r3
|
phosphoinositide-3-kinase regulatory subunit 3 |
| chr7_+_4463686 | 2.21 |
ENSMUST00000167298.2
ENSMUST00000171445.8 |
Eps8l1
|
EPS8-like 1 |
| chr1_+_135693818 | 2.20 |
ENSMUST00000038945.6
|
Phlda3
|
pleckstrin homology like domain, family A, member 3 |
| chr6_-_97125817 | 2.19 |
ENSMUST00000204331.3
ENSMUST00000142116.2 ENSMUST00000113387.8 |
Eogt
|
EGF domain-specific O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr11_+_98727611 | 2.12 |
ENSMUST00000107479.3
|
Rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr1_-_54233207 | 2.11 |
ENSMUST00000120904.8
|
Hecw2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr16_+_21023505 | 2.11 |
ENSMUST00000006112.7
|
Ephb3
|
Eph receptor B3 |
| chr1_+_115612458 | 2.07 |
ENSMUST00000043725.9
|
Cntnap5a
|
contactin associated protein-like 5A |
| chr10_-_108846816 | 2.07 |
ENSMUST00000105276.8
ENSMUST00000064054.14 |
Syt1
|
synaptotagmin I |
| chrX_+_100342749 | 2.03 |
ENSMUST00000118111.8
ENSMUST00000130555.8 ENSMUST00000151528.8 |
Nlgn3
|
neuroligin 3 |
| chr7_+_119701670 | 2.02 |
ENSMUST00000118737.3
|
Tmem159
|
transmembrane protein 159 |
| chr6_-_112924205 | 2.01 |
ENSMUST00000088373.11
|
Srgap3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr2_-_151474391 | 2.00 |
ENSMUST00000137936.2
ENSMUST00000146172.8 ENSMUST00000094456.10 ENSMUST00000148755.8 ENSMUST00000109875.8 ENSMUST00000028951.14 ENSMUST00000109877.10 |
Snph
|
syntaphilin |
| chr19_+_16933471 | 2.00 |
ENSMUST00000087689.5
|
Prune2
|
prune homolog 2 |
| chr2_+_145009625 | 2.00 |
ENSMUST00000110007.8
|
Slc24a3
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 |
| chr3_-_125732255 | 2.00 |
ENSMUST00000057944.12
|
Ugt8a
|
UDP galactosyltransferase 8A |
| chr18_-_35348049 | 1.97 |
ENSMUST00000091636.5
ENSMUST00000236680.2 |
Lrrtm2
|
leucine rich repeat transmembrane neuronal 2 |
| chr4_+_6191084 | 1.96 |
ENSMUST00000029907.6
|
Ubxn2b
|
UBX domain protein 2B |
| chr12_+_84161095 | 1.94 |
ENSMUST00000123491.8
ENSMUST00000046340.9 ENSMUST00000136159.2 |
Dnal1
|
dynein, axonemal, light chain 1 |
| chr7_+_99876515 | 1.84 |
ENSMUST00000084935.11
|
Pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr14_-_26693189 | 1.81 |
ENSMUST00000036570.5
|
Appl1
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 1 |
| chr3_-_59251573 | 1.80 |
ENSMUST00000193455.6
ENSMUST00000039419.12 ENSMUST00000195983.2 |
Igsf10
|
immunoglobulin superfamily, member 10 |
| chr7_+_40547608 | 1.79 |
ENSMUST00000044705.12
|
Vstm2b
|
V-set and transmembrane domain containing 2B |
| chr6_-_29212239 | 1.78 |
ENSMUST00000160878.8
|
Impdh1
|
inosine monophosphate dehydrogenase 1 |
| chr11_-_76400245 | 1.76 |
ENSMUST00000094012.11
|
Abr
|
active BCR-related gene |
| chr9_+_106245792 | 1.76 |
ENSMUST00000172306.3
|
Dusp7
|
dual specificity phosphatase 7 |
| chr2_-_68302612 | 1.74 |
ENSMUST00000102715.4
|
Stk39
|
serine/threonine kinase 39 |
| chr4_-_155430153 | 1.73 |
ENSMUST00000103178.11
|
Prkcz
|
protein kinase C, zeta |
| chr5_+_67125759 | 1.73 |
ENSMUST00000238993.2
ENSMUST00000038188.14 |
Limch1
|
LIM and calponin homology domains 1 |
| chr11_-_100363551 | 1.71 |
ENSMUST00000056665.4
|
Klhl11
|
kelch-like 11 |
| chr2_+_174485327 | 1.70 |
ENSMUST00000059452.6
|
Zfp831
|
zinc finger protein 831 |
| chr9_-_42035560 | 1.69 |
ENSMUST00000060989.9
|
Sorl1
|
sortilin-related receptor, LDLR class A repeats-containing |
| chr4_+_152181155 | 1.69 |
ENSMUST00000105661.10
ENSMUST00000084115.4 |
Plekhg5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
| chr4_-_119349760 | 1.69 |
ENSMUST00000049994.8
|
Rimkla
|
ribosomal modification protein rimK-like family member A |
| chr2_+_65451100 | 1.66 |
ENSMUST00000144254.6
ENSMUST00000028377.14 |
Scn2a
|
sodium channel, voltage-gated, type II, alpha |
| chr1_-_80736165 | 1.65 |
ENSMUST00000077946.12
|
Dock10
|
dedicator of cytokinesis 10 |
| chr2_+_106523532 | 1.63 |
ENSMUST00000111063.8
|
Mpped2
|
metallophosphoesterase domain containing 2 |
| chr5_+_101912939 | 1.61 |
ENSMUST00000031273.9
|
Cds1
|
CDP-diacylglycerol synthase 1 |
| chr9_-_107109108 | 1.60 |
ENSMUST00000044532.11
|
Dock3
|
dedicator of cyto-kinesis 3 |
| chr5_+_118307754 | 1.60 |
ENSMUST00000054836.7
|
Hrk
|
harakiri, BCL2 interacting protein (contains only BH3 domain) |
| chr19_+_56710570 | 1.59 |
ENSMUST00000038949.6
|
Adrb1
|
adrenergic receptor, beta 1 |
| chr9_+_58536386 | 1.58 |
ENSMUST00000176250.2
|
Nptn
|
neuroplastin |
| chr17_+_87415049 | 1.58 |
ENSMUST00000041369.8
ENSMUST00000234803.2 |
Socs5
|
suppressor of cytokine signaling 5 |
| chr18_+_86729184 | 1.58 |
ENSMUST00000068423.10
|
Cbln2
|
cerebellin 2 precursor protein |
| chr3_+_96088467 | 1.58 |
ENSMUST00000035371.9
|
Sv2a
|
synaptic vesicle glycoprotein 2 a |
| chr7_+_100122192 | 1.57 |
ENSMUST00000032958.14
ENSMUST00000107059.2 |
Ucp3
|
uncoupling protein 3 (mitochondrial, proton carrier) |
| chr5_-_135573962 | 1.56 |
ENSMUST00000060311.12
|
Hip1
|
huntingtin interacting protein 1 |
| chr1_+_55445033 | 1.51 |
ENSMUST00000042986.10
|
Plcl1
|
phospholipase C-like 1 |
| chr3_+_121517158 | 1.49 |
ENSMUST00000029771.13
|
F3
|
coagulation factor III |
| chrX_+_97192356 | 1.48 |
ENSMUST00000052837.9
|
Ar
|
androgen receptor |
| chr10_+_40225272 | 1.47 |
ENSMUST00000044672.11
ENSMUST00000095743.4 |
Cdk19
|
cyclin-dependent kinase 19 |
| chr16_+_30418535 | 1.45 |
ENSMUST00000059078.4
|
Fam43a
|
family with sequence similarity 43, member A |
| chr13_+_76532470 | 1.44 |
ENSMUST00000125209.8
|
Mctp1
|
multiple C2 domains, transmembrane 1 |
| chrX_-_156826262 | 1.43 |
ENSMUST00000026750.15
ENSMUST00000112513.2 |
Cnksr2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr11_+_110956980 | 1.42 |
ENSMUST00000042970.3
|
Kcnj2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr3_+_157742299 | 1.42 |
ENSMUST00000072080.10
|
Lrrc40
|
leucine rich repeat containing 40 |
| chr4_+_133097013 | 1.40 |
ENSMUST00000030669.8
|
Slc9a1
|
solute carrier family 9 (sodium/hydrogen exchanger), member 1 |
| chr17_-_27732364 | 1.40 |
ENSMUST00000118161.3
|
Grm4
|
glutamate receptor, metabotropic 4 |
| chr17_-_66384017 | 1.37 |
ENSMUST00000150766.2
ENSMUST00000038116.13 |
Ankrd12
|
ankyrin repeat domain 12 |
| chr15_-_102044658 | 1.37 |
ENSMUST00000154032.2
|
Spryd3
|
SPRY domain containing 3 |
| chr7_-_19043955 | 1.34 |
ENSMUST00000207334.2
ENSMUST00000208505.2 ENSMUST00000207716.2 ENSMUST00000208326.2 ENSMUST00000003640.4 |
Fosb
|
FBJ osteosarcoma oncogene B |
| chrX_+_80114242 | 1.32 |
ENSMUST00000171953.8
ENSMUST00000026760.3 |
Tmem47
|
transmembrane protein 47 |
| chr2_+_145627900 | 1.30 |
ENSMUST00000110005.8
ENSMUST00000094480.11 |
Rin2
|
Ras and Rab interactor 2 |
| chr17_+_73225292 | 1.30 |
ENSMUST00000024857.14
|
Lbh
|
limb-bud and heart |
| chr14_-_66518399 | 1.30 |
ENSMUST00000111121.2
ENSMUST00000022622.14 ENSMUST00000089250.9 |
Ptk2b
|
PTK2 protein tyrosine kinase 2 beta |
| chr12_-_25146078 | 1.29 |
ENSMUST00000222667.2
ENSMUST00000020974.7 |
Id2
|
inhibitor of DNA binding 2 |
| chr2_+_76236870 | 1.29 |
ENSMUST00000077972.11
ENSMUST00000111929.8 ENSMUST00000111930.9 |
Osbpl6
|
oxysterol binding protein-like 6 |
| chr6_+_65648574 | 1.28 |
ENSMUST00000054351.6
|
Ndnf
|
neuron-derived neurotrophic factor |
| chr9_-_81515865 | 1.26 |
ENSMUST00000183482.2
|
Htr1b
|
5-hydroxytryptamine (serotonin) receptor 1B |
| chr6_-_113478779 | 1.25 |
ENSMUST00000101059.4
ENSMUST00000204268.3 ENSMUST00000205170.2 ENSMUST00000205075.2 ENSMUST00000204134.3 |
Prrt3
|
proline-rich transmembrane protein 3 |
| chr12_+_82216193 | 1.25 |
ENSMUST00000166429.9
ENSMUST00000220963.2 |
Sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr11_-_78588194 | 1.24 |
ENSMUST00000142739.8
|
Nlk
|
nemo like kinase |
| chr4_-_25800083 | 1.24 |
ENSMUST00000084770.5
|
Fut9
|
fucosyltransferase 9 |
| chr15_-_77191079 | 1.23 |
ENSMUST00000171751.10
|
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr12_+_29988035 | 1.22 |
ENSMUST00000122328.8
ENSMUST00000118321.3 |
Pxdn
|
peroxidasin |
| chr10_-_25076008 | 1.22 |
ENSMUST00000100012.3
|
Akap7
|
A kinase (PRKA) anchor protein 7 |
| chr2_-_173118315 | 1.20 |
ENSMUST00000036248.13
|
Pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr18_+_37130860 | 1.20 |
ENSMUST00000115659.6
|
Pcdha9
|
protocadherin alpha 9 |
| chr8_+_12807001 | 1.20 |
ENSMUST00000033818.10
ENSMUST00000091237.12 |
Atp11a
|
ATPase, class VI, type 11A |
| chr10_+_128247598 | 1.19 |
ENSMUST00000096386.13
|
Rnf41
|
ring finger protein 41 |
| chr8_-_17585263 | 1.19 |
ENSMUST00000082104.7
|
Csmd1
|
CUB and Sushi multiple domains 1 |
| chr2_+_25070749 | 1.19 |
ENSMUST00000104999.4
|
Nrarp
|
Notch-regulated ankyrin repeat protein |
| chr13_-_13568106 | 1.18 |
ENSMUST00000021738.10
ENSMUST00000220628.2 |
Gpr137b
|
G protein-coupled receptor 137B |
| chr3_-_103645311 | 1.17 |
ENSMUST00000029440.10
|
Olfml3
|
olfactomedin-like 3 |
| chr17_-_66756710 | 1.17 |
ENSMUST00000086693.12
ENSMUST00000097291.10 |
Mtcl1
|
microtubule crosslinking factor 1 |
| chr15_-_58078274 | 1.16 |
ENSMUST00000022986.8
|
Fbxo32
|
F-box protein 32 |
| chr3_-_115508680 | 1.16 |
ENSMUST00000055676.4
|
S1pr1
|
sphingosine-1-phosphate receptor 1 |
| chr2_+_71811526 | 1.15 |
ENSMUST00000090826.12
ENSMUST00000102698.10 |
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr10_+_60113449 | 1.14 |
ENSMUST00000105465.8
ENSMUST00000179238.8 ENSMUST00000177779.8 ENSMUST00000004316.15 |
Psap
|
prosaposin |
| chr8_-_37081091 | 1.14 |
ENSMUST00000033923.14
|
Dlc1
|
deleted in liver cancer 1 |
| chr1_+_105591595 | 1.14 |
ENSMUST00000039173.13
ENSMUST00000086721.10 ENSMUST00000190501.7 |
Relch
|
RAB11 binding and LisH domain, coiled-coil and HEAT repeat containing |
| chr5_+_37208198 | 1.13 |
ENSMUST00000043794.11
|
Jakmip1
|
janus kinase and microtubule interacting protein 1 |
| chr6_-_127128007 | 1.13 |
ENSMUST00000000188.12
|
Ccnd2
|
cyclin D2 |
| chr3_+_107008867 | 1.13 |
ENSMUST00000038695.6
|
Kcna2
|
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr1_+_179788037 | 1.13 |
ENSMUST00000097453.9
ENSMUST00000111117.8 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr8_+_85763534 | 1.12 |
ENSMUST00000093360.12
|
Tnpo2
|
transportin 2 (importin 3, karyopherin beta 2b) |
| chr5_+_137517140 | 1.11 |
ENSMUST00000031727.10
|
Gigyf1
|
GRB10 interacting GYF protein 1 |
| chr15_-_79718423 | 1.11 |
ENSMUST00000109623.8
ENSMUST00000109625.8 ENSMUST00000023060.13 ENSMUST00000089299.6 |
Cbx6
Npcd
|
chromobox 6 neuronal pentraxin chromo domain |
| chr4_-_41695442 | 1.11 |
ENSMUST00000102961.10
|
Cntfr
|
ciliary neurotrophic factor receptor |
| chr12_+_40495951 | 1.10 |
ENSMUST00000037488.8
|
Dock4
|
dedicator of cytokinesis 4 |
| chr8_-_8711211 | 1.10 |
ENSMUST00000001319.15
|
Efnb2
|
ephrin B2 |
| chr3_+_61269059 | 1.09 |
ENSMUST00000049064.4
|
Rap2b
|
RAP2B, member of RAS oncogene family |
| chrX_+_65692924 | 1.09 |
ENSMUST00000166241.2
|
Slitrk2
|
SLIT and NTRK-like family, member 2 |
| chr17_-_56447332 | 1.09 |
ENSMUST00000001256.11
|
Sema6b
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B |
| chr10_+_58649181 | 1.09 |
ENSMUST00000135526.9
ENSMUST00000153031.2 |
Sh3rf3
|
SH3 domain containing ring finger 3 |
| chr2_-_180776920 | 1.07 |
ENSMUST00000197015.5
ENSMUST00000103050.10 ENSMUST00000081528.13 ENSMUST00000049792.15 ENSMUST00000103048.10 ENSMUST00000103047.10 ENSMUST00000149964.9 |
Kcnq2
|
potassium voltage-gated channel, subfamily Q, member 2 |
| chr16_+_58228806 | 1.07 |
ENSMUST00000046663.8
|
Dcbld2
|
discoidin, CUB and LCCL domain containing 2 |
| chr1_+_59802543 | 1.06 |
ENSMUST00000087435.7
|
Bmpr2
|
bone morphogenetic protein receptor, type II (serine/threonine kinase) |
| chr8_+_66070661 | 1.06 |
ENSMUST00000110258.8
ENSMUST00000110256.8 ENSMUST00000110255.8 |
Marchf1
|
membrane associated ring-CH-type finger 1 |
| chr11_+_44409775 | 1.05 |
ENSMUST00000019333.10
|
Rnf145
|
ring finger protein 145 |
| chr5_-_106844685 | 1.04 |
ENSMUST00000127434.8
ENSMUST00000112696.8 ENSMUST00000112698.8 |
Zfp644
|
zinc finger protein 644 |
| chr14_-_24295988 | 1.04 |
ENSMUST00000073687.13
ENSMUST00000090398.11 |
Dlg5
|
discs large MAGUK scaffold protein 5 |
| chr6_-_148797648 | 1.04 |
ENSMUST00000072324.12
ENSMUST00000111569.9 |
Caprin2
|
caprin family member 2 |
| chr19_+_53128861 | 1.04 |
ENSMUST00000111741.10
|
Add3
|
adducin 3 (gamma) |
| chr9_+_25163735 | 1.03 |
ENSMUST00000115272.9
ENSMUST00000165594.4 |
Septin7
|
septin 7 |
| chr10_-_18619439 | 1.03 |
ENSMUST00000019999.7
|
Arfgef3
|
ARFGEF family member 3 |
| chr1_-_126758369 | 1.03 |
ENSMUST00000112583.8
ENSMUST00000094609.10 |
Nckap5
|
NCK-associated protein 5 |
| chr1_-_52766615 | 1.03 |
ENSMUST00000156876.8
ENSMUST00000087701.4 |
Mfsd6
|
major facilitator superfamily domain containing 6 |
| chr2_+_134627987 | 1.03 |
ENSMUST00000131552.5
ENSMUST00000110116.8 |
Plcb1
|
phospholipase C, beta 1 |
| chr8_-_85751897 | 1.03 |
ENSMUST00000064314.10
|
Get3
|
guided entry of tail-anchored proteins factor 3, ATPase |
| chr2_+_14078896 | 1.02 |
ENSMUST00000102960.11
|
Stam
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 |
| chr13_+_48414582 | 1.01 |
ENSMUST00000021810.3
|
Id4
|
inhibitor of DNA binding 4 |
| chr15_-_39807081 | 1.01 |
ENSMUST00000022916.13
|
Lrp12
|
low density lipoprotein-related protein 12 |
| chr14_-_47059694 | 1.01 |
ENSMUST00000111817.8
ENSMUST00000079314.12 |
Gmfb
|
glia maturation factor, beta |
| chr15_-_58695379 | 1.00 |
ENSMUST00000072113.6
|
Tmem65
|
transmembrane protein 65 |
| chr14_+_120513076 | 0.99 |
ENSMUST00000088419.13
|
Mbnl2
|
muscleblind like splicing factor 2 |
| chr9_-_108067552 | 0.99 |
ENSMUST00000035208.14
|
Bsn
|
bassoon |
| chr7_+_51271742 | 0.99 |
ENSMUST00000032710.7
|
Slc17a6
|
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 6 |
| chr10_+_52566616 | 0.98 |
ENSMUST00000105473.3
|
Slc35f1
|
solute carrier family 35, member F1 |
| chr16_-_45563250 | 0.97 |
ENSMUST00000066983.13
|
Abhd10
|
abhydrolase domain containing 10 |
| chr11_-_86648309 | 0.96 |
ENSMUST00000060766.16
ENSMUST00000103186.11 |
Cltc
|
clathrin, heavy polypeptide (Hc) |
| chr10_+_106306122 | 0.95 |
ENSMUST00000029404.17
ENSMUST00000217854.2 |
Ppfia2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr3_+_152052102 | 0.94 |
ENSMUST00000117492.9
ENSMUST00000026507.13 ENSMUST00000197748.5 |
Usp33
|
ubiquitin specific peptidase 33 |
| chr4_-_156319232 | 0.94 |
ENSMUST00000105569.5
|
Klhl17
|
kelch-like 17 |
| chr6_+_49296208 | 0.93 |
ENSMUST00000055559.8
ENSMUST00000114491.2 |
Ccdc126
|
coiled-coil domain containing 126 |
| chr10_-_78080436 | 0.92 |
ENSMUST00000000384.8
|
Trappc10
|
trafficking protein particle complex 10 |
| chr3_-_33136153 | 0.92 |
ENSMUST00000108225.10
|
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr4_-_108637979 | 0.92 |
ENSMUST00000106657.8
|
Zfyve9
|
zinc finger, FYVE domain containing 9 |
| chr8_+_4216556 | 0.92 |
ENSMUST00000239400.2
ENSMUST00000177053.8 ENSMUST00000176149.9 ENSMUST00000176072.9 ENSMUST00000176825.3 |
Evi5l
|
ecotropic viral integration site 5 like |
| chr16_-_20060053 | 0.91 |
ENSMUST00000040880.9
|
Map6d1
|
MAP6 domain containing 1 |
| chr12_+_102095260 | 0.91 |
ENSMUST00000079020.12
|
Slc24a4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chr7_-_109351399 | 0.91 |
ENSMUST00000128043.2
ENSMUST00000033333.13 |
Tmem9b
|
TMEM9 domain family, member B |
| chr6_-_32565127 | 0.90 |
ENSMUST00000115096.4
|
Plxna4
|
plexin A4 |
| chr5_+_32293145 | 0.89 |
ENSMUST00000031017.11
|
Fosl2
|
fos-like antigen 2 |
| chr3_-_32419609 | 0.88 |
ENSMUST00000139660.2
ENSMUST00000168566.3 ENSMUST00000029199.11 |
Zmat3
|
zinc finger matrin type 3 |
| chr16_-_21513317 | 0.87 |
ENSMUST00000231592.2
ENSMUST00000053336.8 |
2510009E07Rik
|
RIKEN cDNA 2510009E07 gene |
| chrX_+_35592006 | 0.86 |
ENSMUST00000016383.10
|
Lonrf3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr6_+_136495784 | 0.84 |
ENSMUST00000032335.13
ENSMUST00000185724.7 |
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr2_+_154498917 | 0.84 |
ENSMUST00000044277.10
|
Chmp4b
|
charged multivesicular body protein 4B |
| chr11_+_78079243 | 0.83 |
ENSMUST00000002128.14
ENSMUST00000150941.8 |
Rab34
|
RAB34, member RAS oncogene family |
| chr5_+_134128543 | 0.83 |
ENSMUST00000016088.9
|
Castor2
|
cytosolic arginine sensor for mTORC1 subunit 2 |
| chr18_+_23885390 | 0.83 |
ENSMUST00000170802.8
ENSMUST00000155708.8 ENSMUST00000118826.9 |
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr18_+_77273510 | 0.82 |
ENSMUST00000075290.8
ENSMUST00000079618.11 |
St8sia5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 |
| chr1_+_140173787 | 0.82 |
ENSMUST00000239229.2
ENSMUST00000120709.8 ENSMUST00000120796.8 ENSMUST00000119786.8 |
Kcnt2
|
potassium channel, subfamily T, member 2 |
| chr3_-_108108113 | 0.82 |
ENSMUST00000106655.2
ENSMUST00000065664.7 |
Cyb561d1
|
cytochrome b-561 domain containing 1 |
| chr10_+_107998219 | 0.82 |
ENSMUST00000070663.6
|
Ppp1r12a
|
protein phosphatase 1, regulatory subunit 12A |
| chr2_+_156038562 | 0.81 |
ENSMUST00000037401.10
|
Phf20
|
PHD finger protein 20 |
| chr7_-_144292257 | 0.81 |
ENSMUST00000121758.8
|
Ano1
|
anoctamin 1, calcium activated chloride channel |
| chr13_+_93444514 | 0.81 |
ENSMUST00000079086.8
|
Homer1
|
homer scaffolding protein 1 |
| chr4_+_49059255 | 0.80 |
ENSMUST00000076670.3
|
Plppr1
|
phospholipid phosphatase related 1 |
| chr12_-_46865709 | 0.80 |
ENSMUST00000021438.8
|
Nova1
|
NOVA alternative splicing regulator 1 |
| chr5_-_124387812 | 0.80 |
ENSMUST00000162812.8
|
Pitpnm2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0060156 | vascular transport(GO:0010232) milk ejection(GO:0060156) |
| 0.9 | 2.8 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.8 | 2.5 | GO:2000769 | regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) |
| 0.8 | 2.4 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.7 | 3.4 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.7 | 2.0 | GO:2000331 | regulation of terminal button organization(GO:2000331) |
| 0.7 | 2.6 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.6 | 2.9 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.6 | 1.7 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.6 | 4.5 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.5 | 1.6 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.5 | 2.6 | GO:0032348 | negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.5 | 3.0 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) |
| 0.5 | 1.5 | GO:0010752 | regulation of cGMP-mediated signaling(GO:0010752) |
| 0.5 | 7.0 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.5 | 1.5 | GO:0060599 | lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) |
| 0.5 | 1.4 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.5 | 2.4 | GO:1904349 | positive regulation of small intestine smooth muscle contraction(GO:1904349) |
| 0.5 | 2.8 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.5 | 1.4 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.4 | 0.9 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.4 | 2.7 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.4 | 1.3 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.4 | 1.7 | GO:2001137 | protein retention in Golgi apparatus(GO:0045053) positive regulation of endocytic recycling(GO:2001137) |
| 0.4 | 1.6 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.4 | 1.2 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.4 | 1.2 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.4 | 1.1 | GO:2000137 | negative regulation of cell proliferation involved in heart morphogenesis(GO:2000137) |
| 0.3 | 1.0 | GO:0071349 | interleukin-12-mediated signaling pathway(GO:0035722) activation of meiosis involved in egg activation(GO:0060466) cellular response to interleukin-12(GO:0071349) response to fluoride(GO:1902617) response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.3 | 1.0 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.3 | 1.0 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.3 | 1.0 | GO:0019389 | glucuronoside metabolic process(GO:0019389) |
| 0.3 | 1.6 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.3 | 1.0 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.3 | 0.9 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.3 | 1.6 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.3 | 1.0 | GO:0045186 | zonula adherens assembly(GO:0045186) |
| 0.3 | 1.8 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.3 | 1.3 | GO:0014063 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) negative regulation of serotonin secretion(GO:0014063) |
| 0.2 | 1.7 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.2 | 0.7 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.2 | 1.2 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.2 | 2.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.2 | 1.6 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.2 | 3.6 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.2 | 2.2 | GO:0046959 | habituation(GO:0046959) |
| 0.2 | 1.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.2 | 1.8 | GO:1902564 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) negative regulation of neutrophil activation(GO:1902564) |
| 0.2 | 0.4 | GO:0060434 | bronchus morphogenesis(GO:0060434) |
| 0.2 | 1.2 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.2 | 1.2 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.2 | 1.1 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.2 | 2.0 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.2 | 0.3 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.2 | 0.7 | GO:0035523 | protein K29-linked deubiquitination(GO:0035523) |
| 0.2 | 1.5 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.2 | 0.7 | GO:1901026 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.2 | 0.8 | GO:1905171 | protein localization to phagocytic vesicle(GO:1905161) regulation of protein localization to phagocytic vesicle(GO:1905169) positive regulation of protein localization to phagocytic vesicle(GO:1905171) |
| 0.2 | 1.1 | GO:1903847 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.2 | 1.4 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.2 | 5.7 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 1.7 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.1 | 1.9 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.6 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.1 | 1.3 | GO:1904672 | regulation of somatic stem cell population maintenance(GO:1904672) |
| 0.1 | 0.4 | GO:0006407 | rRNA export from nucleus(GO:0006407) |
| 0.1 | 0.8 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 2.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 0.5 | GO:0003360 | brainstem development(GO:0003360) |
| 0.1 | 0.4 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.1 | 0.4 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.1 | 1.0 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 | 0.8 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.9 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.1 | 0.3 | GO:0046038 | GMP catabolic process(GO:0046038) hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.1 | 1.6 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.1 | 0.8 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 1.2 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.1 | 1.2 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.1 | 0.1 | GO:0044830 | modulation by host of viral RNA genome replication(GO:0044830) |
| 0.1 | 1.3 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 1.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 1.0 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.1 | 4.0 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 1.2 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) |
| 0.1 | 1.2 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.4 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.1 | 2.3 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.1 | 0.9 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.1 | 1.0 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.1 | 0.4 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.1 | 0.3 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.1 | 2.6 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.1 | 0.3 | GO:0015881 | creatine transport(GO:0015881) |
| 0.1 | 2.1 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.1 | 0.3 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.1 | 1.2 | GO:0036065 | fucosylation(GO:0036065) |
| 0.1 | 1.9 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.1 | 1.1 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.1 | 0.5 | GO:1900451 | positive regulation of glutamate receptor signaling pathway(GO:1900451) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.1 | 0.8 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 0.6 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.1 | 0.7 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 0.9 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 0.4 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.1 | 1.6 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.1 | 0.2 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.1 | 2.9 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.1 | 2.0 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.1 | 0.7 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.1 | 0.7 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 0.9 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 1.1 | GO:0060742 | epithelial cell differentiation involved in prostate gland development(GO:0060742) |
| 0.1 | 0.8 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 0.3 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.1 | 0.5 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.1 | 1.3 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.1 | 0.9 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 1.1 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 0.3 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 | 0.9 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.1 | 3.1 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.1 | 1.1 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.1 | 0.6 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 0.8 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 0.7 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.2 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.0 | 0.2 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.0 | 0.7 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.3 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 | 0.5 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.6 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 4.0 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.0 | 0.5 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.0 | 0.9 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.5 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 1.7 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.0 | 0.3 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.1 | GO:0072194 | ureter smooth muscle development(GO:0072191) ureter smooth muscle cell differentiation(GO:0072193) kidney smooth muscle tissue development(GO:0072194) |
| 0.0 | 0.9 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.6 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.5 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.7 | GO:0036035 | osteoclast development(GO:0036035) |
| 0.0 | 0.3 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 1.8 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.1 | GO:1904093 | regulation of autophagic cell death(GO:1904092) negative regulation of autophagic cell death(GO:1904093) |
| 0.0 | 0.4 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.0 | 1.1 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.1 | GO:0071211 | protein targeting to vacuole involved in autophagy(GO:0071211) |
| 0.0 | 1.8 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 2.9 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 1.2 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.3 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 1.7 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.4 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 1.3 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 1.5 | GO:0001782 | B cell homeostasis(GO:0001782) |
| 0.0 | 0.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 1.0 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.0 | 0.3 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.5 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 3.0 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 1.2 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.1 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.0 | 1.1 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.7 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.1 | GO:0014877 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 1.5 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.8 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.3 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 3.0 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 2.3 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.5 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.0 | 0.5 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0046881 | positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.0 | 0.3 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.1 | GO:1904415 | dendritic cell proliferation(GO:0044565) regulation of xenophagy(GO:1904415) positive regulation of xenophagy(GO:1904417) |
| 0.0 | 0.1 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.6 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.6 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.0 | 0.5 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.0 | GO:1900248 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.0 | 0.6 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.0 | GO:0060220 | camera-type eye photoreceptor cell fate commitment(GO:0060220) negative regulation of neural retina development(GO:0061076) negative regulation of retina development in camera-type eye(GO:1902867) negative regulation of amacrine cell differentiation(GO:1902870) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.5 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.9 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.9 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.4 | GO:0003338 | metanephros morphogenesis(GO:0003338) |
| 0.0 | 0.8 | GO:0030071 | regulation of mitotic metaphase/anaphase transition(GO:0030071) |
| 0.0 | 0.5 | GO:1904037 | positive regulation of epithelial cell apoptotic process(GO:1904037) |
| 0.0 | 0.3 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 2.9 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.5 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0016014 | dystrobrevin complex(GO:0016014) |
| 0.8 | 2.5 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.7 | 4.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.7 | 4.0 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.3 | 1.6 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.3 | 1.2 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.3 | 0.8 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.2 | 2.6 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.2 | 1.1 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.2 | 1.9 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.2 | 4.6 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.2 | 3.8 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.2 | 0.5 | GO:1903095 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.2 | 4.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 1.0 | GO:0031523 | Myb complex(GO:0031523) |
| 0.2 | 1.7 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 1.9 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 1.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 2.3 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) |
| 0.1 | 0.4 | GO:0097635 | Atg12-Atg5-Atg16 complex(GO:0034274) extrinsic component of autophagosome membrane(GO:0097635) |
| 0.1 | 5.6 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 3.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.9 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.1 | 7.6 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 1.4 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 0.4 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 3.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.4 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.1 | 1.4 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.8 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.6 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 2.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 0.8 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 1.7 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 1.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 2.4 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 0.7 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 1.8 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 2.0 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 0.5 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.3 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 1.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.4 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 1.3 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 1.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.7 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 1.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 3.4 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.2 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.9 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 3.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.7 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 2.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 2.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 3.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.2 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.6 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.5 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.5 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 1.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.0 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.9 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 1.0 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.4 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 7.6 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 1.2 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.3 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.9 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 1.1 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 3.1 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 2.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 3.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.2 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.8 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.2 | GO:0000145 | exocyst(GO:0000145) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.0 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.7 | 2.2 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.7 | 2.8 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.7 | 2.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.5 | 3.8 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.5 | 1.6 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.5 | 2.9 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.4 | 4.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.4 | 1.6 | GO:0031694 | beta-adrenergic receptor activity(GO:0004939) alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.4 | 1.2 | GO:0005135 | interleukin-3 receptor binding(GO:0005135) |
| 0.4 | 2.5 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.3 | 1.7 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 0.3 | 2.3 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.3 | 2.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) ankyrin repeat binding(GO:0071532) |
| 0.3 | 1.8 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.3 | 1.8 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.3 | 1.9 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.3 | 2.4 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.3 | 1.6 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.2 | 3.0 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.2 | 1.5 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.2 | 0.9 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.2 | 0.9 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.2 | 2.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.2 | 2.5 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.2 | 1.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.2 | 1.4 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.2 | 1.9 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 0.7 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.2 | 4.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 2.0 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.2 | 1.7 | GO:0022821 | potassium ion antiporter activity(GO:0022821) |
| 0.2 | 2.9 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.2 | 1.7 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.2 | 0.9 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 0.4 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.1 | 0.7 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 0.8 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.1 | 5.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.5 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.1 | 0.7 | GO:0019961 | interferon binding(GO:0019961) |
| 0.1 | 1.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.1 | 2.6 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.1 | 2.6 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 2.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 2.3 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 1.1 | GO:0036122 | BMP binding(GO:0036122) BMP receptor activity(GO:0098821) |
| 0.1 | 1.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.1 | 1.8 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 2.9 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 2.8 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.1 | 1.7 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 1.7 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 1.4 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.1 | 2.6 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 2.9 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 2.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 1.3 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.1 | 0.2 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 0.3 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.1 | 2.2 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 2.9 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 3.8 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 2.0 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 0.4 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 2.5 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 0.4 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.1 | 0.8 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.1 | 1.0 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 1.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.6 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 0.8 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 0.3 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.1 | 0.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 5.7 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 1.2 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.1 | 0.4 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 0.4 | GO:0016433 | rRNA (adenine) methyltransferase activity(GO:0016433) |
| 0.1 | 0.4 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 0.3 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.1 | 0.2 | GO:0070051 | fibrinogen binding(GO:0070051) collagen V binding(GO:0070052) |
| 0.1 | 0.8 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 0.4 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 0.5 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.3 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.2 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 1.2 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.6 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.6 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.2 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.0 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.3 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 2.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 12.6 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 1.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.3 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.5 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 1.0 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 2.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.1 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.0 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 1.5 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.1 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 1.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.6 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.0 | 0.8 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 1.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 0.6 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.5 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 1.7 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.9 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.7 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.4 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.1 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.0 | 0.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.9 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.2 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.6 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.3 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 1.0 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 1.5 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.4 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 6.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 2.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 2.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 1.0 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 1.2 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.1 | 4.0 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 2.1 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 2.9 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 2.9 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 3.4 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 2.1 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 2.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.7 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 2.3 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 1.1 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.6 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 2.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 2.2 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.8 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.9 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.5 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 1.1 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.4 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.5 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.7 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.5 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 2.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.7 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.8 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.2 | 4.8 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.2 | 2.6 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 2.1 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 2.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 2.3 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 4.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 0.4 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 2.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 1.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 1.8 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 1.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 5.9 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 1.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 2.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 1.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 3.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 2.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.6 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 1.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 1.5 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.5 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.9 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 1.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.9 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 1.1 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.9 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 3.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.6 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 3.9 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.4 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 1.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 0.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.3 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 0.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.6 | REACTOME G1 PHASE | Genes involved in G1 Phase |